Abstract
Sequence-specific recognition of DNA can be achieved by triple helix-forming oligonucleotides that bind to the major groove of double-helical DNA. These oligonucleotides have been used as sequence-specific DNA ligands for various purposes, including sequence-specific gene regulation in the so-called ‘antigene strategy’. In particular, (G,A)-containing oligonucleotides can form stable triple helices under physiological conditions. However, triplex formation may be in competition with self-association of these oligonucleotides. For biological applications it would be interesting to identify the conditions under which one structure is favoured as compared to the other(s). Here we have directly studied competition between formation of a parallel (G,A) homoduplex and that of a triple helix by a 13 nt (G,A)-containing oligonucleotide. Temperature gradient gel electrophoresis allows simultaneous detection of competition between the two structures, because of their different temperature dependencies and gel electrophoretic mobilities, and characterisation of this competition.
INTRODUCTION
Short synthetic oligodeoxyribonucleotides can bind to oligopyrimidine·oligopurine structures in a sequence-specific manner by forming triple helices in the major groove of the DNA duplex (1,2). Such triplex-forming oligonucleotides (TFOs) provide a rational basis for the design of sequence-specific DNA ligands for various purposes (reviewed in 3). At least three structural classes of triple helices exist that differ in sequence composition and relative orientations of the phosphate–deoxyribose backbone of the third strand (4). In the pyrimidine motif a (C,T)-containing oligonucleotide binds parallel to the target oligopurine strand through Hoogsteen hydrogen bonds and forms isomorphous C·GC+ and T·AT base triplets. Due to the requirement for cytosine protonation, the stability of a (C,T) motif triple helix is pH dependent. In the purine motif (G,A)-containing oligonucleotides bind in an antiparallel orientation to the oligopurine strand of the duplex by forming C·GG and T·AA base triplets through reverse Hoogsteen hydrogen bonding interactions. (G,T)-containing oligonucleotides can form triplexes involving either Hoogsteen or reverse Hoogsteen hydrogen bonds and have a parallel or antiparallel orientation with respect to the target oligopurine strand sequence, respectively. It has been shown that the preferred strand orientation depends on the sequence of (G,T)-containing oligonucleotides (5). The formation of both (G,A) and (G,T) motif triple helices is pH independent but requires the presence of divalent cations in the millimolar concentration range (6,7). Interestingly, triple helices stable at high temperature have been observed with (G,A)-containing oligonucleotides (8–10). Recently it has been shown that the enthalpy of formation of several short (G,A) motif triplexes is very low, implying that their stability is little affected by an increase in temperature (11). However, these G-rich oligonucleotides are particularly prone to self-association, forming inter- or intramolecular structures that may compete with triplex formation, such as parallel or antiparallel (G,A) homoduplexes or G quadruplexes (12–21). Moreover, it has been shown that, in certain cases, the sequence-specific biological effects of G-rich oligonucleotides were due to mechanisms other than triple helix formation and rather involved the formation of G tetrad structures (22). It is therefore important to characterise the competition between formation of the self-associated structures and that of the triple-helical complex, in order to understand the mechanism of action of these oligonucleotides. We have used the thermal gradient gel electrophoresis (TGGE) method, which was initially introduced to analyse the conformational transitions of double-stranded RNA (23,24) and tertiary structures in tRNAs, introns and ribozymes (25–27) and has recently been used to separate and analyse DNA topoisomers (28).
In the present study the TGGE method allowed us to visualise and characterise the competition between self-association of a 13 nt (G,A)-containing TFO, which forms a parallel homoduplex structure (19), and triplex formation by the TFO on its DNA oligopyrimidine·oligopurine target sequence, as well as to analyse the temperature dependence of triplex stability.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were purchased from Eurogentec and purified using Sephadex G-25 quick spin columns (Boehringer, Mannheim). Concentrations were determined spectrophotometrically at 25°C using molar extinction coefficients at 260 nm calculated according to a nearest neighbour model (29).
