Abstract
The title compound, C14H11BrN2O5·2H2O, crystallizes as hydrogen-bonded sheets. The 2-hydroxy group on the benzylidene group forms an intramolecular hydrogen bond to the N atom of the C=N double bond. The amino N atom is a hydrogen-bond donor to a water molecule. The hydroxy group on the benzohydrazide group is a hydrogen-bond donor to one acceptor site, whereas each water molecule is a hydrogen-bond donor to two acceptor sites.
Related literature
For the structure of a similar Schiff-base ligand, 5-bromosalicylaldehyde benzoylhydrazone, see: Liu et al. (2006 ▶).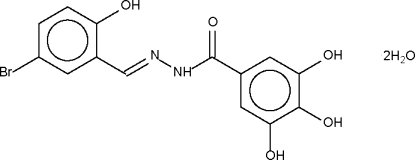
Experimental
Crystal data
C14H11BrN2O5·2H2O
M r = 403.19
Monoclinic,
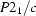
a = 30.8424 (8) Å
b = 3.7999 (1) Å
c = 12.8484 (4) Å
β = 90.280 (2)°
V = 1505.79 (7) Å3
Z = 4
Mo Kα radiation
μ = 2.77 mm−1
T = 100 (2) K
0.30 × 0.03 × 0.03 mm
Data collection
Bruker SMART APEX diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.658, T max = 0.921
9964 measured reflections
3424 independent reflections
2914 reflections with I > 2σ(I)
R int = 0.039
Refinement
R[F 2 > 2σ(F 2)] = 0.064
wR(F 2) = 0.155
S = 1.22
3424 reflections
241 parameters
10 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 1.08 e Å−3
Δρmin = −1.82 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶); software used to prepare material for publication: publCIF (Westrip, 2008 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808022708/bt2751sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808022708/bt2751Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1o⋯N1 | 0.84 (1) | 1.91 (5) | 2.616 (6) | 141 (7) |
| O3—H3o⋯O2w | 0.84 (1) | 1.96 (4) | 2.736 (6) | 153 (7) |
| O4—H4o⋯O2wi | 0.84 (1) | 1.81 (3) | 2.623 (8) | 163 (9) |
| O5—H5o⋯O2ii | 0.84 (1) | 1.93 (2) | 2.764 (5) | 171 (7) |
| O1w—H1w1⋯O2iii | 0.84 (1) | 1.98 (2) | 2.812 (5) | 170 (6) |
| O1w—H1w2⋯O1ii | 0.84 (1) | 2.09 (2) | 2.914 (6) | 167 (6) |
| O2w—H2w1⋯O3iv | 0.84 (1) | 2.13 (5) | 2.845 (9) | 142 (8) |
| O2w—H2w2⋯O4v | 0.84 (1) | 2.12 (4) | 2.900 (8) | 154 (8) |
Symmetry codes: (i) 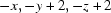 ; (ii)
; (ii) 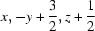 ; (iii)
; (iii) 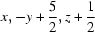 ; (iv)
; (iv) 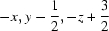 ; (v)
; (v) 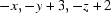 .
.
Acknowledgments
We thank the Science Fund (12–02-03–2031, 12–02-03–2051) and the University of Malaya (PJP) for supporting this study. We are grateful to the University of Malaya for the purchase of the diffractometer.
supplementary crystallographic information
Comment
This study extends the structural study on the Schiff base, 5-bromosalicylaldehyde benzoylhydrazone (Liu et al., 2006) as the title compound (Scheme I, Fig. 1) has several hydroxy groups on one of the aromatic rings. The compound crystallizes with two lattice water molecules. Hydrogen bonding interactions (Table 1) give rise to a layer motif.
Experimental
3,4,5-Trihydroxybenzoylhydrazide (0.65 g, 3.5 mmol) and 5-bromo-2-hydroxybenzaldehyde (0.70 g, 3.5 mmol) were heated for 12 h in ethanol. The solvent was removed and the product recrystallized from ethanol.
Refinement
Carbon and nitrogen-bound H-atoms were placed in calculated positions (C—H 0.95 Å; N–H 0.88 Å) and were included in the refinement in the riding model approximation, with Uiso(H) 1.2 Ueq(C). The hydroxy and water H-atoms were located in a difference Fourier map, and were refined with distance restraints of O–H 0.84±0.01 Å and H···H 1.37±0.01 Å.
The final difference Fourier map had a peak of 1.37eÅ-3 at 0.69Å from Br1 and a hole of -1.81eÅ-3 at 1.33Å from C2.
