Abstract
ERCC1–XPF is a structure-specific nuclease with two subunits, ERCC1 and XPF. The enzyme cuts DNA at junctions where a single strand moves 5′ to 3′ away from a branch point with duplex DNA. This activity has a central role in nucleotide excision repair (NER), DNA cross-link repair and recombination. To dissect the activities of the nuclease it is necessary to investigate the subunits individually, as studies of the enzyme so far have only used the heterodimeric complex. We produced recombinant ERCC1 and XPF separately in Escherichia coli as soluble proteins. Activity was monitored by a sensitive dual incision assay for NER by complementation of cell extracts. XPF and ERCC1 are unstable in mammalian cells in the absence of their partners but we found, surprisingly, that ERCC1 alone could confer some repair to extracts from ERCC1-defective cells. A version of ERCC1 lacking the first 88 non-conserved amino acids was also functional. This indicated that a small amount of active XPF was present in ERCC1 extracts, and immunoassays showed this to be the case. Some repair in XPF-defective extracts could be achieved by adding ERCC1 and XPF proteins together, but not by adding only XPF. The results show for the first time that functional ERCC1–XPF can be formed from separately produced subunits. Protein sequence comparison revealed similarity between the ERCC1 family and the C-terminal region of the XPF family, including the regions of both proteins that are necessary for the ERCC1–XPF heterodimeric interaction. This suggests that the ERCC1 and XPF families are related via an ancient duplication.
INTRODUCTION
The human ERCC1–XPF complex is one of the two DNA endonucleases essential for the dual incision step of nucleotide excision repair (NER), a DNA repair pathway that removes from the genome a wide spectrum of lesions induced by UV and chemicals (1). This complex is found in human cells as a tight association of the ERCC1 and XPF proteins with predicted molecular masses of 31 and 103 kDa, respectively. ERCC1–XPF shows a remarkable structure-specific nuclease activity as it can cut DNA at junctions between a duplex and a single strand, where the single strand moves 5′ to 3′ away from the junction (2,3). This property allows the enzyme to cut the damaged strand during NER specifically on the 5′ side of a lesion once the DNA double helix has been locally unwound at the site of damage.
In addition to its role in NER, ERCC1–XPF is involved in a pathway of recombinational repair of DNA interstrand cross-links. Both ERCC1- and XPF-defective cells show not only UV sensitivity but also high sensitivity to DNA cross-linking agents. In budding yeast, the homologous Rad1–Rad10 complex is involved in mitotic recombination (4) and in a pathway for resolving recombination intermediates in repair of double-strand breaks (5). The Drosophila homologue of XPF, Mei9, is involved in meiosis (6) and the fission yeast homologues of XPF and ERCC1 (rad16 and swi10, respectively) in mating type switching (7,8). Indications of the involvement of ERCC1 in other DNA metabolism pathways in addition to NER are also provided by ERCC1 knock-out mice (9,10). These show an unusual phenotype of growth retardation, early death and liver and kidney abnormalities. This severe phenotype cannot be explained only by a deficiency in NER and it reveals an essential role for the ERCC1 protein.
An early finding that suggested ERCC1 and XPF proteins are in a complex was that ERCC1 and XPF (ERCC4) mutant cell extracts do not complement one another for DNA repair when mixed together in vitro (11–13). The suggested explanation for this is that, in mammalian cells, ERCC1 and XPF are each unstable in the absence of their partner (11,12,14). In agreement with this is the fact that a low content of ERCC1 protein is a frequent feature of XP group F cells (11,15) despite normal levels of ERCC1 mRNA. Furthermore, ERCC1 protein levels can be increased in XPF-defective cells by transfection with an expression vector encoding wild-type XPF (16). Similarly, undetectable levels of XPF in ERCC1 mutant cells can be restored to normal by transfection with an expression vector encoding wild-type ERCC1 (17).
There are many unanswered questions about the ERCC1–XPF nuclease, including which of the subunits mediates DNA binding and cleavage. To answer such questions it is desirable to produce the subunits individually, but studies of the enzyme so far have only used the heterodimeric complex. In this study we produced recombinant ERCC1 and XPF individually in Escherichia coli and obtained activity with individual subunits. We detected significant sequence similarity between the C-terminal regions of both ERCC1 and XPF, including the area in which the two subunits interact.
MATERIALS AND METHODS
Expression of recombinant proteins in E.coli
All proteins were expressed from pET30b(+) vectors (Novagen). ERCC1 and Δ88ERCC1 had a C-terminal His tag while XPF had an N-terminal His tag. For expression of ERCC1 and Δ88ERCC1, E.coli BL21 (DE3) recA host cells were grown at 37°C to an OD600 of 0.5, cooled on ice to 30°C before addition of IPTG (1 mM final) and further incubated for 5 h at 30°C. For expression of XPF, E.coli BL21 (DE3) recA host cells were grown at 37°C to an OD600 of 0.5, cooled on ice to 20°C and further incubated for 20 h at 20°C without IPTG. This helped to avoid extensive proteolytic degradation as well as accumulation in inclusion bodies. After expression, the cells were pelleted, washed in cold PBS and resuspended in a lysis buffer containing 500 mM NaCl, 50 mM Tris–Cl (pH 8.0), 5% glycerol, 1 mM PMSF. Cells were disrupted by sonication and the lysate centrifuged (20 min, 11 000 g). For purification of ERCC1 and Δ88ERCC1, the supernatant was directly loaded on to a Talon Co++ charged resin (Clontech) pre-equilibrated in lysis buffer. Washing and elution were with lysis buffer containing 20 and 500 mM imidazole, respectively. Eluted proteins were dialysed against buffer containing 250 mM KCl, 25 mM HEPES–KOH (pH 7.9), 1 mM EDTA, 10% glycerol, aliquoted and stored at –80°C. For purification of XPF, the supernatant was diluted five times with 50 mM Tris–Cl (pH 8.0), 5% glycerol to bring the salt concentration to 100 mM NaCl. The diluted supernatant was loaded on a phosphocellulose column equilibrated in 100 mM NaCl, 50 mM Tris–Cl (pH 8.0), 5% glycerol. The column was washed twice with 10 column vol of 250 and 500 mM NaCl buffers before eluting the protein fraction containing XPF in 1 M NaCl, 50 mM Tris–Cl (pH 8.0), 5% glycerol. The eluate was directly loaded on a Ni++-Agarose column (Qiagen) prequilibrated in the phosphocellulose elution buffer. The column was washed with 10 column vol of 30 mM imidazole, 1 M NaCl, 50 mM Tris–Cl (pH 8.0), 5% glycerol before eluting XPF in 500 mM imidazole, 1 M NaCl, 50 mM Tris–Cl (pH 8.0), 5% glycerol. The eluate was dialysed against a buffer containing 250 mM KCl, 25 mM HEPES–KOH (pH 7.9), 1 mM EDTA, 10% glycerol, and aliquots stored at –80°C. The ERCC1–XPF complex was purified using a plasmid that expresses both subunits simultaneously (18).
