Abstract
Enterococcus faecalis produces a specific penicillin-binding protein (PBP5) that mediates high-level resistance to the cephalosporin class of β-lactam antibiotics. Deletion of a locus encoding a previously uncharacterized two-component regulatory system of E. faecalis (croRS) led to a 4,000-fold reduction in the MIC of the expanded-spectrum cephalosporin ceftriaxone. The cytoplasmic domain of the sensor kinase (CroS) was purified and shown to catalyze ATP-dependent autophosphorylation followed by transfer of the phosphate to the mated response regulator (CroR). The croR and croS genes were cotranscribed from a promoter (croRp) located in the rrnC-croR intergenic region. A putative seryl-tRNA synthetase gene (serS) located immediately downstream from croS did not appear to be a target of CroRS regulation or to play a role in ceftriaxone resistance. A plasmid-borne croRp-lacZ fusion was trans-activated by the CroRS system in response to the presence of ceftriaxone in the culture medium. The fusion was also induced by representatives of other classes of β-lactam antibiotics and by inhibitors of early and late steps of peptidoglycan synthesis. The croRS null mutant produced PBP5, and expression of an additional copy of pbp5 under the control of a heterologous promoter did not restore ceftriaxone resistance. Deletion of croRS was not associated with any defect in the synthesis of the nucleotide precursor UDP-MurNAc-pentapeptide or of the d-Ala4→l-Ala-l-Ala-Lys3 peptidoglycan cross-bridge. Thus, the croRS mutant was susceptible to ceftriaxone despite the production of PBP5 and the synthesis of wild-type peptidoglycan precursors. These observations constitute the first description of regulatory genes essential for PBP5-mediated β-lactam resistance in enterococci.
Enterococcus faecalis and E. faecium are opportunistic pathogens that are common causes of urinary tract infections, bacteremia, and endocarditis (20). Enterococcal infections are difficult to treat, as enterococci are intrinsically resistant to various antibiotics and can acquire, mainly by horizontal gene transfer, high-level resistance to virtually all antimicrobial agents. The complete genome sequence of E. faecalis strain V583 revealed an unusually high (25%) content of mobile elements and exogenously acquired DNA, including virulence factors and antibiotic resistance genes (22). The plasticity of the genome correlates with the facility of enterococci to acquire novel resistance mechanisms and to transfer the corresponding genes to other genera, as exemplified by the emergence of high-level glycopeptide resistance in E. faecalis and E. faecium in the late 1980s and the dissemination of the same gene cluster in Staphylococcus aureus 15 years later (8).
Enterococci are resistant to the newer cephalosporins which have been developed to treat infections due to gram-negative bacteria producing β-lactamases. Treatment with cephalosporins is one of the risk factors for colonization and infection by multidrug-resistant enterococci. Cephalosporin resistance is mediated by a specific class B penicillin-binding protein (PBP) commonly referred to as low-affinity PBP5 (7, 27). Production of PBP5 also confers moderate-level resistance to ampicillin (MIC, 2 to 16 μg/ml). Acquisition of higher levels of ampicillin resistance, seen mainly in E. faecium, results from overproduction of PBP5 (11, 14), amino acid substitutions that further decrease interaction of PBP5 with β-lactams (26, 32), and modification of as-yet-unidentified non-PBP factors (18, 29).
In this report, we show that a two-component regulatory system (designated CroRS [for “ceftriaxone resistance”]) is essential for intrinsic β-lactam resistance in E. faecalis. This system, designated RR05-HK05 in the classification of Hancock and Perego (15), was initially chosen because of sequence similarity with two-component systems that control acquired enterococcal resistance to the glycopeptide antibiotics vancomycin and teicoplanin (2, 15). We report the resistance phenotype associated with deletions from the croRS locus, purification of the CroR response regulator and of a soluble fragment of the CroS histidine protein kinase to test their activity, and transcriptional analysis of the croRS locus. Since defects in the assembly of peptidoglycan precursors are associated with impaired expression of methicillin resistance in S. aureus (10, 25), we also analyzed the impact of the croRS deletion on the assembly of cytoplasmic precursors and on peptidoglycan cross-bridge formation.
MATERIALS AND METHODS
Growth conditions and susceptibility tests.
Bacterial strains were grown in brain heart infusion (BHI) broth or agar (Becton Dickinson, le Pont de Claix, France) at 37°C. MICs of ampicillin (Bristol-Myers, Paris, France) and ceftriaxone (Laboratoires Roche, Neuilly, France) were determined with 105 CFU per spot on BHI agar after 48 h of incubation.
Deletion of the croR and croS genes.
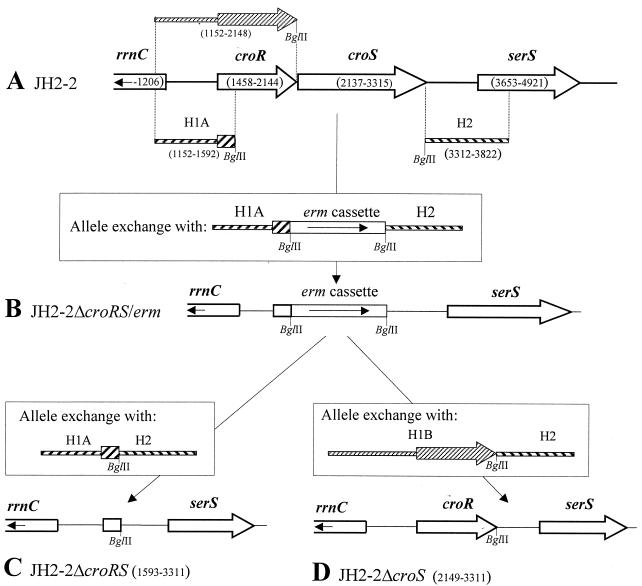
Deletions were made from the chromosome of E. faecalis JH2-2 by homologous recombination using derivatives of the suicide vector pHS1, which is thermosensitive for replication and confers gentamicin resistance (A. Arbeloa and M. Arthur, unpublished data). Briefly, DNA fragments (H1A, H1B, and H2) flanking the sequences targeted for deletion were amplified with primers (for H1A, primers 5′-ATTGATTTCTGAATCGC-3′ and 5′-AGATCTATCTGGTGTTGTGTGC-3′; for H1B, primers 5′-ATTGATTTCTGAATCGC-3′ and 5′-TTTTAGATCTTTAACGAGCATCGATCTTAT-3′; and for H2, primers 5′-AGATCTGAGTTAATTGACATCCC-3′ and 5′-GCAGACACATCATTCCG-3′) containing BglII restriction sites (underlined) to facilitate subsequent cloning steps. The fragments were cloned (with or without an intervening BglII erythromycin resistance cassette [erm]) into pHS1 to generate the inserts (H1A-erm-H2, H1A-H2, and H1B-H2) as shown in the insets in Fig. 1. To replace croRS by erm (ΔcroRS/erm), the derivative of plasmid pHS1 carrying H1A-erm-H2 (pHS1ΩH1A-erm-H2) was introduced by electrotransformation into E. faecalis JH2-2. Replacement of croRS by erm was selected on agar containing erythromycin (10 μg/ml) at the nonpermissive temperature (42°C) for plasmid replication (Fig. 1B). One clone resistant to erythromycin and susceptible to gentamicin was identified by replica plating on BHI agar containing gentamicin (128 μg/ml) and was designated JH2-2ΔcroRS/erm.
