Abstract
Mycobacterium tuberculosis is one of the strongest reducers of nitrate in the genus Mycobacterium. Under microaerobic conditions, whole cells exhibit upregulation of activity, producing approximately eightfold more nitrite than those of aerobic cultures of the same age. Assays of cell extracts from aerobic cultures and hypoxic cultures yielded comparable nitrate reductase activities. Mycobacterium bovis produced only low levels of nitrite, and this activity was not induced by hypoxia. M. tuberculosis has two sets of genes, narGHJI and narX of the narK2X operon, that exhibit some degree of homology to prokaryotic dissimilatory nitrate reductases. Each of these were knocked out by insertional inactivation. The narG mutant showed no nitrate reductase activity in whole culture or in cell-free assays, while the narX mutant showed wild-type levels in both assays. A knockout of the putative nitrite transporter narK2 gene produced a strain that had aerobic levels of nitrate reductase activity but failed to show hypoxic upregulation. Insertion of the M. tuberculosis narGHJI into a nitrate reductase Escherichia coli mutant allowed anaerobic growth in the presence of nitrate. Under aerobic and hypoxic conditions, transcription of narGHJI was constitutive, while the narK2X operon was induced under hypoxia, as measured with a lacZ reporter system and by quantitative real-time reverse PCR. This indicates that nitrate reductase activity in M. tuberculosis is due to the narGHJI locus with no detectable contribution from narX and that the hypoxic upregulation of activity is associated with the induction of the nitrate and nitrite transport gene narK2.
The ability of Mycobacterium tuberculosis to persist in a host for long periods despite acquired immunity without producing obvious symptoms, along with its capacity to reactivate and cause clinical tuberculosis, has been an impediment to the complete control and eradication of this disease. The grim result is that approximately one third of the world's population is asymptomatically infected with M. tuberculosis and thus carries latent disease (14, 16).
Information has recently begun to emerge on the physiological state of the tubercle bacilli during latent infection. In the course of early infection, they are surrounded in granulomas by activated macrophages which limit access to some nutrients including oxygen (7, 35, 52). Among the various systems that have been used to model disease latency in vitro, one involves the cultivation of tubercle bacilli under conditions leading to the controlled gradual depletion of oxygen (49). This model has been used to study dormancy in both Mycobacterium tuberculosis and the closely related Mycobacterium bovis BCG (24). Both species are obligate aerobes unable to grow without oxygen, but when subjected to gradual oxygen depletion, they can survive for extended periods. The growth of cultures in sealed, slowly stirred tubes results in the gradual and uniform controlled depletion of oxygen until the bacilli enter a microaerobic stage of nonreplicating persistence (NRP-1) and then proceed into the anaerobic state (NRP-2), in which they may survive for many months (24, 49).
In an anaerobic environment, many bacteria are able to use nitrate as a final electron acceptor in place of oxygen for the maintenance of a proton motive gradient to continue growing. Historically, M. tuberculosis has been differentiated from M. bovis by the fact that only M. tuberculosis can reduce significant amounts of nitrate (NO3−) to nitrite (NO2−) (44, 48). Nitrate reductase activity occurs at a low level during the aerobic growth of M. tuberculosis and increases significantly upon entry into the microaerobic NRP-1 stage (50). This, along with the fact that neither M. tuberculosis nor M. bovis reduce nitrite (44, 48), suggests that the reduction of nitrate serves as an alternate energy source rather than nitrogen source during the adaptation to hypoxic conditions.
The best characterized nitrate reductase system is that of Escherichia coli where there are two membrane-bound respiratory enzymes. The four-gene narGHJI operon is induced 4-fold under anaerobic conditions and an additional 19-fold by nitrate (40) and permits anaerobic growth in the presence of nitrate. The second very similar operon, narZYWV, encodes a nitrate reductase enzyme expressed at a low level aerobically and induced during stationary phase but not regulated by either oxygen or nitrate levels (8).
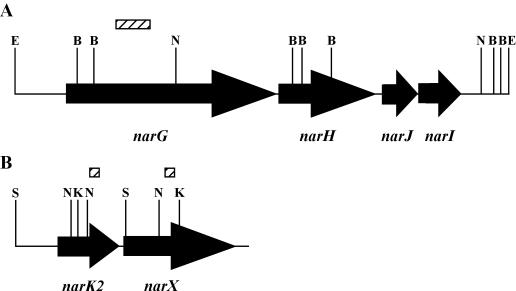
M. tuberculosis has an narGHJI locus (Fig. 1) able to produce nitrate reductase activity when cloned into Mycobacterium smegmatis (53). Also identified in the M. tuberculosis H37Rv genome during sequencing was a gene designated narX, which has been proposed to code for a “fused nitrate reductase” (11). This proposal was made because the predicted product of narX would be a protein with homology to parts of the NarG, NarJ, and NarI proteins, although its actual function is unknown.