Cell lines and whole cell extracts
CHO cell line 43-3B (ERCC1–) was isolated after treatment of CHO-9 cells with the point mutagen ethylnitrosourea (19). ERCC1– CHO cell lines UV20 and CHO UV203 were isolated after treatment of CHO AA8 cells with ethylmethane sulfonate (20,21). CHO UV41 (XPF–) was derived from CHO AA8 cells treated with the frameshift mutagen ICR-170. All cell lines with the UV prefix were provided by Dr D. Busch. The CHO 43-3B His-ERCC1 cells were obtained after transfection into CHO 43-3B cells of a pSVL–ERCC1–His construct as described by Sijbers et al. (2). HeLa cells and the XP-F cell line GM8437 (patient XP2YO) were obtained from the Human Genetic Mutant Cell Repository (Coriell Institute, Camden, NJ). Whole cell extracts were prepared as previously described (22).
DNA repair assay
Reaction mixtures (10 µl) included 50 ng of plasmid DNA molecules containing a single cisplatin adduct, 60 µg of extract protein, 10 mM HEPES–KOH (pH 7.9), 70 mM KCl, 2.5 mM MgCl2, 0.4 mM EDTA, 0.4 mM DTT, 2 mM ATP, 0.02% NP-40, 3.4% glycerol. When recombinant proteins were included in the reaction they were pre-incubated at 30°C with all the components of the reaction except the DNA. The reaction was started by addition of the DNA substrate and further incubated for 45 min at 30°C. Repair was detected as previously described (23). Briefly, 5 ng of a 34mer oligonucleotide complementary to the region around the adduct was annealed with excised repair products, creating an overhang of four G residues. The repair products were labelled with 0.1 U Sequenase v2.0 polymerase (USB) and 1 µCi [α-32P]dCTP (Amersham Pharmacia Biotech, 3000 Ci/mmol) and separated on a 14% (w/v) polyacrylamide gel. The dried gel was then exposed to a phosphorimager screen for 2 days and analysed using Image Quant Software. In order to detect complementation of XPF-defective cell extracts by rΔ88ERCC1 and rXPF, repair reactions were scaled up 10-fold to 100 µl. The reaction was terminated after 45 min by addition of 100 µl of a Stop Mix (30 mM EDTA, 0.7% SDS) and 10 µl of Proteinase K (2 mg/ml), and incubated for 30 min at 37°C. After phenol/chloroform extraction, DNA was ethanol-precipitated and resuspended in 20 µl of a 1× Sequenase v2.0 polymerase reaction buffer. The excised oligonucleotides were labelled with 0.2 U Sequenase v2.0 polymerase and 1 µCi [α-32P]dCTP after they were annealed to 10 ng of the 34mer described above. The labelled repair products were then processed as above.
Immunoblotting and immunoprecipitation
Detection of recombinant proteins was carried out with a monoclonal anti-6-His antibody (Clontech) and a secondary anti-mouse IgG antibody conjugated to horseradish peroxidase (Sigma). The peroxidase activity was detected with the ECL system (Amersham Pharmacia Biotech). XPF was detected with rabbit polyclonal serum KJ (directed against a C-terminal fragment containing the last 391 residues of human XPF, kindly provided by Dr N. G. J. Jaspers, Erasmus University, Rotterdam) and with a secondary anti-rabbit IgG antibody conjugated to horseradish peroxidase (Sigma). For immunoprecipitation of XPF from extract, M-280 anti-rabbit IgG magnetic beads (Dynal) coated with polyclonal antibody RA1 (directed against the last 334 C-terminal residues of human XPF) were incubated with extract for 6 h at 20°C. Proteins recovered with the magnetic beads were detected by immunoblotting as described above for immunodetection of XPF in extract.
RESULTS
Production of ERCC1 and XPF independently in E.coli
Human ERCC1 and XPF were engineered to contain fusion domains that included a stretch of six histidines to facilitate their purification through metal chelate affinity chromatography. Expression conditions in E.coli were optimised for each protein by adjusting the temperature, time of culture, and concentration of IPTG. Substantial purification of ERCC1 could be achieved on a Co++ charged resin (Fig. 1A). In contrast, recombinant XPF bound very weakly to Co++ or Ni++ charged resins unless the E.coli lysate was first cleared by chromatography on phosphocellulose. Two smaller polypeptides co-purified with full-length XPF (Fig. 1B). The smaller was identified as an N-terminal fragment using an anti-6-His antibody, while the larger fragment was identified as a C-terminal XPF fragment by mass spectrometry fingerprinting and by using affinity-purified polyclonal antibody RA1 (data not shown).
Figure 1.
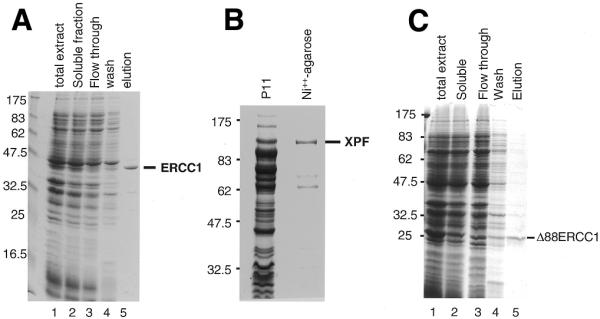
Expression in E.coli and purification of soluble recombinant proteins. (A) Purification of ERCC1 on a Talon Co++ charged resin. Proteins were separated on a 12% SDS–PAGE gel and stained with Coomassie blue. Lane 1, total cellular proteins; lane 2, soluble proteins; lane 3, flow through from a Talon column; lane 4, 10 mM imidazole wash; lane 5, ERCC1 eluted with 500 mM imidazole. Size markers (kDa) are on the left. (B) Purification of XPF. Proteins were separated on an 8% SDS–PAGE gel and stained with Coomassie blue. Sizes (kDa) are shown on the left. The protein lanes show the high salt eluate from P11 phosphocellulose (P11) and the Ni++-agarose eluate. (C) Purification of Δ88ERCC1 on Talon. Proteins were separated on a 12% SDS–PAGE gel and stained with Coomassie blue. Lane 1, total cellular proteins; lane 2, soluble proteins, loaded on Talon; lane 3, flow through; lane 4, 10 mM imidazole wash; lane 5, Δ88ERCC1 eluted with 500 mM imidazole.