FIG. 1.
Deletions from the croRS locus. (A) Map of the wild-type locus of E. faecalis JH2-2 and location of the DNA fragments (H1A, H1B, and H2) used for allele exchange by homologous recombination. Numbers in parentheses indicate the coordinates of the extremities of the croR, croS, and serS open reading frames (open arrows), of the H1A, H1B, and H2 DNA fragments (hatched), and of a portion of the rrnC rRNA gene cluster (open box). BglII restriction sites were introduced at one extremity of H1A, H1B, and H2. (B) Replacement of croRS by an erythromycin resistance gene cassette (erm). JH2-2ΔcroRS/erm was obtained by homologous recombination between the wild-type croRS locus of JH2-2 and a derivative of the thermosensitive plasmid pHS1 carrying the H1A-erm-H2 DNA insert depicted in the inset. (C) To construct JH2-2ΔcroRS, the erm cassette was deleted from the chromosome of JH2-2ΔcroRS/erm by allele exchange with H1A directly linked to H2. (D) To construct JH2-2ΔcroS, the erm cassette of JH2-2ΔcroRS/erm was replaced (using H1B linked to H2) by the croR open reading frame. Numbers in parentheses indicate the extremities of the deletions from JH2-2ΔcroRS and from JH2-2ΔcroS. Coordinate 1 corresponds to position 3,169,253 of the assembled E. faecalis genome at www.tigr.org.
Deletion of the erm cassette of JH2-2ΔcroRS/erm was obtained in two steps using a derivative of pHS1 carrying H1A directly fused to H2 (H1A-H2; Fig. 1C). In the first step, integration of plasmid pHS1ΩH1A-H2 by homologous recombination was selected at 42°C on agar containing gentamicin (128 μg/ml), generating a partial duplication of the locus, since the sequence of the pHS1 vector was flanked by the H1A-H2 and H1A-erm-H2 alleles. Serial subcultures at the permissive (28°C) and nonpermissive (42°C) temperatures in the absence of antibiotic were used to stimulate the excision and loss of pHS1ΩH1A-erm-H2, leaving the H1A-H2 allele in the chromosome. One clone (designated JH2-2ΔcroRS) was obtained by screening for gentamicin and erythromycin susceptibility.
Replacement of the erm cassette of JH2-2ΔcroRS/erm by the croR open reading frame was obtained by the same two-step procedure with a derivative of pHS1 carrying croR as a part of the H1B-H2 insert (Fig. 1D). The resulting clone, JH2-2ΔcroS, lacked the precise croS open reading frame. PCR and Southern blot hybridization were used to confirm that the expected deletions from and gene replacements in JH2-2ΔcroRS/erm, JH2-2ΔcroRS, and JH2-2ΔcroS had taken place.
Shuttle plasmids for croS, croRS, serS, and pbp5 expression.
The croS open reading frame was amplified with primers P80 and P87. Primer P80 (5′-ATCGAGGTACCGGATCCTAAAATATCGGAGGGTTTATTATGCTCGTTAAACCTAAAAA-3′) contained a KpnI restriction site (underlined), an artificial ribosome binding site (italicized), and 20 bases complementary to the 5′ end of croS that included the translation initiation codon (italicized). Primer P87 (5′-ATCGATCTAGAAGATCTTTAACTCTCTGATTTCTTGT-3′) contained an XbaI restriction site (underlined) and 20 bases complementary to the 3′ end of croS that included the stop codon (italicized). The croS open reading frame was cloned under the control of the aphA-3p promoter (1) in the shuttle vector pAT18 (30) to generate pRQ12(croS). The serS open reading frame was amplified with primers DS (5′-AAGAGCTCTCATTTCGTCCCAAGAATATT-3′) and ES (5′-TTTTGGTACCTTATTTAATAACTG-3′), digested with SacI and KpnI (underlined), and cloned under the control of the aphA-3p promoter to generate pYC5(serS). A DNA fragment containing the rrnC-croR intergenic region, croR, and croS (coordinates 1191 to 3315) was amplified with primer P25 (5′-AGTTCGGTACCTAAGACATGTAATAATATACCAA-3′) and P87 (described above) and cloned into pAT18 using BamHI (underlined) and XbaI to generate pRQ13(croRS). Plasmid pAA15 (Arbeloa and Arthur, unpublished) contains the PBP5 open reading frame cloned downstream from aphA-3p. DNA sequencing was performed for all recombinant plasmids used in this study to check the accuracy of the PCRs.
Purification and analysis of mRNA.
Strains of E. faecalis were grown in 6 ml of BHI broth to an optical density at 600 nm (OD600) of 0.6. Bacteria were collected by centrifugation (12,000 × g for 30 s at 4°C) and treated with lysozyme and lysostaphin (GramCracker kit; Ambion Inc., Austin, Tex.), and total RNA was extracted with phenol and chloroform (RNAwiz; Ambion Inc.). RNA concentration and purity were determined by the absorbance at 260 nm (A260) and the A260/A280 ratio, respectively.
Mapping of the 5′ extremity of mRNA isolated in vivo was performed by primer extension with oligonucleotides P49 (5′-AATACTCAATAGTTCTACAATTTC-3′), P51 (5′-CACGCCACGGGTTTGTAGCTTTGC-3′), and P76 (5′-GACTTCTTTATAGATGAATGTTT-3′). Primers (10 pmol) were end labeled with [γ-32P]ATP (Perkin Elmer Life Sciences Inc., Boston, Mass.) (3,000 Ci/mmol) by using 10 U of polynucleotide kinase (primer extension system; Promega Corp., Madison, Wis.) and annealed to 30 μg of total RNA for 20 min at 58°C followed by 10 min at 20°C. Avian myeloblastosis virus reverse transcriptase (primer extension system; Promega) (1 U) was added, and incubation was continued for 30 min at 42°C. The reverse transcription products were analyzed in 6% denaturing polyacrylamide gels. DNA sequencing reactions were performed with the same primers (Sequenase version 2.0 DNA; USB Corp., Cleveland, Ohio).