Oligonucleotide-directed triple helix formation was investigated using a 31 bp DNA target containing a 15 bp oligopyrimidine·oligopurine tract, which is part of the HIV-nef gene (located at position 8571–8585) and has been previously described (19).
Gel retardation assay
TGGE experiments were performed with a Biometra TGGE System (kindly lent to us for testing by Vysis, France). A non-denaturing 12% polyacrylamide gel, 1 mm thick, soaked in 10 mM MgCl2 and 50 mM HEPES, pH 7.2, was attached to a Polybond film to avoid deformation at high temperature and was set on a gradient block so that the temperature gradient was perpendicular to the electric field. Either the purine-rich strand of the 31 bp duplex or the TFO was 5′-end-labelled with [γ-32P]ATP (Amersham, IL) using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). Fixed concentrations (2 µM) of the TFO or the corresponding duplex target were added to 20 nM labelled TFO in the presence of 10 mM MgCl2, 100 mM NaCl, 50 mM HEPES, pH 7.2, 10% sucrose and 0.5 µg/µl tRNA. The samples were incubated overnight at the indicated temperature (4 or 37°C). The gel was loaded with 30 µl samples. It was then electrophoresed for 5 min at the incubation temperature at 150 V to ensure entry of the samples into the gel matrix. The slots were rinsed, the gel covered to thermally insulate it, a linear thermal gradient applied and the samples electrophoresed for 35 min at 205 V. The gels were scanned with a Molecular Dynamics 445SI Phosphorimager.
UV absorption spectroscopy
Thermal denaturation and renaturation studies of the target duplex or the homoduplex (2 µM) were carried out on a Kontron Uvikon 940 spectrophotometer with 1 cm optical path length quartz cuvettes. The cell holder was thermoregulated with circulating 80% water/20% ethylene glycol. The sample temperature was decreased from 80 to 0°C and increased back to 80°C at 0.1°C/min with absorption readings at 260 and 440 nm taken every 1–1.2°C. Samples were kept for an additional 10 min at the lowest and highest temperatures. All samples were prepared in a buffer containing 10 mM sodium cacodylate, pH 7.0, 100 mM NaCl and 10 mM MgCl2. For melting temperature (Tm) analysis the drift in baseline was corrected by subtracting absorption at 440 nm from that at 260 nm and plotted against temperature (°C). The maximum of the first derivative ∂A/∂T versus T was taken as an estimation of the Tm value.
RESULTS
The competition between self-association and triplex formation was investigated using a 31 bp DNA target containing a 15 bp oligopyrimidine·oligopurine tract (31Y·31R) and a 13 nt (G,A)-containing TFO (called 13GA) (Fig. 1A). Triplex formation has already been characterised for this system and it has been observed that the 13mer (G,A)-containing oligonucleotide forms a stable triple helix (19). Previous spectroscopic and PAGE experiments have shown that the 13GA TFO forms a Mg2+-dependent intermolecular parallel homoduplex at low temperature and high oligonucleotide concentration, which can compete with triplex formation (Fig. 1B).
Figure 1.
(A) Sequence of the 31 bp DNA target duplex (31R·31Y) and the 13 nt (G,A)-containing TFO (13GA). (B) The possible structures formed by the 13GA oligonucleotide are here schematised.