It is known that a truncated ERCC1 gene lacking the coding information for the first 91 non-conserved residues can still correct the UV- and MMC-sensitivity of ERCC1-defective Chinese hamster ovary (CHO) cells (14). We produced a similar truncated version of ERCC1 (Δ88ERCC1) in E.coli. Expression levels of Δ88ERCC1 were much higher than for full-length ERCC1, but most of the expressed protein accumulated in inclusion bodies and so the amounts recovered in the soluble fraction were comparable to those obtained with ERCC1 (Fig. 1C).
Repair activity can be conferred to an ERCC1-defective cell extract by ERCC1 alone
The activity of the recombinant proteins was monitored by their capacity to correct the NER defect of an ERCC1 mutant extract in a cell-free repair assay. The dual incision step of NER was followed on a closed circular plasmid bearing a single 1,3 intrastrand d(GpTpG) cisplatin–DNA cross-link. Damaged oligonucleotides excised during NER were detected by end-labelling.
ERCC1 is found in low amounts in XPF-defective cells, presumably because in the absence of its protein partner, XPF, it is unstable and undergoes proteolytic degradation (14). It was anticipated that XPF in turn would also be unstable in the absence of ERCC1, and that complementation of ERCC1-defective cell extracts would require the addition of both subunits. Various amounts of recombinant ERCC1 and recombinant XPF were added to an extract from ERCC1-defective 43-3B CHO cells. Surprisingly, addition of ERCC1 alone conferred some dual incision activity to the extract (Fig. 2A, lane 2). As expected, XPF alone did not correct the defect (Fig. 2A, lane 6). The level of complementation reached with ERCC1 alone was comparable to the level reached with XPF and ERCC1 added together, indicating that XPF did not contribute to the reaction (lanes 3–5). These data suggested that the extract from 43-3B cells contained residual amounts of wild-type XPF that could associate in vitro with recombinant ERCC1. The amount of repair was relatively low, as a 25-fold stronger complementation was obtained when the extract was supplemented with a limited amount of recombinant ERCC1–XPF complex (18) produced in E.coli from a dicistronic vector encoding both subunits (Fig. 2A, lane 7). The fact that XPF did not enhance the level of complementation obtained with ERCC1 alone contrasted with the high complementation activity of the preformed complex. This suggested that XPF produced independently of ERCC1 might be misfolded and unable to easily associate with ERCC1. The weak complementation observed with ERCC1 alone might be due to the fact that only a small amount of hamster XPF is present in the extract. We tested the activity of the shorter version of ERCC1, Δ88ERCC1, and found that it could also confer repair activity to a 43-3B whole cell extract (Fig. 2B). The complementation levels reached with Δ88ERCC1 were higher than with full-length ERCC1 (Fig. 2C), suggesting that this version of ERCC1 might be better folded. In fact, Δ88ERCC1 could be refolded from inclusion bodies in a similarly active form (data not shown). The rest of the experiments were carried out with Δ88ERCC1.
Figure 2.
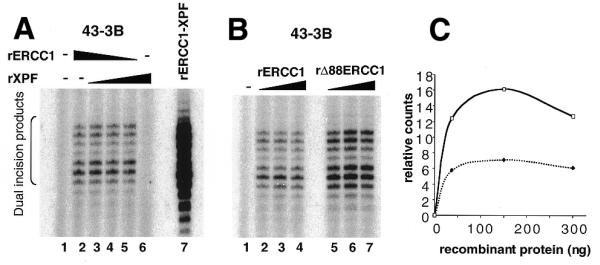
Complementation of a CHO 43-3B extract with ERCC1 protein. (A) Extract (60 µg protein) derived from the CHO 43-3B ERCC1 mutant cell line was incubated with plasmid DNA molecules containing a single cisplatin lesion (lane 1) and various amounts of recombinant ERCC1 and/or recombinant XPF (lanes 2–6). Reactions in lanes 2–5 received 300, 262.5, 150 and 37.5 ng of ERCC1, respectively. Reactions in lanes 3–6 received 37.5, 150, 262.5 and 300 ng of XPF, respectively. Lane 7 is a control reaction with 18.5 ng of the ERCC1–XPF complex produced in E.coli with a dicistronic vector encoding both subunits. Excised oligonucleotides are indicated by the bracket on the left. (B) The activity of Δ88ERCC1 was compared to the activity of full-length ERCC1. Reactions in lanes 2–4 received 38, 150 and 300 ng of rERCC1, respectively, and in lanes 5–7 they received 38, 150 and 300 ng of Δ88ERCC1, respectively. (C) Quantification of the relative complementation levels reached with ERCC1 (dotted line) and Δ88ERCC1 (solid line).
Presence of XPF in ERCC1 mutant cell extracts
The finding that ERCC1 alone could confer some NER activity to 43-3B ERCC1-defective cell extracts suggested that these extracts retain a significant amount of XPF. The 43-3B cells have a point mutation in the ERCC1 gene (24,25) that leads to the replacement of a Val by a Glu at position 98 (see Fig. 5). This mutation affects the binding of ERCC1 to XPA but does not prevent binding to XPF (24). In addition, 43-3B cells contain very low amounts of mutant ERCC1 (11,24,25). Therefore, 43-3B cells might harbour low amounts of wild-type XPF stabilised by the presence of low amounts of mutant ERCC1. Recombinant human ERCC1 added to a 43-3B extract could then form a complex with the residual XPF after subunit exchange with the mutant hamster ERCC1. Consequently, we made extracts from two other ERCC1-defective cell lines, CHO UV203 and CHO UV20, which both result from mutations that lead to truncated ERCC1 proteins (25) lacking the entire XPF binding domain (see Fig. 5). Dual incisions were formed readily by a CHO UV203 extract after addition of Δ88ERCC1 (Fig. 3A, lanes 1–6). As for 43-3B, co-addition of XPF did not significantly enhance the level of complementation obtained with Δ88ERCC1 alone (Fig. 3A, lanes 7–9). Identical results were obtained with a CHO UV20 extract. These results suggest that low levels of XPF protein are present in cells even in the complete absence of its partner ERCC1.