Northern blot hybridization was performed with commercial denaturing and running buffers (NorthernMax; Ambion Inc.). Briefly, total RNA (30 μg) was denatured in formaldehyde loading dye for 15 min at 65°C. Electrophoresis was performed in formaldehyde denaturing gel at 5 V/cm for 3 h, and RNA was transferred by vacuum onto a nylon membrane (BrightStar-Plus; Ambion Inc.). RNA was cross-linked, using UV light at 254 nm (Stratalinker UV Cross-linker 1800; Stratagene, La Jolla, Calif.) (120,000 microjoules/cm2), to the membrane. Prehybridization was performed overnight at 68°C (ULTRAhyb solution; Ambion Inc.). Double-stranded DNA fragments used as probes (50 ng) were labeled with [α-32P]dCTP (Ready-to-Go DNA labeling beads; Amersham Pharmacia Biotech, Piscataway, N.J.) (3,000 Ci/mmol), denatured at 90°C for 10 min, and added to the prehybridization solution. Hybridization was performed overnight at 42°C. The membranes were washed (using washing solutions from a NorthernMax kit [Ambion Inc.]) twice with a low-stringency solution for 5 min at room temperature and twice with a high-stringency solution for 15 min at 90°C.
Purification of CroRH.
The croR open reading frame of E. faecalis JH2-2 was amplified with primers 1R61 (5′-TCATGAAAATTTTAGTTGC-3′) and 2R61His (5′-AGATCTACGAGCATCGATCTTAT-3′) containing BspHI and BglII restriction sites (underlined), respectively. The BspHI-BglII fragment was cloned into the expression vector pTRCHis60 (23) digested with NcoI and BglII. For protein production, Escherichia coli JM83 harboring plasmid pTRCHis60ΩcroRH was grown at 37°C to an OD600 of 0.7 in 1 liter of BHI broth containing ampicillin (100 μg/ml). Isopropyl-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and incubation was continued for 3 h at 30°C. Bacteria were harvested by centrifugation (8,000 × g for 20 min at 4°C), washed in 180 ml of 50 mM Tris-HCl (pH 7.5), and resuspended in 10 ml of the same buffer containing 100 mM NaCl. Bacteria were disrupted by sonication for 2 min with cooling, the extract was centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was mixed with 4 ml of Ni2+-nitrilotriacetate-agarose resin (Amersham Pharmacia Biotech, Saclay, France) previously equilibrated with 50 mM Tris-HCl (pH 7.8). After 1 h of incubation at 4°C, the resin was recovered by centrifugation and washed with 50 mM Tris-HCl (pH 7.8) containing increasing concentrations of imidazole (20, 25, 40, 100, and 250 mM). The protein fraction eluting at 250 mM was dialyzed with 50 mM Tris-HCl (pH 7.4). Gel filtration was performed on a Superdex HR10/30 column (Amersham Pharmacia Biotech) equilibrated with 50 mM Tris-HCl (pH 7.5) containing 300 mM NaCl at a flow rate of 0.5 ml/min.
Purification of CroSS.
A portion of the croS open reading frame of E. faecalis JH2-2 was amplified with primers S61int4 (5′-GGTGGTCATATGCTGGTGGATAGTACTGTCG-3′) and S61int3 (5′-GGTGGTTGCTCTTCCGCAACTCTCTGATTTCTTGTTG-3′), and the PCR product was digested with NdeI and SapI (underlined) and cloned into pTYB1 (New England Biolabs, Frankfurt am Main, Germany) digested with the same enzymes. The resulting plasmid, pTYB1ΩcroSS, encoded a fusion protein consisting of a methionine specified by the ATG initiation codon of pTYB1, residues 145 to 393 of croS, and the self-cleavable C-terminal intein tag. E. coli ER2566 (New England Biolabs) harboring pTYB1ΩcroSS was grown at 37°C to an OD650 of 0.5 in 3 liters of BHI broth containing ampicillin (300 μg/ml). IPTG was added to achieve a final concentration of 0.5 mM, and incubation was continued for 17 h at 16°C. CroSS was purified from a clarified lysate by affinity chromatography on chitin beads followed by cleavage of the fusion protein with 2-mercaptoethanol (50 mM) for 18 h at 20°C (IMPACT-CN kit; New England Biolabs). Gel filtration was performed as described above for CroRH.
Protein phosphorylation assays.
The kinetics of CroSS autophosphorylation was tested at 20°C in a total volume of 64 μl containing the purified protein (30 μM), [γ-32P]ATP triethylammonium salt (Amersham Pharmacia Biotech) (3.4 μM; 0.37 TBq/mmol), and buffer A (50 mM Tris-HCl, 25 mM KCl, 0.5 mM MgCl2, pH 7.4). Samples (12 μl) were taken at 0, 5, 10, 30, and 60 min, and the reaction was quenched by the addition of 5 μl of a solution containing 125 mM Tris-HCl (pH 6.8), 2.5% sodium dodecyl sulfate (SDS), 2 mM EDTA, 0.025% bromophenol blue, and 25% glycerol. Samples were applied directly to SDS-13.5% polyacrylamide gels. Gels were dried and subjected to autoradiography without Coomassie blue staining.
To test the transfer of the phosphate group from CroSS to CroRH, phosphorylated CroSS (phospho-CroSS) was prepared by incubating the protein (24 μM) with [γ-32P]ATP for 60 min in a total volume of 64 μl as described above. Phospho-CroSS was separated from [γ-32P]ATP by ultrafiltration (Microcon YM10; Millipore Corporation, Bedford, Mass.). CroRH (24 μM) was incubated with phospho-CroSS in buffer A (64 μl), and samples (15 μl) were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Assay for in vivo promoter activity.
DNA fragments were cloned upstream from the promoterless lacZ reporter gene of the promoter probing vector pTCV-lac (24). Strains of E. faecalis harboring derivatives of pTCV-lac were grown to an OD600 of 0.55 in broth containing erythromycin (10 μg/ml) in addition to the drug tested for induction. Mueller-Hinton broth (Bio-Rad, Marnes-la-Coquettes, France) was used for trimethoprim, and BHI broth was used for all other drugs. Bacteria were collected by centrifugation and permeabilized with toluene. The β-galactosidase activity was expressed in arbitrary units calculated according to the equation 103 × {(the OD420 value of the reaction mixture) − (1.75 × the OD550 value)/[the time of the reaction (in minutes) × the OD600 value of the quantity of cells used in the assay]}, as described previously (24).
Analysis of PBPs.
The technique used for the analysis of PBPs of the different strains was employed as previously described (31) except that labeling was performed with 40 μg of benzyl[14C]penicillin potassium (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England)/ml (2.11 GBq/mmol).