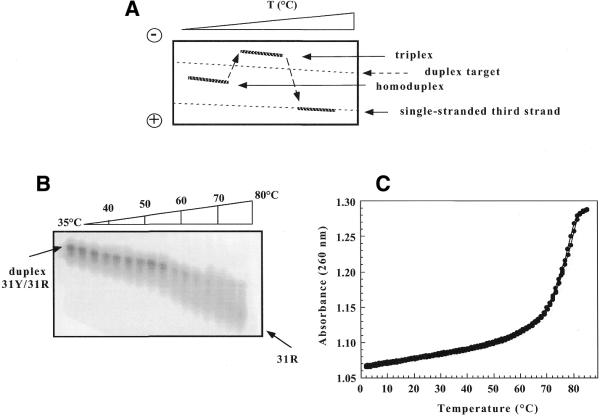
Non-denaturing PAGE is a technique that relies on the difference in mobility of the nucleic acids in a polyacrylamide gel according to their shape/structure, charge and molecular weight under the action of an electric field and it thus allows us to study triplex formation and to detect the different structures adopted by an oligonucleotide. In TGGE a linear temperature gradient (0.6–0.8°C/mm) is applied by Peltier elements, perpendicular to the direction of the electrophoretic run (Fig. 2A). The TGGE method adds an additional parameter for separating molecules, namely temperature-dependent structural changes. Even if not fully quantitative, this technique is complementary to other techniques such as UV melting, as interconversion between different complexes is directly visualised. Figure 2A show a schematic TGGE gel pattern indicating the position of the different structures that we expected to be present in solution in our system.
Figure 2.
(A) Model TGGE gel: a linear temperature gradient was applied perpendicular to the direction of the electrophoresis run. The arrows indicate the different species and their expected mobilities. (B) TGGE assay carried out with the 31R·31Y target duplex radiolabelled on the oligopurine strand at 2 µM in 50 mM HEPES (pH 7.2), 100 mM NaCl and 10 mM MgCl2. The linear temperature gradient covered the range 35–80°C. The arrows indicate the corresponding species. (C) UV melting and renaturation profile recorded at 260 nm of the 31R·31Y target duplex at 2 µM in a buffer containing 20 mM sodium cacodylate (pH 7.2), 100 mM NaCl and 10 mM MgCl2.
First, we tested the TGGE technique to follow melting of the 31 bp DNA target duplex (at 2 µM) radiolabelled on the oligopurine strand (Fig. 2B). At high temperature the resolution of the gel is lost because of heat-accelerated diffusion. For comparison the UV cooling and heating profiles recorded at 260 nm of the same duplex at 2 µM are reported. There is good consistency between the two techniques, confirming the validity of the TGGE method (Tm = 78°C by absorption measurements, 75°C by TGGE analysis).
In Figure 3A melting of the intermolecular parallel homoduplex formed by the 13GA TFO is observed (20 nM radiolabelled 13GA was mixed at 10°C with 2 µM unlabelled 13GA). Quantitative analysis of fusion of the homoduplex observed by TGGE is reported in Figure 3B and is compared to the melting profile obtained by UV absorbance measurements at 260 nm (circles). A transition midpoint of 25°C was obtained with both techniques.
Figure 3.
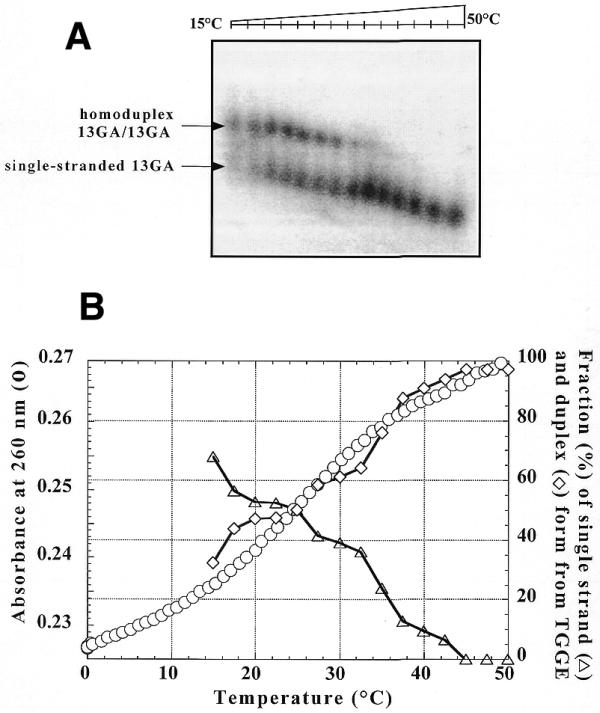
(A) TGGE assay carried out with 20 nM radiolabelled 13GA oligonucleotide in the presence of 2 µM unlabelled 13GA in 50 mM HEPES (pH 7.2), 100 mM NaCl and 10 mM MgCl2. After overnight incubation at 10°C the sample was run on a TGGE gel with a linear temperature gradient between 15 and 50°C. The arrows indicate the corresponding species. (B) UV melting profile recorded at 260 nm of the homoduplex at 2 µM (circles) in a buffer containing 20 mM sodium cacodylate (pH 7.2), 100 mM NaCl and 10 mM MgCl2 was compared to the quantitative analysis of (A). Triangles, percentage of homoduplex as a function of the temperature; diamonds, percentage of single-stranded 13GA.