Figure 5.
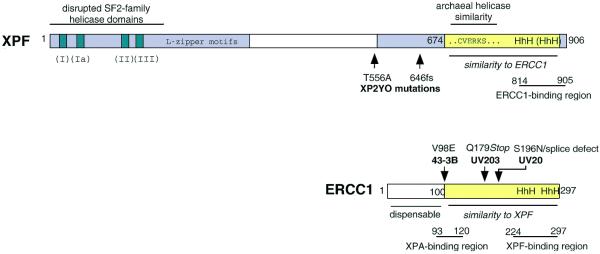
Domain structure of XPF and ERCC1. Human XPF and ERCC1 are shown as examples. XPF consists of two conserved areas (grey) separated by a less conserved region in the middle. The N-terminal area includes homology to SF2-family helicase domains I–III, and a predicted leucine zipper region. In the C-terminal region there is an area of similarity (yellow) to ERCC1. Sequence details for this region are shown in Figure 6. Part of this region shows a high similarity to a region near the C-terminus of archaeal helicases (36). In ERCC1, approximately the first 100 amino acids are dispensable and the remainder of the protein (yellow) shows similarity to XPF. Positions of known mutations in the ERCC1 and XPF cells used in this study are indicated; no mutation has been reported for UV41. The ERCC1- and XPF-binding regions are from de Laat et al. (31) and the XPA-binding region is from Li et al. (44).
Figure 3.
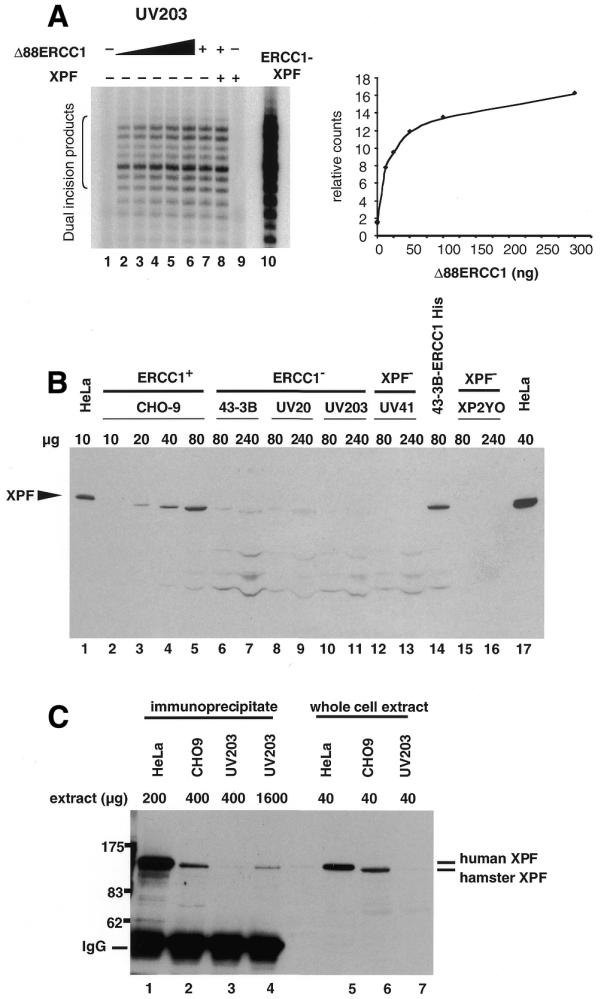
Some XPF remains in cells lacking ERCC1. (A) Extract (60 µg protein) derived from CHO UV203 ERCC1 mutant cells was used in a repair assay and complemented with increasing amounts of rΔ88ERCC1 ranging from 12.5 to 300 ng (lanes 2–6) and 38 ng in lanes 7 and 8. Reactions in lanes 8 and 9 received 260 ng of XPF. A control reaction (lane 10) was carried out with the rERCC1–XPF complex. Excised oligonucleotides are indicated by the bracket on the left. Right panel: quantification of the relative complementation levels reached with increasing amounts of rΔ88ERCC1 in lanes 1–6. (B) Extract as indicated (µg protein), from repair-proficient and mutant human and CHO cells was separated on an 8% SDS–PAGE gel. Immunodetection was carried out with XPF antibody KJ. (C) Immunoprecipitation of XPF from cell extracts. XPF antibody RA1 was used to immunoprecipitate XPF from 200 µg protein of HeLa extract (lane 1), 400 µg of normal CHO-9 extract (lane 2) and 400 (lane 3) and 1600 µg (lane 4) of ERCC1 mutant CHO UV203 extract. Immunodetection was carried out with XPF antibody KJ.
To search for XPF protein in these extracts, immunoblotting of ERCC1 and XPF mutant extracts was carried out. Hamster XPF could be detected in a repair-proficient CHO-9 cell extract (Fig. 3B, lanes 2–5). As another positive control, human XPF was detected in HeLa cells and in an extract derived from 43-3B cells transfected with a cDNA coding for human ERCC1 (Fig. 3B, lanes 14 and 17). Only very weak XPF signals, if any, were detected in 240 µg of any ERCC1- or XPF-defective extract (Fig. 3B, lanes 6–13), whereas it was detected in as little as 20 µg of CHO-9 extract. Because non-specific bands were also visible, it was difficult to be certain that the band at the XPF position was specific. Therefore, an immunoprecipitation assay was used. Larger amounts (400–1600 µg protein) of normal CHO-9 and mutant UV203 extract were incubated with XPF antibody RA1. HeLa cell extract was used as a control. The precipitated protein was detected with XPF antibody KJ. When sufficient CHO UV203 extract was used (1600 µg protein), a unique band migrating at the position of XPF was detected (Fig. 3C, lane 4), confirming that a small amount of XPF persists in cells even in the absence of complex formation with ERCC1. We tested directly for the ability of the added Δ88ERCC1 to physically interact with this residual XPF. A polyclonal antibody against ERCC1 (26) was used to immunoprecipitate Δ88ERCC1 after it was incubated for 1 h at 30°C with UV203 extract as in Figure 3A. XPF from the UV203 extract was found to co-immunoprecipitate (data not shown) indicating association of Δ88ERCC1 with endogenous hamster XPF.