Peptidoglycan structure analysis.
Preparation and structure assignment of muropeptides by mass spectrometry were performed as previously described (6). Briefly, bacteria were grown at 37°C in BHI broth to an optical density of 0.8. Peptidoglycan was extracted with 8% SDS at 100°C, treated with pronase (200 μg/ml) and trypsin (200 μg/ml), and digested with lysozyme (200 μg/ml) and mutanolysin (200 μg/ml). Muropeptides were reduced with sodium borohydrate and separated by reverse-phase high-performance liquid chromatography (rpHPLC) on a C18 column (Interchrom, Monluçon, France) (3 μm; 4.6 by 250 mm) at a flow rate of 0.5 ml/min with a 0 to 20% gradient applied at between 10 and 90 min (buffer A, 0.05% trifluoroacetic acid in water; buffer B, 0.035% trifluoroacetic acid in acetonitrile [per volume]). The relative abundance of muropeptides was estimated according to the percentage of the integrate area of peaks detected by the absorbance at 210 nm. Mass spectral data were collected with an electrospray time-of-flight mass spectrometer operating in the positive mode (Qstar Pulsar I; Applied Biosystems, Courtaboeuf, France) directly connected to the C18 column (flow rate, 0.5 ml/min). The data were acquired with a capillary voltage of 5,200 V and a declustering potential of 20 V. The mass scan range was from m/z 400 to m/z 2,500, and the scan cycle was 1 s.
Preparation and analysis of the cytoplasmic peptidoglycan precursors.
Bacteria were grown to an OD650 of 0.7 and treated with vancomycin (100 μg/ml) for 15 min. Peptidoglycan precursors were extracted with formic acid (1.1 M) as previously described (4) and analyzed by rpHPLC with a μ-Bondapak C18 column (Waters, Milford, Mass.) (3.0 by 250 mm) at a flow rate of 0.5 ml/min with 50 mM ammonium formiate (pH 3.8). A methanol gradient (0 to 20%) was applied between 24 and 44 min, and elution with 20% methanol was continued for 10 min. The relative abundance of the UDP-MurNAc-peptide was estimated according to the percentage of the integrate area of peaks detected with the absorbance at 262 nm. For mass spectral analysis, products isolated by rpHPLC were lyophilized and dissolved in a solution containing acetonitrile (49.5%) and formic acid (0.5%). The samples were desalted by rpHPLC with isocratic elution (50% acetonitrile) at a flow rate of 10 μl/min. Tandem mass spectrometry analyzes were performed with a cone voltage of 55V and with argon as the collision gas at a pressure of 15 lb/in2 (energy, 20 to 50 eV) as previously described (6).
RESULTS
Role of the croRS locus in ceftriaxone resistance.
The croRS locus of E. faecalis encoded a putative response regulator (CroR) belonging to the OmpR-PhoB subclass and a putative sensor kinase (CroS) containing two clusters of hydrophobic amino acid residues that might correspond to transmembrane segments (Fig. 1A). The croRS locus was flanked by a copy of an rRNA gene cluster (rrnC) and a putative seryl-tRNA synthetase gene (serS). Deletions were made (using the suicide vector pHS1) from the croRS locus by allele exchange. In the first mutant, JH2-2ΔcroRS/erm (Fig. 1B), the sequence encoding a large C-terminal portion of CroR and the entire croS open reading frame was replaced by an erm erythromycin resistance cassette. The mutant retained the first 45 codons of croR. Deletion of a larger portion of croR by homologous recombination was not attempted, since this would have required the use of a significant portion of rrnC which is repeated in the three other rRNA clusters. JH2-2ΔcroRS was obtained by removing the erm cassette from the chromosome of JH2-2ΔcroRS/erm (Fig. 1C). Replacement of the cassette by the croR open reading frame generated JH2-2ΔcroS, which differed from wild-type JH2-2 by a precise deletion of the croS gene (Fig. 1D).
The croRS and croS deletions led to a 4,000-fold decrease in the MIC of ceftriaxone and a 4-fold decrease in the MIC of ampicillin (Table 1). Deletion of croRS also led to similarly large (>100-fold) decreases in the MICs of expanded-spectrum cephalosporins (e.g., cefuroxime and cefepime) and moderate (2- to 8-fold) decreases in the MICs of other β-lactams (e.g., cephalothin, imipenem, amdinocillin, and oxacillin). A similar phenotype was observed following deletion of the pbp5 gene from the chromosome of JH2-2 (JH2-2Δpbp5; Arbeloa and Arthur, unpublished). A trans-complementation of the croRS deletion was obtained with a DNA fragment containing the rrnC-croR intergenic region, croR, and croS cloned into the replicative vector pAT18 (Table 1). Expression of croS alone under the control of a heterologous promoter (aphA-3p) complemented the croS deletion. Thus, the croR and croS genes were both required for intrinsic β-lactam resistance.
TABLE 1.
Susceptibility of E. faecalis strains to β-lactam antibiotics
| Strain | Plasmida | MIC (μg/ml) of:
|
|
|---|---|---|---|
| Ceftriaxone | Ampicillin | ||
| JH2-2 | None | 1,000 | 2 |
| JH2-2Δpbp5 | None | 0.25 | 0.5 |
| JH2-2ΔcroRS | None | 0.25 | 0.5 |
| JH2-2ΔcroS | None | 0.25 | 0.5 |
| JH2-2ΔcroRS | pRQ13 (croRS) | 256 | 2 |
| JH2-2ΔcroS | pRQ13 (croRS) | 512 | 2 |
| JH2-2ΔcroRS | pRQ12 (croS) | 0.25 | 0.5 |
| JH2-2ΔcroS | pRQ12 (croS) | 1,000 | 2 |
| JH2-2ΔcroRS | pYC5 (serS) | 0.25 | 0.5 |
| JH2-2ΔcroRS | pAA15 (pbp5) | 0.5 | 0.5 |
| JH2-2Δpbp5 | pAA15 (pbp5) | 1,000 | 2 |
The croRS genes were expressed under the control of the native croRp promoter in pRQ13. The croS, serS, and pbp5 open reading frames were expressed under the control of the heterologous aphA-3p promoter in pRQ12, pYC5, and pAA15, respectively.
Deletion of croS and croRS was not associated with modification of the pattern of PBPs labeled with benzyl[14C]penicillin (data not shown). Expression of pbp5 under the control of aphA-3p did not restore β-lactam resistance in JH2-2ΔcroRS (Table 1) despite overproduction of PBP5. These results show that low-affinity PBP5 was produced but could not mediate ceftriaxone resistance in JH2-2ΔcroRS. Expression of the serS seryl tRNA synthetase gene under the control of aphA-3p did not restore ceftriaxone resistance in JH2-2ΔcroRS (Table 1). The role of the latter gene was investigated in the present study, since two-component regulatory systems frequently regulate adjacent genes.