When 2 µM 31R/31Y duplex target was added to the 13GA solution depicted in Figure 3A at room temperature competition between self-association and triplex formation is clearly seen (Fig. 4A). At low temperature 13GA is trapped in the homoduplex structure and the small amount of free single-stranded TFO is complexed to the target duplex, forming the triple-helical structure. Upon increasing the temperature the homoduplex dissociates and single-stranded 13GA binds to the duplex forming a triple helix, until the latter starts to dissociate and the signal corresponding to the single-stranded TFO appears. The TGGE method therefore allows us to directly monitor the switch of the third strand from the self-associated structure to the triple-helical structure and to determine the melting temperature of this very stable (G,A) triple helix, which was not possible by other techniques (19). Figure 4B shows melting of the triple-helical structure formed by 20 nM radiolabelled 13GA oligonucleotide in the presence of 2 µM target duplex. In contrast to Figure 4A, the total concentration of the 13GA TFO was only 20 nM (as compared to 2 µM in Fig. 4A). From analysis of this curve it was determined that half-dissociation of the 13GA third strand from the triple helix occurred at ∼63°C, below that of duplex melting (∼75°C).
Figure 4.

(A) Radiolabelled 13GA oligonucleotide (20 nM) was incubated overnight at room temperature in the presence of 2 µM unlabelled 13GA and 2 µM 31R·31Y target duplex in 50 mM HEPES (pH 7.2), 100 mM NaCl and 10 mM MgCl2. The sample was run on a TGGE gel with a linear temperature gradient between 20 and 65°C. The arrows indicate the corresponding species. (B) Radiolabelled 13GA oligonucleotide (20 nM) was incubated overnight at 37°C in the presence of 2 µM 31R·31Y target duplex in the same buffer as in (A) and was run with a temperature gradient ranging between 30 and 75°C. The arrows indicate the corresponding species.