Complementation of XPF-defective extracts by addition of both subunits
Since ERCC1 or Δ88ERCC1 could alone confer repair on an ERCC1-defective extract, it was still uncertain whether the recombinant XPF had any activity. To test this, XPF-defective extracts derived from XP-F rodent cells (CHO UV41) and XP-F human cells (XP2YO) were supplemented with XPF and Δ88ERCC1. Detection of the dual incision products was facilitated by scaling up the repair reactions from 10 to 100 µl and purifying the DNA after the repair reaction, before end-labelling the excised oligonucleotides. Some correction of the NER defect of XPF-defective extracts was in fact achieved by co-addition of both proteins (Fig. 4A and B, lanes 3–5). Interestingly, whereas no NER activity could be detected in a non-complemented CHO UV41 extract (Fig. 4A, lane 1), we detected low dual incision activity in a non-complemented XP2YO extract (Fig. 4B, lane 1). This confirms previous suggestions that XP2YO cells have a residual NER activity that could explain the mild clinical symptoms of the donor patient (27,28). One allele of XP2YO is a frameshift change that would remove the critical C-terminus of the protein (see Fig. 5), whereas the other allele is an amino acid substitution at a poorly conserved residue (29).
Figure 4.
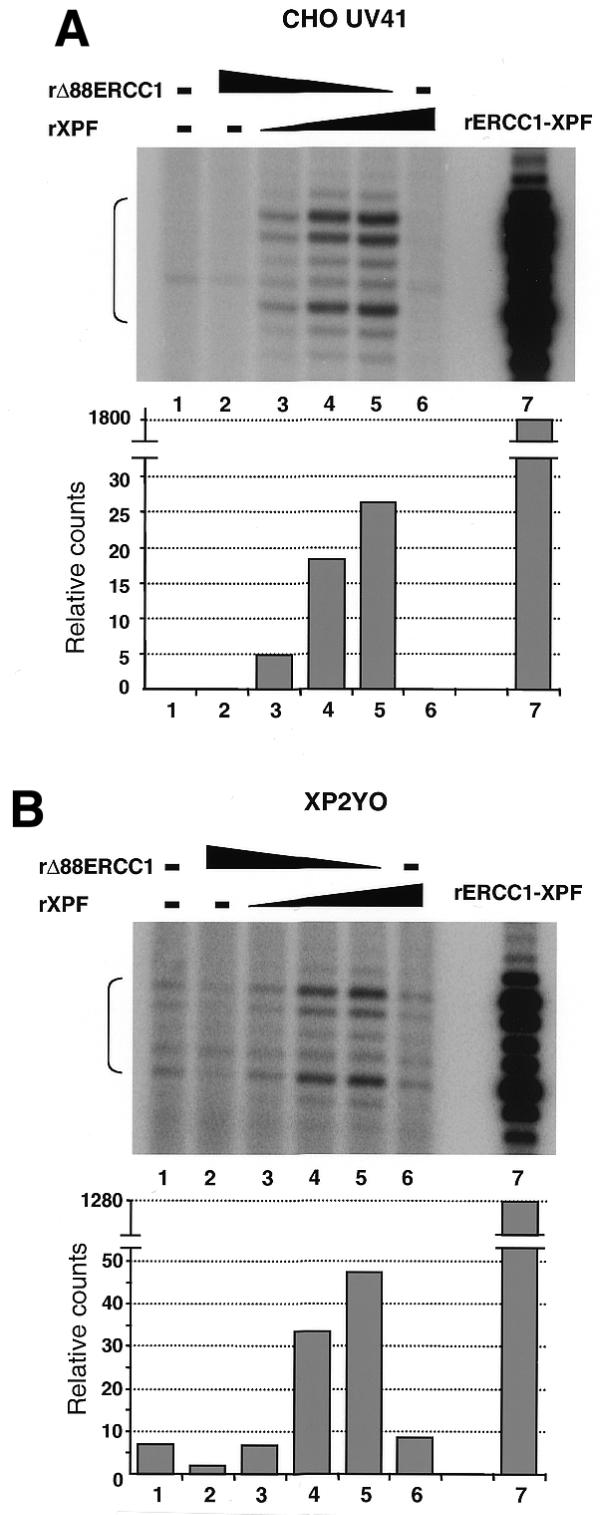
Complementation of XPF mutant extract with rΔ88ERCC1 and rXPF. Repair reactions (100 µl) were set up with XP-F extract (180 µg protein) and 250 ng of plasmid DNA with a single cisplatin lesion (lane 1) and various amounts of recombinant ERCC1 and/or recombinant XPF (lanes 2–6). Reactions in lanes 2–5 received 3000, 2625, 1500 and 375 ng of ERCC1, respectively. Reactions in lanes 3–6 received 375, 1500, 2625 and 3000 ng of XPF, respectively. Lane 7 is a control reaction with 90 ng of the ERCC1–XPF complex produced in E.coli with a vector encoding both subunits. Excised oligonucleotides are indicated by the brackets on the left. (A) Reactions with extract derived from the CHO UV41 XPF mutant cell line. (B) Reactions with extract derived from the human XP2YO XPF mutant cell line. Dual incision signals are quantified in the lower panels.
These results indicate that functional XPF can be produced independently from ERCC1 and show reconstitution of ERCC1–XPF activity with subunits produced separately. However, the complementation reached with independently produced subunits was very low compared to that observed in control reactions with preformed ERCC1–XPF complex (Fig. 4A and B, lane 7). This suggested that only a minor fraction of recombinant Δ88ERCC1 and XPF associate to form an active complex.