Figures
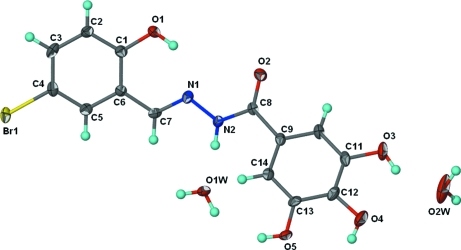
Fig. 1.
View (Barbour, 2001) of N-(5-bromo-2-hydroxybenzylidene)-3,4,5-trihydroxybenzohydrazide with displacement ellipsoids at the 70% probability level. Hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C14H11BrN2O5·2H2O | F000 = 816 |
| Mr = 403.19 | Dx = 1.779 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2758 reflections |
| a = 30.8424 (8) Å | θ = 3.2–27.4º |
| b = 3.7999 (1) Å | µ = 2.77 mm−1 |
| c = 12.8484 (4) Å | T = 100 (2) K |
| β = 90.280 (2)º | Needle, colorless |
| V = 1505.79 (7) Å3 | 0.30 × 0.03 × 0.03 mm |
| Z = 4 |
Data collection
| Bruker SMART APEX diffractometer | 3424 independent reflections |
| Radiation source: fine-focus sealed tube | 2914 reflections with I > 2σ(I) |
| Monochromator: graphite | Rint = 0.039 |
| T = 100(2) K | θmax = 27.5º |
| ω scans | θmin = 1.3º |
| Absorption correction: Multi-scan(SADABS; Sheldrick, 1996) | h = −38→40 |
| Tmin = 0.658, Tmax = 0.921 | k = −4→4 |
| 9964 measured reflections | l = −15→16 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.064 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.155 | w = 1/[σ2(Fo2) + (0.0499P)2 + 10.2476P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.22 | (Δ/σ)max = 0.001 |
| 3424 reflections | Δρmax = 1.08 e Å−3 |
| 241 parameters | Δρmin = −1.82 e Å−3 |
| 10 restraints | Extinction correction: none |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.467910 (16) | 0.31017 (16) | 0.89799 (4) | 0.01787 (17) | |
| O1 | 0.32358 (12) | 0.6446 (12) | 0.5944 (3) | 0.0201 (9) | |
| H1O | 0.3020 (15) | 0.745 (18) | 0.621 (5) | 0.030* | |
| O2 | 0.20078 (11) | 0.9755 (11) | 0.6545 (3) | 0.0182 (8) | |
| O3 | 0.05472 (13) | 1.3768 (13) | 0.7962 (4) | 0.0295 (11) | |
| H3O | 0.0335 (16) | 1.36 (2) | 0.836 (5) | 0.044* | |
| O4 | 0.05208 (14) | 1.1036 (15) | 0.9894 (4) | 0.0353 (12) | |
| H4O | 0.048 (3) | 0.933 (15) | 1.030 (6) | 0.053* | |
| O5 | 0.12324 (12) | 0.7692 (12) | 1.0737 (3) | 0.0182 (9) | |
| H5O | 0.1471 (12) | 0.682 (18) | 1.092 (5) | 0.027* | |
| O1W | 0.25912 (12) | 1.3152 (13) | 0.9988 (3) | 0.0209 (9) | |
| H1W1 | 0.2444 (17) | 1.382 (18) | 1.050 (3) | 0.031* | |
| H1W2 | 0.2806 (14) | 1.199 (17) | 1.020 (4) | 0.031* | |
| O2W | −0.02887 (15) | 1.3435 (17) | 0.8657 (5) | 0.0506 (16) | |
| H2W1 | −0.042 (3) | 1.30 (2) | 0.810 (4) | 0.076* | |
| H2W2 | −0.038 (3) | 1.534 (14) | 0.890 (7) | 0.076* | |
| N1 | 0.27850 (13) | 0.8683 (12) | 0.7531 (3) | 0.0139 (9) | |
| N2 | 0.24174 (13) | 0.9859 (13) | 0.8027 (3) | 0.0138 (9) | |