DISCUSSION AND CONCLUSIONS
The perpendicular TGGE curve allows us to follow temperature-dependant structural changes of nucleic acids. Figure 2 shows denaturation of the target duplex and is compared with a UV absorbance denaturing experiment. The temperature range of the mid-transition is the same in the two experiments. It is important to note that the experimental conditions are quite different in the two methods (but similar in ionic conditions) and that in several cases, especially with (G,A)-containing oligonucleotides, it was possible to follow triplex formation by gel electrophoresis experiments at a fixed temperature but not by UV absorbance melting experiments (19,30). It is thus important to have access to the thermal behaviour of the species in gel electrophoresis experiments. As has already been reported (19), the 13mer TFO investigated here forms an intermolecular parallel (G,A) homoduplex, which is Mg2+ dependent and melts at 25°C (2 µM concentration, 20 mM sodium cacodylate, 10 mM MgCl2, 100 mM NaCl). Figure 3A shows the melting profile of this structure in a TGGE experiment. Again, the transition midpoint determined by this method (T1/2 = 25 ± 1°C) is in good agreement with that obtained by UV melting experiments (Fig. 3B). According to this work the homoduplex structure is present under conditions where the triple helix could form, therefore, we were interested in determining which species were present in solution at each temperature. Figure 4A shows a TGGE analysis, in the temperature range 20–65°C, of 20 nM radiolabelled 13GA and 2 µM unlabelled 13GA mixed and incubated for 12 h at 20°C in the presence of 2 µM 31R/31Y target duplex. The profile clearly shows that at low temperature the homoduplex structure is favoured as compared to the triple-helical one and only when the first structure melts does the single-stranded TFO bind to the target duplex to form the corresponding triple helix. At higher temperatures the triplex melts and only single-stranded 13GA is observed. This is the only reported example of direct visualisation of competition between self-association and triplex formation. The change in mobility seen for the triplex between 35 and 50°C might reflect either a conformational change within the triplex structure or a shift due to a change in concentration of the 13GA single strand arising from progressive dissociation of the homoduplex structure. If the temperature range of the experiment is shifted to higher temperature (30–75°C) and only the triple helix is preformed, melting of the triple-helical structure is obtained, with a Tm value of 63°C (Fig. 4B). This is the only technique that allowed us to determine the melting temperature of this (G,A) triplex; thermal denaturation experiments followed by UV spectroscopy failed, because of a lack of hyperchromism associated with dissociation of the (G,A)-containing third strand and the close proximity of the melting temperatures for the triplex and duplex species (19,30,31). The observation of bands of intermediate mobility during melting of the triple helix might indicate that an equilibrium between the triplex and duplex + single strand occurrs within the gel matrix (32), as previously observed when the 13GA oligonucleotide was electrophoresed at different concentrations (19). In the latter case, instead of observing radioactive bands corresponding only to monomer and dimer, bands of intermediate mobility were observed when the concentration was raised from 20 nM to 20 µM. A similar phenomenon, called ‘cyclic capture and dissociation’ (CCD), was previously described by Belotserkovskii and Johnston for short-lived complexes when the faster migrating component A of the complex AB was labelled and the slower migrating component B was at a high concentration (32). Under our conditions the situation is even more complex due to partial dissociation of the duplex 31Y·31R (see Fig. 2B).
To visualise competition between formation of the parallel homoduplex and the triple helix we used the fact that the two structures have different temperature dependencies and gel electrophoretic mobilities (19). This difference in behaviour with temperature has recently been explained by calculation of the enthalpy of formation for (G,A) triplexes (11). These triplexes have an enthalpy of formation close to 0, which implies that these structures are practically temperature independent, and it was shown that they dissociate only when the duplex target dissociates. This explains why stable (G,A) triplexes have been observed at high temperatures even though the association constants are not as high as expected (8,11).
In conclusion, classical electrophoresis and TGGE rely on differences in mobility due to the shape/charge of the structure, while TGGE adds a new parameter, a temperature gradient that allows evolution of the structures with temperature to be followed. To obtain equivalent data by classical electrophoresis a gel for each temperature scanned by the gradient in the TGGE experiment should be done, which is tedious and time consuming. However, in order to validate the TGGE technique we have also analysed the same samples by classical electrophoresis at three temperatures (10, 37 and 55°C); similar results were obtained. The TGGE method allowed us to directly monitor in a single experiment dissociation of the TFO from the self-associated homoduplex structure and its binding to the target duplex to form a triple helix. At higher temperatures the TFO dissociates from the triple-helical structure and only the single-stranded TFO is observed. Finally, TGGE allowed us to determine the Tm value of this triplex, which cannot be obtained by other methods.
UV absorbance melting experiments, which can give similar information, are limited to analysis of structures that have different absorption coefficients. In the case studied here, (G,A) triplex formation, UV spectroscopy failed because of a lack of hyperchromism associated with dissociation of the (G,A)-containing third strand.