Primary sequence similarity between ERCC1 and XPF proteins
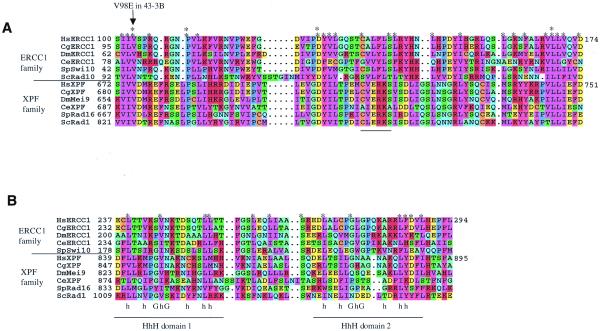
In an iterated PSI-BLAST search (30) with the human XPF sequence, we were intrigued to find that members of the ERCC1 family were also detected (E-value = 10–6 in the second iteration). To explore the significance of this, alignments were carried out with the sequences of ERCC1 and XPF homologues from human, Chinese hamster, Drosophila, Caenorhabditis elegans and yeast. We found a region of similarity that includes two areas of the C-terminal domain of XPF and much of ERCC1. The similarity (Figs 5 and 6) begins with residue 100 of human ERCC1, just at the point where residues of ERCC1 are known to be essential [a deletion of the first 88–91 residues retains function in vivo (14) and in vitro (this study), whereas deletion of the first 102 residues inactivates the protein (14)]. Members of the ERCC1 family have a variable, non-conserved N-terminal extension before this point.
Figure 6.
Regions of similarity between XPF and ERCC1. Results of a Clustal X alignment performed between XPF and ERCC1 family members starting at the positions numbered in (A). This comparison used sequences from Homo sapiens (Hs), the Chinese hamster Cricetulus griseus (Cg), the fruitfly Drosophila melanogaster (Dm), the nematode C.elegans, the fission yeast Schizosaccharomyces pombe, and the budding yeast S.cerevisiae. Residues were coloured in MacBoxshade using similarity groupings as follows: {K, R, H}, {D, E}, {I, L, V, M}, {F, Y, W}, {Q, N}, {G, A}, {S, T}, {P} and {C}. Asterisks indicate positions highly conserved throughout the ERCC1 and XPF family alignment, and double asterisks highlight residues identical throughout both families. (A) The area of proteins most conserved between the two families. (B) The region including the HhH domains, along with the consensus sequence for HhH domains (42), where h represents a hydrophobic residue. Saccharomyces cerevisiae Rad10 does not contain this region and so is absent from this portion of the alignment.
In a region of about 90 amino acids the alignment shows a number of identical or highly-conserved residues found throughout the XPF and ERCC1 families (Fig. 6A). This area of similarity between ERCC1 and XPF family members is also the most conserved region within the XPF family and within the ERCC1 family. A second region of similarity is found near the C-terminus of both proteins (Fig. 6B). In ERCC1, this region has been predicted to include two helix–hairpin–helix (HhH) motifs (14). These are domains of 20 amino acids containing hydrophobic residues (h) at key positions surrounding a hairpin, usually centred at a GhG sequence. The alignment with the XPF sequence indicates a number of residues conserved throughout both families and by comparison with the HhH consensus we predict that XPF also has an HhH in domain 1 (Fig. 1B). The presence of a domain 2 HhH sequence in the XPF family seems unlikely because of a gap in the alignment. Nevertheless, in this area there are significantly conserved residues between the ERCC1 and XPF families.
By using truncated proteins and immunoprecipitation of in vitro translated proteins, the major regions of interaction between ERCC1 and XPF have been localised to residues 224–297 of human ERCC1 and 814–905 of human XPF (31). The region in Figure 6B covers most of these areas.
DISCUSSION
The first indication that the ERCC1 and XPF gene products are biochemically connected was the observation that extracts of ERCC1- and XPF-defective mammalian cells do not complement one another in NER assays in vitro (11–13). Subsequently, ERCC1 and XPF were found to be bound to one another in a heterodimeric complex. Each subunit has been undetectable by standard methods in cell extracts where the corresponding protein partner was mutated or absent. Thus, it has been assumed that each protein in the heterodimer is absolutely necessary for stability. A main result of the present study is that ERCC1-defective cells in fact harbour a small amount of XPF protein. It is interesting that in Saccharomyces cerevisiae, the situation is significantly different. Extracts of Rad1 and Rad10 mutant cells can readily complement one another, to nearly normal levels (32). This indicates that although there are many similarities, budding yeast is not the optimal model for understanding the multifunctional ERCC1–XPF complex in mammalian cells.
A related finding is that for the first time we have produced ERCC1 and XPF individually in a form active for repair. This should facilitate identification of individual functions of the two subunits. The activity of XPF protein could only be detected when both ERCC1 and XPF were added to XPF-defective cell extracts, using more protein than in normal assays. The amount of repair in this situation is considerably lower than in reaction mixtures where ERCC1 alone was added to ERCC1-defective cell extracts. The weak activity of recombinant XPF explains why it has no easily detectable effect in the experiments with ERCC1-defective extracts.
An interaction between ERCC1 and the XPA protein is also critical for the NER reaction (33). ERCC1 produced individually by in vitro translation, or in E.coli with affinity tags, can readily interact with XPA (24,33–35). It is likely that our recombinant ERCC1 also interacts adequately with XPA, although we did not directly test this. A weak interaction between individually produced ERCC1 and XPF seems sufficient to explain the low complementation activity in the present experiments.
So far, however, we have only been able to assess function of ERCC1 and XPF when they are mixed together in mammalian cell extracts, and have not been able to reconstitute NER or nuclease activity with purified factors including the two individual subunits (data not shown). These results suggest that the independently produced proteins in E.coli do not easily associate in vitro. A possible explanation is that each subunit tends to aggregate in the absence of its partner because of exposed hydrophobic surfaces usually involved in heterodimeric interactions. In fact, we find that XPF, ERCC1 and Δ88ERCC1 all have a strong tendency to self-aggregate. Gel exclusion chromatography showed that each protein independently produced in E.coli was in a large protein complex recovered in the void volume of a Superose 12 column even in 0.5 M NaCl buffer. It seems likely that proper folding requires other cellular factors. Limited complex formation between ERCC1 and XPF has been achieved by mixing reticulocyte lysates in which each subunit was synthesized by in vitro translation (31). Reticulocyte lysates are crude systems containing protein chaperones that may encourage folding. The fact that ERCC1, Δ88ERCC1 and XPF can confer repair to defective mammalian cell extracts suggests that these extracts may also contain protein chaperones that allow association of the subunits.
ERCC1–XPF structure and function
Although both ERCC1 and XPF protein families are evolutionarily conserved among eukaryotes, neither subunit has bacterial orthologs. However, the XPF protein is related to presumed archaeal RNA-helicases belonging to superfamily 2 (36,37). Alignments within the XPF family (2,38) show that it is composed of two conserved regions in the N-terminal and C-terminal thirds, separated by a poorly conserved domain in the middle (Fig. 5). The N-terminal domain of XPF is related to superfamily 2 DNA and RNA helicases in the region covered by domains I, Ia, II and III (36). Because XPF is not a helicase and critical residues that would be required for helicase function are not present, this resemblance is thought to reflect an overall structural fold. It may reflect the mode of polynucleotide binding, in that both superfamily 2 helicases and the XPF family would be expected to bind to a DNA strand at junctions between single-stranded and duplex DNA with the same polarity.