| H2N | 0.2431 | 1.0511 | 0.8684 | 0.017* | |
| C1 | 0.35563 (16) | 0.5778 (15) | 0.6648 (4) | 0.0144 (10) | |
| C2 | 0.39381 (16) | 0.4251 (16) | 0.6295 (4) | 0.0173 (11) | |
| H2 | 0.3970 | 0.3730 | 0.5576 | 0.021* | |
| C3 | 0.42746 (17) | 0.3479 (16) | 0.6985 (4) | 0.0193 (12) | |
| H3 | 0.4535 | 0.2431 | 0.6743 | 0.023* | |
| C4 | 0.42245 (16) | 0.4256 (16) | 0.8025 (4) | 0.0160 (11) | |
| C5 | 0.38470 (16) | 0.5781 (15) | 0.8398 (4) | 0.0143 (10) | |
| H5 | 0.3820 | 0.6294 | 0.9118 | 0.017* | |
| C6 | 0.35065 (15) | 0.6562 (15) | 0.7712 (4) | 0.0126 (10) | |
| C7 | 0.31105 (16) | 0.7988 (15) | 0.8138 (4) | 0.0130 (10) | |
| H7 | 0.3089 | 0.8415 | 0.8865 | 0.016* | |
| C8 | 0.20343 (16) | 1.0010 (13) | 0.7503 (4) | 0.0108 (10) | |
| C9 | 0.16507 (16) | 1.0460 (15) | 0.8180 (4) | 0.0130 (10) | |
| C10 | 0.12793 (16) | 1.2046 (16) | 0.7771 (4) | 0.0163 (11) | |
| H10 | 0.1281 | 1.2984 | 0.7086 | 0.020* | |
| C11 | 0.09101 (17) | 1.2242 (15) | 0.8368 (4) | 0.0176 (12) | |
| C12 | 0.09010 (17) | 1.0775 (17) | 0.9364 (4) | 0.0204 (12) | |
| C13 | 0.12713 (17) | 0.9146 (15) | 0.9777 (4) | 0.0152 (11) | |
| C14 | 0.16479 (17) | 0.9040 (15) | 0.9188 (4) | 0.0149 (11) | |
| H14 | 0.1903 | 0.8006 | 0.9468 | 0.018* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0094 (2) | 0.0195 (3) | 0.0247 (3) | −0.0001 (2) | −0.00302 (17) | −0.0020 (3) |
| O1 | 0.0160 (18) | 0.032 (3) | 0.0125 (17) | 0.0031 (18) | −0.0028 (14) | −0.0016 (17) |
| O2 | 0.0127 (17) | 0.027 (2) | 0.0148 (18) | −0.0039 (16) | −0.0015 (14) | 0.0021 (17) |
| O3 | 0.0142 (19) | 0.041 (3) | 0.034 (2) | 0.0120 (19) | −0.0051 (17) | −0.005 (2) |
| O4 | 0.016 (2) | 0.060 (4) | 0.030 (2) | 0.009 (2) | 0.0035 (18) | −0.013 (2) |
| O5 | 0.0129 (17) | 0.031 (2) | 0.0110 (17) | −0.0007 (17) | 0.0027 (14) | −0.0045 (16) |
| O1W | 0.0162 (18) | 0.035 (2) | 0.0113 (17) | 0.0027 (18) | 0.0015 (14) | −0.0044 (18) |
| O2W | 0.016 (2) | 0.049 (4) | 0.087 (5) | 0.010 (2) | 0.010 (2) | 0.013 (3) |
| N1 | 0.0107 (19) | 0.014 (3) | 0.017 (2) | −0.0016 (17) | 0.0010 (16) | −0.0008 (18) |
| N2 | 0.0096 (19) | 0.019 (3) | 0.013 (2) | −0.0004 (18) | −0.0007 (15) | −0.0028 (18) |
| C1 | 0.014 (2) | 0.016 (3) | 0.014 (2) | −0.002 (2) | −0.0013 (19) | −0.002 (2) |
| C2 | 0.013 (2) | 0.025 (3) | 0.014 (2) | −0.001 (2) | 0.0024 (19) | 0.000 (2) |
| C3 | 0.013 (2) | 0.020 (3) | 0.025 (3) | 0.001 (2) | 0.006 (2) | −0.003 (2) |
| C4 | 0.012 (2) | 0.014 (3) | 0.021 (3) | −0.001 (2) | −0.003 (2) | 0.003 (2) |
| C5 | 0.015 (2) | 0.013 (3) | 0.015 (2) | −0.001 (2) | 0.0005 (19) | −0.002 (2) |