Relying on differences in mobility of structures in a polyacrylamide gel under the action of an electric field, the TGGE technique is applicable to all structures formed by oligonucleotides (duplexes, triplexes, quadruplexes, etc.) and competition between structures having different thermal dependencies can be analysed and resolved. As further validation of the technique we have applied TGGE to follow a (T,C) triplex→(G,T) triplex interconversion previously studied by other techniques (11).
Acknowledgments
ACKNOWLEDGEMENTS
The authors kindly thank Vysis for the loan of the TGGE System and Loïc Perrouault for technical help.
References
- 1.Le Doan T., Perrouault,L., Praseuth,D., Habhoub,N., Decout,J.-L., Thuong,N.T., Lhomme,J. and Hélène,C. (1987) Sequence specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo α thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res., 15, 7749–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moser H.E. and Dervan,P.B. (1987) Sequence specific cleavage of double helical DNA by triple helix formation. Science, 238, 645–650. [DOI] [PubMed] [Google Scholar]
- 3.Giovannangeli C. and Hélène,C. (1997) Progress in developments of triplex-based strategies. Antisense Nucleic Acid Drug Dev., 7, 413–421. [DOI] [PubMed] [Google Scholar]
- 4.Thuong N.T. and Hélène,C. (1993) Sequence-specific recognition and modification of double-helical DNA by oligonucleotides. Angew. Chem. Int. Ed. Engl., 32, 666–690. [Google Scholar]
- 5.Sun J.S., de Bizemont,T., Duval-Valentin,G., Montenay-Garestier,T. and Hélène,C. (1991) Extension of the range of recognition for triple helix formation by oligonucleotides containing guanines and thymines. C. R. Acad. Sci. Paris III, 313, 585–590. [PubMed] [Google Scholar]
- 6.Potaman V.N. and Sinden,R.R. (1995) Stabilization of triple-helical nucleic acids by basic oligopeptides. Biochemistry, 34, 14885–14892. [DOI] [PubMed] [Google Scholar]
- 7.Potaman V.N. and Soyfer,V.N. (1998) Stabilization of the purine.purine.pyrimidine DNA base triplets by divalent metal cations J. Biomol. Struct. Dyn., 16, 145–146. [DOI] [PubMed] [Google Scholar]
- 8.Svinarchuk F., Bertrand,J.R. and Malvy,C. (1994) A short purine oligonucleotide forms a highly stable triple helix with the promoter of the murine c-pim-1 proto-oncogene. Nucleic Acids Res., 22, 3742–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svinarchuk F., Monnot,M., Merle,A., Malvy,C. and Fermandjian,S. (1995) The high stability of the triple helices formed between short purine oligonucleotides and SIV/HIV-2 vpx genes is determined by the targeted DNA structure. Nucleic Acids Res., 23, 3831–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debin A., Malvy,C. and Svinarchuk,F. (1997) Investigation of the formation and intracellular stability of purine(purine/pyrimidine) triplexes. Nucleic Acids Res., 25, 1965–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills M., Arimondo,P.B., Lacroix,L., Garestier,T., Hélène,C., Klump,H. and Mergny,J.L. (1999) Energetics of strand-displacement reactions in triple helices: a spectroscopic study. J. Mol. Biol., 291, 1035–1054. [DOI] [PubMed] [Google Scholar]
- 12.Roy C. (1993) Inhibition of gene transcription by purine rich triplex forming oligodeoxyribonucleotides. Nucleic Acids Res., 21, 2845–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noonberg S.B., Francois,J.C., Garestier,T. and Hélène,C. (1995) Effect of competing self-structure on triplex formation with purine-rich oligodeoxynucleotides containing GA repeats. Nucleic Acids Res., 23, 1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivas W.M. and Maher,L.J. (1995) Competitive triplex/quadruplex equilibria involving guanine-rich oligonucleotides. Biochemistry, 34, 278–284. [DOI] [PubMed] [Google Scholar]