Phosphotransfer reactions catalyzed by purified CroSS and CroRH.
A soluble fragment of CroS lacking the two putative trans-membrane segments of the protein was produced in E. coli as a translational fusion containing a C-terminal intein tag. Following affinity purification on chitin beads and cleavage of the tag, the soluble fragment of the sensor kinase (designated CroSS) was expected to differ from CroS by the absence of the first 144 amino acid residues and the presence of an additional methionine introduced for translation initiation. Analysis of purified CroSS (28,183 Da, 250 residues) by SDS-PAGE showed a 31-kDa protein band estimated to be 95% pure (3 mg of protein per liter of culture). Gel filtration under the conditions described in Materials and Methods revealed a protein peak with an estimated mass of 60 kDa, indicating that CroSS eluted as a dimer. No protein was detected at the elution volume expected for the monomer.
Full-length CroR fused to a C-terminal six-histidine tag (Ser-Arg-His6) was purified by affinity chromatography on a nickel column. The protein (designated CroRH) was judged using SDS-PAGE to be more than 95% pure and had an estimated mass of 27 kDa (30 mg per liter of culture). CroRH eluted as a monomer in the gel filtration column.
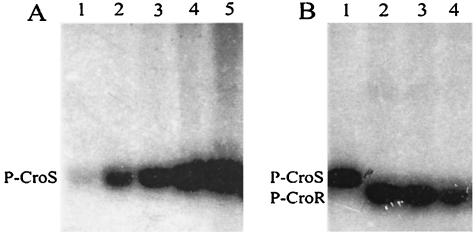
Autophosphorylation of CroSS was assayed by incubating the purified protein with [γ32-P]ATP (Fig. 2). A radioactive protein band corresponding to phospho-CroSS was detectable after 5 min of incubation and increased up to 60 min (Fig. 2A). The phospho-CroSS adduct was sufficiently stable to allow for removal of [γ-32P]ATP by ultrafiltration (Fig. 2B, lane 1). Upon addition of purified CroRH, the radiolabeled phosphate group was entirely transferred from CroSS to CroRH in less than 2 min. Upon further incubation, the intensity of the phospho-CroRH protein band slowly decreased due to dephosphorylation of the protein.
FIG. 2.
Phosphotransfer reactions catalyzed by CroSS and CroRH. (A) Kinetics of CroSS autophosphorylation. CroSS was incubated with [γ32-P]ATP for 0, 5, 10, 30, and 60 min (lanes 1 to 5, respectively) and applied to an SDS-13.5% polyacrylamide gel. (B) Transfer of the phosphate group from the phosphorylated form of CroSS (P-CroS) to CroRH. Phospho-CroSS was prepared (lane 1) and incubated with CroRH for 2, 5, and 20 min (lanes 2, 3, and 4, respectively).
Mapping of mRNA isolated in vivo.
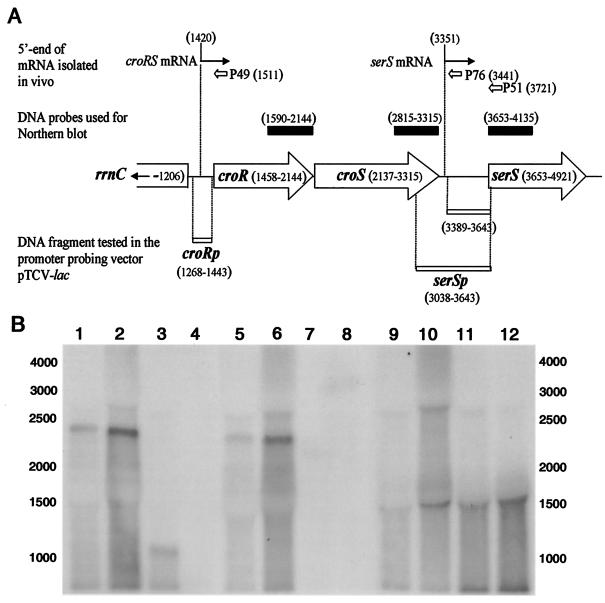
Reverse transcription of mRNA isolated (using primer P49 [Fig. 3A]) from E. faecalis JH2-2 revealed a single putative transcriptional start site upstream from croR (data not shown). The product of reverse transcription was more abundant for mRNA isolated from bacteria grown in the presence of ceftriaxone at 1,000 μg/ml, and additional products were not detected. The putative transcription start site detected for both growth conditions was preceded by two hexanucleotides separated by 18 bases (TTGTCC-N18-TAAAAT) resembling the −35 and −10 consensus sequences of vegetative promoters. Primer extension with oligonucleotide P76 (Fig. 3A) revealed a putative transcription start downstream from croS that was also preceded by sequences similar to the consensus sequence (TTGACA-N17-TATTCT). The corresponding transcript extended into the serS open reading frame, as a ca. 370-base reverse-transcription product was observed with primer P51 (data not shown). The other two extension products (182 and 131 bases) were unlikely to correspond to transcription initiation at additional sites since they may be accounted for by mRNA processing and a reverse transcription stop at a putative transcriptional terminator, respectively, as was shown for the serS gene of Bacillus subtilis (9). In addition, the DNA fragment delineated by coordinates 3,389 to 3,643 (Fig. 3A) did not display promoter activity in the promoter probing vector pTCV-lac (data not shown).
FIG. 3.
Transcriptional analysis of the croRS locus. (A) The positions of oligonucleotides P49, P51, and P76 used for primer extension and of the DNA fragments used as probes for the Northern blot hybridizations (solid boxes) are indicated above the map of the cluster. Open boxes below the map represent the DNA fragments tested for promoter activity in pTCV-lac. (B) Northern blot analysis of total RNA with probes generated from internal fragments of croR (lanes 1 to 4), croS (lanes 5 to 8), and serS (lanes 9 to 12). Total RNA was extracted from E. faecalis JH2-2 grown in the absence (lanes 1, 5, and 9) or presence (lanes 2, 6, and 10) of ceftriaxone (1,000 μg/ml). RNA from JH2-2ΔcroS (lanes 3, 7, and 11) and JH2-2ΔcroRS (lanes 4, 8, and 12) was extracted from bacteria grown in the absence of ceftriaxone.