| C6 | 0.010 (2) | 0.015 (3) | 0.013 (2) | −0.003 (2) | 0.0008 (18) | 0.001 (2) |
| C7 | 0.013 (2) | 0.015 (3) | 0.012 (2) | −0.006 (2) | 0.0022 (18) | −0.003 (2) |
| C8 | 0.013 (2) | 0.004 (3) | 0.015 (2) | −0.0007 (18) | −0.0019 (18) | 0.0009 (19) |
| C9 | 0.011 (2) | 0.013 (3) | 0.015 (2) | 0.000 (2) | −0.0027 (19) | −0.003 (2) |
| C10 | 0.013 (2) | 0.016 (3) | 0.020 (3) | 0.001 (2) | −0.0034 (19) | 0.000 (2) |
| C11 | 0.017 (2) | 0.013 (3) | 0.022 (3) | 0.008 (2) | −0.007 (2) | −0.007 (2) |
| C12 | 0.012 (2) | 0.027 (3) | 0.022 (3) | 0.002 (2) | 0.001 (2) | −0.012 (2) |
| C13 | 0.016 (2) | 0.017 (3) | 0.013 (2) | 0.001 (2) | 0.0025 (19) | −0.008 (2) |
| C14 | 0.013 (2) | 0.016 (3) | 0.015 (2) | 0.001 (2) | −0.0022 (19) | −0.006 (2) |
Geometric parameters (Å, °)
| Br1—C4 | 1.909 (5) | C1—C6 | 1.408 (7) |
| O1—C1 | 1.361 (6) | C2—C3 | 1.393 (8) |
| O1—H1O | 0.840 (10) | C2—H2 | 0.9500 |
| O2—C8 | 1.236 (6) | C3—C4 | 1.378 (8) |
| O3—C11 | 1.362 (6) | C3—H3 | 0.9500 |
| O3—H3O | 0.838 (10) | C4—C5 | 1.388 (7) |
| O4—C12 | 1.362 (7) | C5—C6 | 1.400 (7) |
| O4—H4O | 0.840 (10) | C5—H5 | 0.9500 |
| O5—C13 | 1.357 (7) | C6—C7 | 1.446 (7) |
| O5—H5O | 0.841 (10) | C7—H7 | 0.9500 |
| O1W—H1W1 | 0.839 (10) | C8—C9 | 1.482 (7) |
| O1W—H1W2 | 0.838 (10) | C9—C10 | 1.395 (7) |
| O2W—H2W1 | 0.838 (10) | C9—C14 | 1.403 (7) |
| O2W—H2W2 | 0.839 (10) | C10—C11 | 1.378 (7) |
| N1—C7 | 1.296 (7) | C10—H10 | 0.9500 |
| N1—N2 | 1.378 (6) | C11—C12 | 1.396 (8) |
| N2—C8 | 1.359 (6) | C12—C13 | 1.401 (8) |
| N2—H2N | 0.8800 | C13—C14 | 1.390 (7) |
| C1—C2 | 1.391 (7) | C14—H14 | 0.9500 |
| C1—O1—H1O | 113 (5) | C5—C6—C7 | 118.3 (4) |
| C11—O3—H3O | 111 (6) | C1—C6—C7 | 122.9 (5) |
| C12—O4—H4O | 112 (6) | N1—C7—C6 | 120.1 (4) |
| C13—O5—H5O | 110 (5) | N1—C7—H7 | 120.0 |
| H1W1—O1W—H1W2 | 109.6 (18) | C6—C7—H7 | 120.0 |
| H2W1—O2W—H2W2 | 109.5 (18) | O2—C8—N2 | 122.9 (4) |
| C7—N1—N2 | 115.1 (4) | O2—C8—C9 | 123.0 (4) |
| C8—N2—N1 | 120.0 (4) | N2—C8—C9 | 114.1 (4) |
| C8—N2—H2N | 120.0 | C10—C9—C14 | 120.3 (5) |
| N1—N2—H2N | 120.0 | C10—C9—C8 | 119.0 (5) |
| O1—C1—C2 | 118.3 (5) | C14—C9—C8 | 120.4 (5) |
| O1—C1—C6 | 121.6 (5) | C11—C10—C9 | 119.5 (5) |
| C2—C1—C6 | 120.1 (5) | C11—C10—H10 | 120.2 |
| C1—C2—C3 | 120.6 (5) | C9—C10—H10 | 120.2 |
| C1—C2—H2 | 119.7 | O3—C11—C10 | 119.3 (5) |
| C3—C2—H2 | 119.7 | O3—C11—C12 | 120.1 (5) |
| C4—C3—C2 | 119.1 (5) | C10—C11—C12 | 120.6 (5) |
| C4—C3—H3 | 120.5 | O4—C12—C11 | 116.7 (5) |
| C2—C3—H3 | 120.5 | O4—C12—C13 | 123.0 (5) |
| C3—C4—C5 | 121.5 (5) | C11—C12—C13 | 120.3 (5) |
| C3—C4—Br1 | 119.2 (4) | O5—C13—C14 | 124.1 (5) |