- 15.Mishima Y., Suda,T. and Kominami,R. (1996) Formation of a triple-stranded DNA between d(GGA:TCC) repeats and d(GGA) repeat oligonucleotides. J. Biochem. (Tokyo), 119, 805–810. [DOI] [PubMed] [Google Scholar]
- 16.Cheng A.J., Wang,J.C. and Van Dyke,M.W. (1998) Self-association of G-rich oligodeoxyribonucleotides under conditions promoting purine-motif triplex formation. Antisense Nucleic Acid Drug Dev., 8, 215–225. [DOI] [PubMed] [Google Scholar]
- 17.Cheng A.J. and Van Dyke,M.W. (1994) Oligodeoxyribonucleotide length and sequence effects on intermolecular purine.purine.pyrimidine triple-helix formation. Nucleic Acids Res., 22, 4742–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng A.J. and Van Dyke,M.W. (1997) Oligodeoxyribonucleotide length and sequence effects on intramolecular and intermolecular G-quartet formation. Gene, 197, 253–260. [DOI] [PubMed] [Google Scholar]
- 19.Arimondo P.B., Barcelo,F., Sun,J.S., Maurizot,J.C., Garestier,T. and Hélène,C. (1998) Triple helix formation by (G,A)-containing oligonucleotides: asymmetric sequence effect. Biochemistry, 37, 16627–16635. [DOI] [PubMed] [Google Scholar]
- 20.Hud N.V., Smith,F.W., Anet,F.A.L. and Feigon,J. (1996) The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by H-1 NMR. Biochemistry, 35, 15383–15390. [DOI] [PubMed] [Google Scholar]
- 21.Williamson J.R. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 22.Simonsson T., Pecinka,P. and Kubista,M. (1998) DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res., 26, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbaum V. and Riesner,D. (1987) Temperature-gradient gel electrophoresis. Thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys. Chem., 26, 235–246. [DOI] [PubMed] [Google Scholar]
- 24.Schafer S., Heumann,H. and Gross,H.J. (1997) Mammalian tRNA(Lys)3 and pre-tRNA(Lys)3 variants as primers and inhibitors of viral cDNA synthesis by HIV reverse transcriptase in vitro. Nucleic Acids Symp. Ser., 37, 291–292. [PubMed] [Google Scholar]
- 25.Szewczak A.A., Podell,E.R., Bevilacqua,P.C. and Cech,T.R. (1998) Thermodynamic stability of the P4-P6 domain RNA tertiary structure measured by temperature gradient gel electrophoresis. Biochemistry, 37, 11162–11170. [DOI] [PubMed] [Google Scholar]
- 26.Aphasizhev R., Theobald-Dietrich,A., Kostyuk,D., Kochetkov,S.N., Kisselev,L., Giege,R. and Fasiolo,F. (1997) Structure and aminoacylation capacities of tRNA transcripts containing deoxyribonucleotides. RNA, 3, 893–904. [PMC free article] [PubMed] [Google Scholar]
- 27.Brion P., Michel,F., Schroeder,R. and Westhof,E. (1999) Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucleic Acids Res., 27, 2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viglasky V., Antalik,M., Bagel’ova,J., Tomori,Z. and Podhradsky,D. (2000) Heat-induced conformational transition of cytochrome c observed by temperature gradient gel electrophoresis at acidic pH. Electrophoresis, 21, 850–858. [DOI] [PubMed] [Google Scholar]
- 29.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotides interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 30.Faucon B., Mergny,J.-L. and Hélène,C. (1996) Effect of third strand composition on triple helix formation: purine versus pyrimidine oligodeoxynucleotides. Nucleic Acids Res., 24, 3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tinoco I. (1960) Hypochromism in Polynucleotides J. Am. Chem. Soc., 82, 4785–4790. [Google Scholar]
- 32.Belotserkovskii B.P. and Johnston,B.H. (1997) A random-walk model for retardation of interacting species during gel electrophoresis: implications for gel-shift assays. Biophys. J., 73, 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]