Northern blot analysis of total RNA from JH2-2, JH2-2ΔcroRS, and JH2-2ΔcroS was performed with probes generated from internal fragments of croR, croS, and serS (Fig. 3B). The croR and croS probes detected the same ca. 2,400-base band in RNA preparations from JH2-2, indicating that the two genes were cotranscribed (lanes 1 and 5). Growth in the presence of ceftriaxone (1,000 μg/ml) led to the same hybridization pattern (lanes 2 and 6). As expected, the 1,161-bp croS deletion resulted in a decrease in the size of the RNA band detected by the croR probe in JH2-2ΔcroS (lane 3). A unique 1,500-base band was detected by the serS probe in RNA preparations from the three strains (lanes 9 to 12), indicating that croRS and serS were transcribed independently.
In vivo activity of the croRp and serSp promoters.
DNA fragments carrying the croR and serS promoters (Fig. 3A) were cloned upstream from the lacZ reporter gene of plasmid pTCV-lac and introduced into E. faecalis JH2-2, JH2-2ΔcroRS, and JH2-2ΔcroS. Control experiments were also performed with the vector alone and the heterologous aphA-3p promoter previously characterized in this system (24). Determination of β-galactosidase activity (Table 2) showed that the croRp promoter in JH2-2 was inducible by ceftriaxone. The basal level of expression of the croRp-lacZ transcriptional fusion appeared reduced in JH2-2ΔcroRS (fivefold) and in JH2-2ΔcroS (threefold) in comparison to the level seen with the JH2-2 host. The serSp promoter in JH2-2 did not respond to ceftriaxone, and similar β-galactosidase activity was detected in the ΔcroS and ΔcroRS mutants.
TABLE 2.
Promoter activities in various derivatives of E. faecalis JH2-2
| Promoter fused to lacZ | β-Galactosidase activities (mean arbitrary units ± SD) for straina:
|
|||
|---|---|---|---|---|
| JH2-2
|
JH2-2ΔcroRS | JH2-2ΔcroS | ||
| Not induced | Inducedb | Not induced | Not induced | |
| croRp | 5.8 ± 1.5 | 35.0 ± 8.0 | 1.3 ± 0.6 | 2.1 ± 0.7 |
| serSp | 6.4 ± 1.9 | 7.5 ± 1.1 | 13.5 ± 1.4 | 4.3 ± 0.6 |
| aphA-3p | 56.6 ± 14.0 | 66.4 ± 17.3 | 71.5 ± 4.0 | 76.7 ± 2.4 |
| None | <1 | <1 | <1 | <1 |
Experiments were performed in triplicate, and the mean values ± standard deviation (SD) of arbitrary units of β-galactosidase-specific activities are indicated.
Induction was performed with ceftriaxone at 1,000 μg/ml.
Representatives of various antibiotic classes were also tested for their capacity to induce the croRp-lacZ fusion in JH2-2 (Table 3). All β-lactam antibiotics that were tested acted as inducers, including narrow-, expanded-, and broad-spectrum cephalosporins, imipenem, ampicillin, oxacillin, and amdinocillin. Induction also occurred with inhibitors of early (phosphomycin and d-cycloserine) and late (vancomycin, moenomycin, ramoplanin, and bacitracin) steps of peptidoglycan synthesis. In contrast, no induction was observed with the aminoglycoside gentamicin (a ribosome inhibitor) and the fluoroquinolone ofloxacin (a DNA gyrase and topoisomerase IV inhibitor). The dihydrofolate reductase inhibitor trimethoprim was a weak inducer.
TABLE 3.
Induction of the croRp-lacZ fusion in E. faecalis JH2-2 by various antibioticsa
| Antibiotic | Tested range (μg/ml) | Drug concn (μg/ml) for maximum induction | Induction ratio |
|---|---|---|---|
| Cephalothin | 1-100 | 20 | 8.6 |
| Cefuroxime | 50-1,000 | 1,000 | 7.3 |
| Ceftriaxone | 20-1,000 | 1,000 | 6.0 |
| Cefepime | 100-800 | 200 | 7.8 |
| Ampicillin | 0.01-2 | 0.5 | 6.4 |
| Amdinocillin | 50-1,000 | 500 | 17.2 |
| Imipenem | 0.01-0.5 | 0.2 | 6.2 |
| Oxacillin | 1-300 | 100 | 8.0 |
| Fosfomycin | 0.01-40 | 40 | 12.9 |
| d-Cycloserine | 50-200 | 200 | 26.6 |
| Moenomycin | 0.01-0.4 | 0.3 | 8.9 |
| Bacitracin | 1-64 | 32 | 12.5 |
| Ramoplanin | 0.01-20 | 20 | 10.5 |
| Vancomycin | 0.01-5 | 2 | 4.7 |
| Gentamicin | 4-32 | 4 | 1.2 |
| Ofloxacin | 0.2-8 | 4 | 1.2 |
| Trimethoprim | 0.001-10 | 5 | 3.9 |
Bacteria were grown to an OD600 of 0.55 in broth containing erythromycin (10 μg/ml) in addition to the drug tested for induction. A minimum of three concentrations were tested, and the induction ratio was calculated for the concentration leading to the highest activity.
Ceftriaxone at low concentrations (0.05 to 0.25 μg/ml) did not induce the croRp-lacZ fusion in JH2-2ΔcroRS and JH2-2ΔcroS (data not shown). The role of CroR and CroS in induction could not be fully investigated with this drug, since the concentrations required in JH2-2 for induction were inhibitory for the mutants. For this reason, the glycosyltransferase inhibitor moenomycin was also tested, revealing induction in JH2-2 but not in JH2-2ΔcroRS or JH2-2ΔcroS (data not shown).
Structure of cytoplasmic peptidoglycan precursors and of muropeptides.
UDP-MurNAc-peptide precursors from E. faecalis JH2-2 and JH2-2ΔcroRS were compared by rpHPLC and mass spectrometry (Table 4). Deletion of the croRS locus was not associated with any defect in the assembly of the nucleotide UDP-MurNAc-pentapeptide, since precursors containing incomplete peptide stems were present in similar low amounts in both strains (UDP-MurNAc-tripeptide) or detected in neither strain (UDP-MurNAc-L-Ala and UDP-MurNAc-L-Ala-D-Glu). Addition of l-Ala to the ɛ-amino group of the pentapeptide stem of the nucleotide by the BppA1 transferase (5) resulted in similar amounts of UDP-MurNAc-hexapeptide in the ΔcroRS mutant and in the parental strain. As expected, the pools of UDP-MurNAc-hexapeptide were small, since the BppA1 transferase preferentially uses lipid intermediates as substrates (5).
TABLE 4.