| C5—C4—Br1 | 119.3 (4) | O5—C13—C12 | 116.7 (5) |
| C4—C5—C6 | 120.0 (5) | C14—C13—C12 | 119.2 (5) |
| C4—C5—H5 | 120.0 | C13—C14—C9 | 120.0 (5) |
| C6—C5—H5 | 120.0 | C13—C14—H14 | 120.0 |
| C5—C6—C1 | 118.7 (5) | C9—C14—H14 | 120.0 |
| C7—N1—N2—C8 | −167.2 (5) | N2—C8—C9—C10 | 154.0 (5) |
| O1—C1—C2—C3 | −179.3 (5) | O2—C8—C9—C14 | 147.8 (5) |
| C6—C1—C2—C3 | 0.1 (9) | N2—C8—C9—C14 | −31.4 (7) |
| C1—C2—C3—C4 | −0.1 (9) | C14—C9—C10—C11 | 0.6 (9) |
| C2—C3—C4—C5 | 0.1 (9) | C8—C9—C10—C11 | 175.2 (5) |
| C2—C3—C4—Br1 | 178.5 (4) | C9—C10—C11—O3 | −179.5 (5) |
| C3—C4—C5—C6 | 0.0 (9) | C9—C10—C11—C12 | −2.0 (9) |
| Br1—C4—C5—C6 | −178.4 (4) | O3—C11—C12—O4 | −1.4 (8) |
| C4—C5—C6—C1 | −0.1 (8) | C10—C11—C12—O4 | −178.9 (6) |
| C4—C5—C6—C7 | 177.0 (5) | O3—C11—C12—C13 | 178.9 (5) |
| O1—C1—C6—C5 | 179.4 (5) | C10—C11—C12—C13 | 1.4 (9) |
| C2—C1—C6—C5 | 0.1 (8) | O4—C12—C13—O5 | 2.2 (9) |
| O1—C1—C6—C7 | 2.5 (9) | C11—C12—C13—O5 | −178.1 (5) |
| C2—C1—C6—C7 | −176.9 (5) | O4—C12—C13—C14 | −179.1 (6) |
| N2—N1—C7—C6 | 176.8 (5) | C11—C12—C13—C14 | 0.6 (9) |
| C5—C6—C7—N1 | −178.7 (5) | O5—C13—C14—C9 | 176.7 (5) |
| C1—C6—C7—N1 | −1.8 (8) | C12—C13—C14—C9 | −1.9 (8) |
| N1—N2—C8—O2 | −13.6 (8) | C10—C9—C14—C13 | 1.3 (8) |
| N1—N2—C8—C9 | 165.7 (5) | C8—C9—C14—C13 | −173.2 (5) |
| O2—C8—C9—C10 | −26.7 (8) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1o···N1 | 0.84 (1) | 1.91 (5) | 2.616 (6) | 141 (7) |
| O3—H3o···O2w | 0.84 (1) | 1.96 (4) | 2.736 (6) | 153 (7) |
| O4—H4o···O2wi | 0.84 (1) | 1.81 (3) | 2.623 (8) | 163 (9) |
| O5—H5o···O2ii | 0.84 (1) | 1.93 (2) | 2.764 (5) | 171 (7) |
| O1w—H1w1···O2iii | 0.84 (1) | 1.98 (2) | 2.812 (5) | 170 (6) |
| O1w—H1w2···O1ii | 0.84 (1) | 2.09 (2) | 2.914 (6) | 167 (6) |
| O2w—H2w1···O3iv | 0.84 (1) | 2.13 (5) | 2.845 (9) | 142 (8) |
| O2w—H2w2···O4v | 0.84 (1) | 2.12 (4) | 2.900 (8) | 154 (8) |
Symmetry codes: (i) −x, −y+2, −z+2; (ii) x, −y+3/2, z+1/2; (iii) x, −y+5/2, z+1/2; (iv) −x, y−1/2, −z+3/2; (v) −x, −y+3, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT2751).
References
- Barbour, L. J. (2001). J. Supramol. Chem.1, 189–191.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Liu, H.-Y., Wang, H.-Y., Gao, F., Lu, Z.-S. & Niu, D.-Z. (2006). Acta Cryst. E62, o4495–o4496.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2008). publCIF In preparation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536808022708/bt2751sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536808022708/bt2751Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report