FIG. 1.
Genes possibly involved in nitrate reductase activity. Schematic diagram of both the narGHJI (A) and narK2X (B) loci in M. tuberculosis. Arrows show the open reading frames with the gene names below the arrows. Relevant restrictions sites are shown: B, BamHI; S, SmaI; E, EcoRV; N, NcoI; K, KpnI. The locations of probes used in Southern blots (hatched boxes) are indicated. The diagram is not drawn to scale.
Here we further characterize the nitrate reducing system of M. tuberculosis and compare it to that of M. bovis. To determine the role of narG and narK2X in the reduction of nitrate by M. tuberculosis, each gene was inactivated. Nitrate reduction and mRNA levels of the various genes were measured during aerobic growth and shiftdown into NRP-1 as well as stationary phase.
MATERIALS AND METHODS
Strains and media.
M. tuberculosis H37Rv and avirulent M. bovis BCG Pasteur from the culture collection of this laboratory were used. Virulent M. bovis Ravenel was provided by Frank Collins. M. smegmatis mc2155 was from William Jacobs (38). The Nar− E. coli mutant JCB4023 [araD139 Δ(argF-lac)U169 rpsL150 relA1 gyrA219 non-9 narG::erm ΔnapAB narZ::Ω] (9, 31) and JCB4018 (ΔnarAB Δnar::Ω ΔnarK ΔnarU::kan) were provided by Jeff Cole (9, 31).
Liquid mycobacterial cultures were grown at 37°C in Dubos Tween-albumin broth (DTA) (Difco, Detroit, Mich.). The cultures were plated on either Dubos oleic albumin agar (DOA) or DTA with the addition of 1.6% agar (DTA agar). Aerobic cultures were incubated on a model G24 rotary shaker-incubator (New Brunswick Scientific, Edison, N.J.). For NRP cultures, conditions included slow magnetic stirring in sealed tubes with a headspace ratio (HSR) of 0.5 as previously described (46, 49). Nitrate, when used, was added at a final concentration of 5 mM unless indicated otherwise. E. coli cultures were grown in Luria-Bertani (LB) or M9 medium. Anaerobic cultures of E. coli were grown in full, tightly sealed tubes containing medium supplemented with Oxyrase (Oxyrase, Inc., Mansfield, Ohio).
Kanamycin was used at 25 μg/ml for M. tuberculosis and at 50 μg/ml for E. coli. Gentamicin was used at 5 μg/ml, and apramycin was used at 30 μg/ml. All antibiotics and chemicals, including sodium azide, sodium molybdate, and tungstic acid, were from Sigma (St. Louis, Mo.). All oligonucleotide primers were from Ransom Hill Bioscience (Ramona, Calif.) and are listed in Table 1.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′)a | Purpose in this studyb |
|---|---|---|
| p11 | ACTACGCCGACAACACCAAGTTCG | Probe for narG |
| p12 | TGTTCTGCTGGGTTCGGTCGTGGTA | Probe for narG |
| p13 | TGCTGCTGGTGGCGACCGCGCTGGTCGCGT | Cloning narX |
| p14 | GCGCGGACTTGTTCGACGAGTAGACGTGT | Cloning narX |
| p19 | GTGAGCGGGTACCAATTTCACACAGG | Cloning lacZ |
| p20 | GGCCTGCCCGGGTATTATTATTTTTG | Cloning lacZ |
| p23 | GGTGATCGCCATGGTCGACCTGCAGG | Cloning aphI |
| p24 | TAATGCTCTGCCATGGTTACAACCAA | Cloning aphI |
| p27 | ATGCGCCTCGGTGCTGCTGACCTACCCGAA | Probe for narX |
| p28 | GGATCGGCACGGCGCAGCTCAGAGACCGTG | Probe for narX |
| p46 | CTGGATTGCGGATCTAGAGCGAACCCTCAA | Construction of pMP101 |
| p47 | ACGGTCACGGGGGTACCCTCCTCGTCATGA | Construction of pMP101 |
| p48 | TCCCCTTTCTAGAGGCGACCAGGCTCAGCT | Construction of pMP102 |
| p49 | TCATCGACAGGTACCGGGGTCTCGGACTCC | Construction of pMP102 |
| p51 | CAGCTCCTGGATCCGGCTGCCGGTCCGTGG | Cloning narK2 |
| p58 | ACTCGAGTGGCGAACGGGTGAGTAACACGT | RT-PCR of 16S rRNA |
| p59 | AGGCCGTCACCCCACCAACAAGCTGATAGG | RT-PCR of 16S rRNA |
| p60 | ACTACGCCGACAACACCAAGTTCGCCGACG | RT-PCR of narG |
| p61 | AGCGGCGCACATAGTCGACAAAGAACGGAA | RT-PCR of narG |
| p62 | ATTGGTGGGACGTGGTGTGGCAATGCGCCT | RT-PCR of narX |
| p63 | GACCGTCGATGTGGGCCAGCAATTCCTCTG | RT-PCR of narX |
| p66 | TGCTTCGTGATGCACCCTACTTTCGGCCCA | RT-PCR of narK2 |
| p67 | CCGCCGAACACGATCGCGTACAGAAACGAC | RT-PCR of narK2 |
| p68 | CCAAGTCGGACAAGCTTCGGGCGACCGAGA | Cloning narK2 |
Mismatches used to create restriction sites are indicated by underlining.