Peptidoglycan precursors from E. faecalis JH2-2 and JH2-2ΔcroRS
| Precursora | RTb | Mass | Relative abundance (%)
|
|
|---|---|---|---|---|
| JH2-2 | ΔcroRS | |||
| UDP-MurNAc-tripeptide | 40.0 | 1,007.2 | 0.3 | 0.2 |
| UDP-MurNAc-pentapeptide | 44.6 | 1,149.3 | 95.7 | 95.6 |
| UDP-MurNAc-hexapeptide | 49.2 | 1,220.3 | 4.0 | 4.2 |
UDP-MurNAc-tripeptide, UDP-N-acetylmuramoyl-l-Ala-γ-d-Glu-l-Lys; UDP-MurNAc-pentapeptide, UDP-N-acetylmuramoyl-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala; UDP-MurNAc-hexapeptide, UDP-N-acetylmuramoyl-l-Ala-γ-d-Glu-l-Lys(Nɛ-l-Ala)-d-Ala-d-Ala.
RT, retention time in minutes.
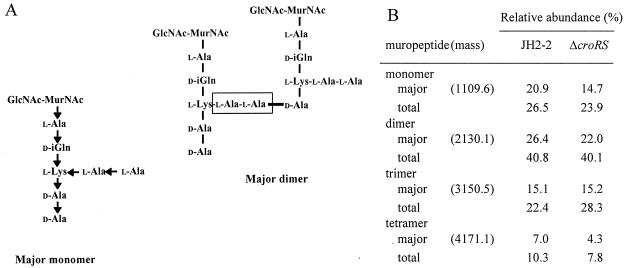
The muropeptide compositions of the peptidoglycans of E. faecalis JH2-2 and JH2-2ΔcroRS were also similar (Fig. 4 and data not shown). The predominant muropeptides contained two d-alanyl residues at the free C-terminal end and two l-alanyl residues both in the cross-bridge and at the free N-terminal end. Muropeptides of lesser abundance differed from the above structures by combinations of MurNAc O-acetylation and loss of the C-terminal d-Ala residues, as previously described (6). These results indicate that deletion of the croRS locus did not affect the extent or mode of peptidoglycan cross-linking in E. faecalis JH2-2.
FIG. 4.
Muropeptides from E. faecalis JH2-2 and JH2-2ΔcroRS. (A) The most abundant muropeptides of both strains contained two d-Ala residues at the free C-terminal end and two l-Ala residues both at the free N-terminal end and in the cross-bridge (boxed). d-Glutamic acid is incorporated into the precursors and secondarily α-amidated (d-iGln, γ-d-glutaminyl residue). GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid. The orientation of the CO→NH peptide bonds is indicated by arrows. (B) Relative abundances of the major monomer, dimer, trimer, and tetramer. The totals include muropeptides of lesser abundance lacking the two C-terminal d-Ala residues and O-acetylated derivatives.
DISCUSSION
The deletions from the chromosomal croRS locus of E. faecalis JH2-2 and complementation analysis indicated that the CroR response regulator and the CroS sensor kinase were required for intrinsic β-lactam resistance (Table 1). A soluble fragment of CroS and full-length CroR were purified to confirm that the proteins were functional with respect to the phosphotransfer reactions commonly catalyzed by mated kinases and response regulators of two-component regulatory systems (Fig. 2). The kinase activity of CroS may be responsible for activation of the response regulator in vivo, as found for several response regulators of the OmpR-PhoB subclass (12), although experimental evidence that phospho-CroR is the active form of the protein was not obtained in the present study. This would imply that CroR cannot be activated by cross talk, since the croS null mutant was susceptible to ceftriaxone.
The croR and croS genes were cotranscribed from a promoter, croRp, which was inducible by inhibitors of peptidoglycan synthesis in E. faecalis JH2-2 (Table 2). The reporter gene was expressed at a constitutive low level in JH2-2ΔcroRS and JH2-2ΔcroS. Thus, the adaptive response elicited by CroR and CroS involved increased transcription of the regulatory genes. This autoregulation mechanism is common to other two-component regulatory systems (16). The sensor kinases encoded by glycopeptide resistance gene clusters appear to specifically respond to vancomycin (VanB-type resistance) or to inhibitors of the transglycosylation reaction (VanA-type resistance) (2). In contrast, the CroRS system did not respond to inhibition of a specific step of peptidoglycan synthesis, since the croRp-lacZ transcriptional fusion was inducible by all peptidoglycan synthesis inhibitors that were tested, including compounds acting on early cytoplasmic steps, the metabolism of the lipid intermediates, transglycosylation, and transpeptidation (Table 3). A recent study of the role of two-component regulatory systems of E. faecalis in the response to environmental stresses suggests that the CroS sensor kinase responds to an even broader spectrum of signals (19).
Sequences flanking the croRS locus were independently transcribed, since the rrnC rRNA operon was in a divergent orientation and Northern blot hybridization revealed a distinct mRNA for the downstream serS seryl-tRNA synthetase gene. The serSp promoter was not inducible by peptidoglycan synthesis inhibitors and was similarly active in JH2-2, JH2-2ΔcroRS, and JH2-2ΔcroS (Table 2). Expression of serS under the control of the heterologous aphA-3p promoter did not restore ceftriaxone resistance in the JH2-2ΔcroRS mutant. Thus, there is apparently no functional link between the croRS locus and the flanking serS locus. Of note, this region of the chromosome does not seem to undergo frequent recombination events, since the relative positions of croRS and serS are conserved in E. faecalis and in E. faecium (data not shown).
Deletion of croRS and pbp5 had the same impact on the MICs of β-lactam antibiotics (Table 1). The patterns of PBPs labeled with benzyl[14C]penicillin were similar for JH2-2, JH2-2ΔcroRS, and JH2-2ΔcroS (data not shown). Introduction of plasmid pAA15, harboring pbp5 under the control of aphA-3p, led to overproduction of PBP5 in JH2-2ΔcroRS, but the MIC of ceftriaxone was only marginally increased (Table 1). These results indicate that susceptibility to ceftriaxone caused by the croRS deletion cannot be attributed to the lack of PBP5 production.