The highest degree of similarity between the XPF family and the archaeal enzymes is found in the C-terminal half of the proteins (36). This region contains a CVERKS consensus sequence of unknown function that is not found in RNA or DNA helicases other than these archaeal examples. The signature domain is found in few other proteins; one example is Mus81 of S.cerevisiae, apparently involved in a cell cycle checkpoint (39).
Several predictions have been made previously regarding functional domains in ERCC1 and XPF. When the DNA sequence of ERCC1 became available, a similarity of the last 60 amino acids with the C-terminus of the E.coli NER nuclease UvrC was quickly noted (40,41). Although it was suggested that this similarity might reflect a nuclease activity of ERCC1 (14), the similarity with UvrC is confined to similar HhH regions, which are found in many DNA-binding proteins and are not necessarily diagnostic of nuclease activity (31,42).
The conserved region of the XPF family (Fig. 6A, bottom sequences), including and surrounding the CVERKS sequence, has many conserved acidic residues and it was predicted that this may be a nuclease domain (37). This is an intriguing possibility but so far it has not been biochemically verified. If it is true, it may suggest that the archaeal helicases also have nuclease activity. In contrast, McCutchen-Maloney et al. (43) have suggested that N-terminal fragments of XPF produced in E.coli have nuclease activity, but C-terminal fragments do not. We have not been able to find specific nuclease activity of XPF alone, or of C- or N-terminal fragments (data not shown).
The N-terminal region of XPF is related to superfamily 2 helicases although key helicase domains are disrupted. The C-terminal region of XPF is related to the functional region of ERCC1, probably via an ancient duplication. This region is the one in which the two proteins interact. This provides one reasonable explanation for the tendency of the subunits to self-aggregate in the absence of their partners. Consequently, for structural and functional studies of the isolated nuclease we suggest that it may be preferable to work with complexes between the subunits, or fragments of them.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Maureen Biggerstaff, Isao Kuraoka and Annabelle Rigaud for discussions and reagents, and John Sgouros, Neil MacDonald and Matthew Newman for valuable advice.
References
- 1.Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 2.Sijbers A.M., de Laat,W.L., Ariza,R.R., Biggerstaff,M., Wei,Y.-F., Moggs,J.G., Carter,K.C., Shell,B.K., Evans,E., de Jong,M.C., Rademakers,S., de Rooij,J., Jaspers,N.G.J., Hoeijmakers,J.H.J. and Wood,R.D. (1996) Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell, 86, 811–822. [DOI] [PubMed] [Google Scholar]
- 3.de Laat W.L., Appeldoorn,E., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1998) DNA structural elements required for ERCC1–XPF endonuclease activity. J. Biol. Chem., 273, 7835–7842. [DOI] [PubMed] [Google Scholar]
- 4.Schiestl R.H. and Prakash,S. (1990) RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol., 10, 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishman-Lobell J. and Haber,J.E. (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- 6.Sekelsky J.J., McKim,K.S., Chin,G.M. and Hawley,R.S. (1995) The Drosophila meiotic recombination gene mei-9 encodes a homolog of the yeast excision repair protein Rad1. Genetics, 141, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodel C., Kirchhoff,S. and Schmidt,H. (1992) The protein sequence and some intron positions are conserved between the switching gene swi10 of Schizosaccharomyces pombe and the human excision repair gene ERCC1. Nucleic Acids Res., 20, 6347–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr A., Schmidt,H., Kirchhoff,S., Muriel,W., Sheldrick,K., Griffiths,D., Basmacioglu,C., Subramani,S., Clegg,M. and Nasim,A. (1994) The rad16 gene of Schizosaccharomyces pombe—a homolog of the Rad1 gene of Saccharomyces cerevisiae. Mol. Cell Biol., 14, 2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McWhir J., Selfridge,J., Harrison,D.J., Squires,S. and Melton,D.W. (1993) Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nature Genet., 5, 217–224. [DOI] [PubMed] [Google Scholar]
- 10.Weeda G., Donker,I., de Wit,J., Morreau,H., Janssens,R., Vissers,C.J., Nigg,A., van Steeg,H., Bootsma,D. and Hoeijmakers,J.H.J. (1997) Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol ., 7, 427–439. [DOI] [PubMed] [Google Scholar]
- 11.Biggerstaff M., Szymkowski,D.E. and Wood,R.D. (1993) Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J., 12, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Vuuren A.J., Appeldoorn,E., Odijk,H., Yasui,A., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1993) Evidence for a repair enzyme complex involving ERCC1, ERCC4, ERCC11 and the xeroderma pigmentosum group F proteins. EMBO J., 12, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reardon J.T., Thompson,L.H. and Sancar,A. (1993) In Stillman,B. and Alberts,B. (eds), Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Press, Cold Spring Harbor, NY, Vol. 58, pp. 605–617. [DOI] [PubMed]
- 14.Sijbers A.M., van der Spek,P.J., Odijk,H., van den Berg,J., van Duin,M., Westerveld,A., Jaspers,N.G.J., Bootsma,D. and Hoeijmakers,J.H.J. (1996) Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res., 24, 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yagi T., Wood,R.D. and Takebe,H. (1997) A low content of ERCC1 and a 120 kDa protein is a frequent feature of group F xeroderma pigmentosum fibroblast cells. Mutagenesis, 12, 41–44. [DOI] [PubMed] [Google Scholar]
- 16.Yagi T., Matsumura,Y., Sato,M., Nishigori,C., Mori,T., Sijbers,A.M. and Takebe,H. (1998) Complete restoration of normal DNA repair characteristics in group F xeroderma pigmentosum cells by over-expression of transfected XPF cDNA. Carcinogenesis, 19, 55–60. [DOI] [PubMed] [Google Scholar]
- 17.Houtsmuller A.B., Rademakers,S., Nigg,A.L., Hoogstraten,D., Hoeijmakers,J.H.J. and Vermeulen,W. (1999) Action of DNA repair endonuclease ERCC1/XPF in living cells. Science, 284, 958–961. [DOI] [PubMed] [Google Scholar]
- 18.Kuraoka I., Kobertz,W.R., Ariza,R.R., Biggerstaff,M., Essigmann,J.M. and Wood,R.D. (2000) Repair of an interstrand DNA crosslink initiated by ERCC1–XPF repair/recombination nuclease. J. Biol. Chem., 275, 26632–26636. [DOI] [PubMed] [Google Scholar]
- 19.Wood R. and Burki,H.J. (1982) Repair capability and the cellular age response for killing and mutation induction after UV. Mutat. Res., 95, 505–514. [DOI] [PubMed] [Google Scholar]
- 20.Thompson L., Rubin,J., Cleaver,J., Whitmore,G. and Brookman,K. (1980) A screening method for isolating DNA repair-deficient mutants of CHO cells. Somat. Cell Genet., 6, 391–405. [DOI] [PubMed] [Google Scholar]
- 21.Busch D., Greiner,C., Lewis,K., Ford,R., Adair,G. and Thompson,L. (1989) Summary of complementation groups of UV-sensitive CHO cell mutants isolated by large scale screening. Mutagenesis, 4, 349–354. [DOI] [PubMed] [Google Scholar]
- 22.Biggerstaff M. and Wood,R.D. (1999) In Henderson,D.S. (ed.), DNA Repair Protocols: Eukaryotic Systems. Humana Press, Totowa, NJ, Vol. 113, pp. 357–372.