RT-PCR, reverse transcriptase PCR.
Sonication.
To make cell-free mycobacterial extracts, cultures were centrifuged and the cells were washed with an equal volume of phosphate buffer (pH 7.2) containing 0.02% Tween 80 and then with phosphate buffer (pH 7.2) without Tween 80. The cell suspensions were sonicated for 15 min in the cup-horn of a W-380 sonicator (Heat Systems-Ultrasonic, Farmingdale, N.Y.) at 4°C and filtered through 0.22-μm-pore-size Spin-X filters (Corning Costar Corp., Cambridge, Mass.). Approximately 30% of the nitrate reductase activity was recovered in the filtrate from both aerobic and NRP cultures. Glycerol was added to the filtrate to 10%, and the protein concentration was determined by a standard Bradford assay. The samples were frozen at −70°C until use.
Nitrate reductase assay.
For the cell-free assay, 100 μg of protein was added to a 0.6-ml tube. Then 50 μl of 1 M NaNO3 and 50 μl of methyl viologen (in 100 mM phosphate buffer [pH 7.2]) were added to the tube, and the volume was adjusted to 450 μl with phosphate buffer, pH 7.2. Argon was bubbled over the samples, which were placed at 37°C for 2 min. To start the reaction, 50 μl of freshly made 29 mM sodium hydrosulfite (dithionite) in 10 mM NaOH was added. At 20-min intervals, 100-μl samples were removed, and the nitrite concentration was determined. All assays were performed in triplicate and replicated at least once with independent cell sonicates.
For whole-cell assays, the nitrite concentration was determined in 200 μl of culture from aerobic cultures or 20 μl from NRP and senescent stationary cultures. Nitrite concentration was determined by the Griess reaction (48).
Reporter analysis.
To create the reporter plasmid, the E. coli lacZ gene was amplified by PCR with primers p19 and p20 (Table 1). These primers created a KpnI site upstream and a SmaI site downstream of the gene. After digestion with these enzymes, the fragment was cloned into DraI-KpnI-digested pMH94 (23) to create pMP100. pMP100 exists as a plasmid in E. coli, but in Mycobacterium species, it integrates into the chromosome as a single copy.
The upstream promoter regions of narGHJI and narK2X were amplified by PCR. A 376-bp fragment directly upstream of the narG start site was amplified with primers p46 and p47. For narK2X, a 282-bp fragment was generated with primers p48 and p49 based on previous promoter analysis of narK2X (21). After digestion with XbaI and KpnI, these fragments were cloned into pMP100 to create pMP101 (narGHJI) and pMP102 (narK2X).
pMP100, pMP101, and pMP102 were electroporated into M. tuberculosis to create strains PMP100, PMP101, and PMP102, respectively. The insertion of the plasmid into the chromosome as a single copy was verified by Southern blot analysis. β-Galactosidase activity was determined by the method of Timm et al. (42) and calculated as follows: 200 × optical density at 420 nm (OD420)/mg of protein/min.
RNA isolation.
To isolate RNA, cultures were quickly mixed with 4 volumes of 5 M GTC lysis solution 1 (5 M guanidine thiocyanate, 0.5% sodium N-lauryl sarcosine, 25 mM trisodium citrate, 0.1 M 2-mercaptoethanol, 0.5% Tween 80 [pH 7.0]) (25) and immediately pelleted by centrifugation at 20,000 × g for 30 min. The pellet was resuspended in 1 ml of Trizol (InVitrogen, Carlsbad, Calif.) and transferred to a 2-ml tube approximately one-third filled with 0.1-mm-diameter zirconia-silica beads. Cells were disrupted by three 1-min pulses at full speed in a Mini-Bead beater (Bio Spec Products, Bartlesville, Okla.). Samples were centrifuged for 5 min at 10,000 × g, and the supernatants were extracted once with CHCl3 and precipitated with isopropanol. After resuspension, DNA was removed by treatment with DNase (Boehringer Mannheim) for 2 h at 37°C. RNA was purified by chromatography with an RNeasy column (Qiagen, Chatsworth, Calif.) and treated again with DNase, which was then inactivated at 70°C for 5 min.