Gram-positive cocci produce branched peptidoglycan precursors containing a side chain consisting of two l-alanyl residues in E. faecalis (Fig. 4A), five glycyl residues in S. aureus, and the sequence l-Ser-l-Ala or l-Ala-l-Ala in S. pneumoniae (28). The genes encoding the transferases for synthesis of the pentaglycine side chain in S. aureus were initially identified as factors essential for methicillin resistance (fem) after random mutagenesis of the S. aureus chromosome (3, 25). More recently, production of incomplete side chains was also reported to lead to impaired expression of acquired resistance to β-lactam antibiotics in S. pneumoniae (13) and of intrinsic resistance to ceftriaxone in E. faecalis (6). The screening for impaired expression of methicillin resistance in S. aureus also identified mutations in genes encoding enzymes for the assembly of the nucleotide precursor UDP-MurNAc-pentapeptide (17, 21). In the latter case, the mutations could affect the amount of precursor produced rather than its structure (10). Since (despite the production of PBP5) deletion of the croRS locus was associated with ceftriaxone susceptibility, the structures of cytoplasmic precursors and muropeptides were analyzed to screen for defects in the production of the substrate of PBP5. Deletion of croRS was not associated with accumulation of nucleotide precursors containing incomplete stem peptides (Table 4). Further, synthesis of the l-alanyl-l-alanine side chain was not impaired since the mutant produced wild-type levels of UDP-MurNAc-hexapeptide (Table 4) and two l-alanyl residues were present both in the cross-bridge and in the free N-terminal end of the muropeptides (Fig. 4). Finally, the relative proportions of monomer, dimer, trimer, and tetramer were similar in JH2-2ΔcroRS and in the parental strain. These results indicate that PBP5 did not mediate ceftriaxone resistance in JH2-2ΔcroRS, despite delivery of an apparently unaltered supply of disaccharide-peptide subunits to the peptidoglycan polymerization complexes.
Acknowledgments
This work was supported by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires (MENRT), the Fondation pour la Recherche Médicale, and Pfizer Inc.
E. faecalis genome sequence data were kindly provided by The Institute for Genomic Research as publicly released at www.tigr.org.
REFERENCES
- 1.Arthur, M., C. Molinas, and P. Courvalin. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., and R. Quintiliani, Jr. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger-Bachi, B., L. Barberis-Maino, A. Strassle, and F. H. Kayser. 1989. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol. Gen. Genet. 219:263-269. [DOI] [PubMed] [Google Scholar]
- 4.Billot-Klein, D., L. Gutmann, E. Collatz, and J. Van Heijenoort. 1992. Analysis of peptidoglycan precursors in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 36:1487-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouhss, A., N. Josseaume, D. Allanic, M. Crouvoisier, L. Gutmann, J.-L. Mainardi, D. Mengin-Lecreulx, J. van Heijenoort, and M. Arthur. 2001. Identification of the UDP-MurNAc-pentapeptide:l-alanine ligase for synthesis of branched peptidoglycan precursors in Enterococcus faecalis. J. Bacteriol. 183:5122-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhss, A., N. Josseaume, A. Severin, K. Tabei, J. E. Hugonnet, D. Shlaes, D. Mengin-Lecreulx, J. Van Heijenoort, and M. Arthur. 2002. Synthesis of the l-alanyl-l-alanine cross-bridge of Enterococcus faecalis peptidoglycan. J. Biol. Chem. 277:45935-45941. [DOI] [PubMed] [Google Scholar]
- 7.Canepari, P., M. del mar Lleo, G. Cornaglia, R. Fontana, and G. Satta. 1986. In Streptococcus faecium penicillin-binding protein 5 alone is sufficient for growth at sub-maximal but not at maximal rate. J. Gen. Microbiol. 132:625-631. [DOI] [PubMed] [Google Scholar]
- 8.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 9.Condon, C., H. Putzer, and M. Grunberg-Manago. 1996. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:6992-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Lencastre, H., S. W. Wu, M. G. Pinho, A. M. Ludovice, S. Filipe, S. Gardete, R. Sobral, S. Gill, M. Chung, and A. Tomasz. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5:163-175. [DOI] [PubMed] [Google Scholar]
- 11.Duez, C., W. Zorzi, F. Sapunaric, A. Amoroso, I. Thamm, and J. Coyette. 2001. The penicillin resistance of Enterococcus faecalis JH2-2r results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology 147:2561-2569. [DOI] [PubMed] [Google Scholar]
- 12.Ellison, D. W., and W. R. McCleary. 2000. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J. Bacteriol. 182:6592-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipe, S. R., M. G. Pinho, and A. Tomasz. 2000. Characterization of the murMN operon involved in the synthesis of branched peptidoglycan peptides in Streptococcus pneumoniae. J. Biol. Chem. 275:27768-27774. [DOI] [PubMed] [Google Scholar]
- 14.Fontana, R., M. Aldegheri, M. Ligozzi, H. Lopez, A. Sucari, and G. Satta. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 38:1980-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock, L., and M. Perego. 2002. Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffer, S. M., H. V. Westerhoff, K. J. Hellingwerf, P. W. Postma, and J. Tommassen. 2001. Autoamplification of a two-component regulatory system results in “learning” behavior. J. Bacteriol. 183:4914-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolly, L., S. Wu, J. van Heijenoort, H. de Lencastre, D. Mengin-Lecreulx, and A. Tomasz. 1997. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J. Bacteriol. 179:5321-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klare, I., A. C. Rodloff, J. Wagner, W. Witte, and R. Hakenbeck. 1992. Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 36:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Breton, Y., G. Boel, A. Benachour, H. Prevost, Y. Auffray, and A. Rince. 2003. Molecular characterization of Enterococcus faecalis two-component signal transduction pathways related to environmental stresses. Environ. Microbiol. 5:329-337. [DOI] [PubMed] [Google Scholar]
- 20.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ornelas-Soares, A., H. de Lencastre, B. L. de Jonge, and A. Tomasz. 1994. Reduced methicillin resistance in a new Staphylococcus aureus transposon mutant that incorporates muramyl dipeptides into the cell wall peptidoglycan. J. Biol. Chem. 269:27246-27250. [PubMed] [Google Scholar]
- 22.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 23.Pompeo, F., J. van Heijenoort, and D. Mengin-Lecreulx. 1998. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J. Bacteriol. 180:4799-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 25.Rohrer, S., and B. Berger-Bächi. 2003. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and β-lactam resistance in gram-positive cocci. Antimicrob. Agents Chemother. 47:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybkine, T., J. L. Mainardi, W. Sougakoff, E. Collatz, and L. Gutmann. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J. Infect. Dis. 178:159-163. [DOI] [PubMed] [Google Scholar]
- 27.Sauvage, E., F. Kerff, E. Fonze, R. Herman, B. Schoot, J. P. Marquette, Y. Taburet, D. Prevost, J. Dumas, G. Leonard, P. Stefanic, J. Coyette, and P. Charlier. 2002. The 2.4-A crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 59:1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sifaoui, F., M. Arthur, L. Rice, and L. Gutmann. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 31.Williamson, R., C. le Bouguenec, L. Gutmann, and T. Horaud. 1985. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J. Gen. Microbiol. 131:1933-1940. [DOI] [PubMed] [Google Scholar]
- 32.Zorzi, W., X. Y. Zhou, O. Dardenne, J. Lamotte, D. Raze, J. Pierre, L. Gutmann, and J. Coyette. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J. Bacteriol. 178:4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]