- 23.Shivji M.K.K., Moggs,J.G., Kuraoka,I. and Wood,R.D. (1999) In Henderson,D.S. (ed.), DNA Repair Protocols: Eukaryotic Systems. Humana Press, Totowa, NJ, Vol. 113, pp. 373–392.
- 24.Hayashi T., Takao,M., Tanaka,K. and Yasui,A. (1998) ERCC1 mutations in UV-sensitive Chinese hamster ovary (CHO) cell lines. Mutat. Res., 407, 269–276. [DOI] [PubMed] [Google Scholar]
- 25.Rolig R.L., Lowery,M.P., Adair,G.M. and Nairn,R.S. (1998) Characterization and analysis of Chinese hamster ovary cell ERCC1 mutant alleles. Mutagenesis, 13, 357–365. [DOI] [PubMed] [Google Scholar]
- 26.Köberle B., Masters,J.R.W., Hartley,J.A. and Wood,R.D. (1999) Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr. Biol., 9, 273–276. [DOI] [PubMed] [Google Scholar]
- 27.Hayakawa H., Ishizaki,K., Inoue,M., Yagi,T., Sekiguchi,M. and Takebe,H. (1981) Repair of ultraviolet radiation damage in xeroderma pigmentosum cells belonging to complementation group F. Mutat. Res., 80, 381–388. [DOI] [PubMed] [Google Scholar]
- 28.Zelle B., Berends,F. and Lohman,P.H.M. (1980) Repair of damage by ultraviolet radiation in xeroderma pigmentosum cell strains of complementation groups E and F. Mutat. Res., 73, 157–169. [DOI] [PubMed] [Google Scholar]
- 29.Matsumura Y., Nishigori,C., Yagi,T., Imamura,S. and Takebe,H. (1998) Characterization of molecular defects in xeroderma-pigmentosum group-F in relation to its clinically mild symptoms. Hum. Mol. Genet ., 7, 969–974. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S.F., Madden,T., Schaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped blast and PSI-BLAST—a new-generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Laat W.L., Sijbers,A.M., Odijk,H., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1998) Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res., 26, 4146–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z.G., Wu,X.H. and Friedberg,E.C. (1993) Nucleotide-excision repair of DNA in cell-free-extracts of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L., Peterson,C.A., Lu,X.Y. and Legerski,R.J. (1995) Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision-repair. Mol. Cell. Biol ., 15, 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park C.H. and Sancar,A. (1994) Formation of a ternary complex by human XPA, ERCC1 and ERCC4(XPF) excision-repair proteins. Proc. Natl Acad. Sci. USA, 91, 5017–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagai A., Saijo,M., Kuraoka,I., Matsuda,T., Kodo,N., Nakatsu,Y., Mimaki,T., Mino,M., Biggerstaff,M., Wood,R.D., Sijbers,A., Hoeijmakers,J.H.J. and Tanaka,K. (1995) Enhancement of damage-specific DNA binding of XPA by interaction with the ERCC1 DNA repair protein. Biochem. Biophys. Res. Commun., 211, 960–966. [DOI] [PubMed] [Google Scholar]
- 36.Sgouros J., Gaillard,P.-H.L. and Wood,R.D. (1999) A relationship between a DNA-repair/recombination nuclease family and archaeal helicases. Trends Biochem. Sci., 24, 95–97. [DOI] [PubMed] [Google Scholar]
- 37.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brookman K.W., Lamerdin,J.E., Thelen,M.P., Hwang,M., Reardon,J.T., Sancar,A., Zhou,Z.Q., Walter,C.A., Parris,C.N. and Thompson,L.H. (1996) ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol. Cell. Biol., 16, 6553–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Interthal H. and Heyer,W.D. (2000) MUS81 encodes a novel Helix–hairpin–Helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet., 263, 812–827. [DOI] [PubMed] [Google Scholar]
- 40.Doolittle R.F., Johnson,M.S., Husain,I., Van Houten,B., Thomas,D.C. and Sancar,A. (1986) Domainal evolution of a prokaryotic DNA repair protein and its relationship to active transport proteins. Nature, 323, 451–453. [DOI] [PubMed] [Google Scholar]
- 41.van Duin M., van den Tol,J., Warmerdam,P., Odijk,H., Meijer,D., Westerveld,A., Bootsma,D. and Hoeijmakers,J.H.J. (1988) Evolution and mutagenesis of the mammalian excision repair gene ERCC-1. Nucleic Acids Res., 16, 5305–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCutchen-Maloney S.L., Giannecchini,C.A., Hwang,M.H. and Thelen,M.P. (1999) Domain mapping of the DNA binding, endonuclease and ERCC1 binding properties of the human DNA repair protein XPF. Biochemistry, 38, 9417–9425. [DOI] [PubMed] [Google Scholar]
- 44.Li L., Elledge,S.J., Peterson,C.A., Bales,E.S. and Legerski,R.J. (1994) Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl Acad. Sci. USA, 91, 5012–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]