Quantitation of mRNA levels.
For each sample, 500 μg of RNA was added to a mixture of antisense primers (total concentration of 1 μM) and all four deoxynucleoside triphosphates (total concentration of 500 μM) in a total volume of 16 μl. The resulting mixture was heated to 80°C for 3 min. Subsequently, 2 μl of 10× PCR buffer, 1 μl of Moloney murine leukemia virus reverse transcriptase (RT), and 1 μl of placental RNase inhibitor from a RETROscript kit (Ambion, Austin, Tex.) were added. In every case, a duplicate sample was prepared without Moloney murine leukemia virus RT for the no-RT control. The reaction was stopped, and the enzyme was inactivated by heating at 92°C for 10 min.
Real-time quantitative PCR was performed with the Brilliant SYBR green QPCR Master Mix kit (Stratagene, La Jolla, Calif.). Reactions were performed in a volume of 50 μl, and the reaction mixtures consisted of a 0.1 μM concentration of both primers (Table 1), 25 μl of 2× master mix, and 5 μl of cDNA. The control with no RT was included in each run. An additional sample with RNA diluted 1:10 was also included to measure 16S rRNA. Amplification was performed in the ICycler (Bio-Rad, Hercules, Calif.) with sampling during elongation. The samples were subjected to PCR as follows: (i) an initial denaturation step of 10 min at 95°C; (ii) 30 cycles, with 1 cycle consisting of 30 s at 95°C and 1 min at 68°C; (iii) an extension step of 7 min at 68°C. A melting curve analysis was then performed. All samples were run on a 2% agarose gel to verify that only a single band was produced. Each gene was analyzed from three independent RNA samples.
Construction of knockouts.
Knockouts were created by cloning either narG, narX, or narK2 into pJQ200KS (32) followed by the insertion of aphI, a kanamycin resistance marker. For narG, a BamHI-ApaI fragment was cloned from cosmid I65 (6) into the same sites of pJQ200KS. For narX and narK2, the entire genes were amplified by PCR performed with the Advantage-GC 2 PCR kit with primers p13 and p14 (narX) or p51 and p68 (narK2) and cloned with the TA cloning kit (InVitrogen) to produce pNarX2 or pNarK2. narX was subcloned into pJQ200KS by cutting both plasmids with XhoI and SacI. narK2 was subcloned with BamHI and HindIII to make pTSJ1.
The aphI gene from Tn903 was amplified with primers p23 and p24, which created NcoI sites at both ends of the 946-bp fragment. This fragment was cloned into the unique NcoI sites of the narG fragment to create plasmid pNarG3.1 or into narX to make pNarX3.1. Due to the presence of two NcoI sites, insertion of aphI to create pTSJ2 resulted in a small deletion in the narK2 gene.
pNarG3.1, pNarX3.1, and pTSJ2 were electroporated into M. tuberculosis followed by selection on DTA agar plates with 25 μg of kanamycin per ml and 5 μg of gentamicin per ml. (DTA agar produced soft colonies that were easy to pick.) Colonies that were the results of single-crossover events having either pNarG3.1, pNarX3.1, or pTSJ2 inserted into the chromosome copy of narG, narX, or narK2, respectively, were identified by Southern blot analysis. This confirmed the presence of both the wild-type gene and the additional gene with the aphI insertion. Digoxigenin-labeled probes for Southern blot analysis were created by PCR (Roche Diagnostics, Indianapolis, Ind.) using p11 and p12 as primers for narG, p27 and p28 for narX, and p66 and p67 for narK2.
Three single-crossover mutants were chosen and replated on DTA agar with 2% sucrose and kanamycin. Possible double-crossover mutants, containing aphI inserted into the NcoI site of the chromosome copy of narG or narX, were initially identified by sensitivity to gentamicin and confirmed by Southern blot analysis. This showed both the presence of the gene with the aphI insertion and the loss of the wild-type gene.
Cloning of narGHJI, narX, and narK2.
The M. tuberculosis narGHJI operon was cloned as an EcoRV fragment from the I65 cosmid (6) into the DraI sites of pPE207 (28). This was electroporated into M. smegmatis, and after selection for apramycin resistance, clones were screened for nitrate reductase activity. The plasmid obtained was named pNarGHJI1. This plasmid was electroporated into E. coli JCB4023 and M. smegmatis and maintained aerobically with apramycin but without nitrate.
To clone narX, the narX gene was cut from pNarX2 with BamHI and cloned into the same site of pPE207 to create pNarX5.
To clone narK2, BamHI and HindIII were used to subclone the narK2 gene into the same sites in pPE207 to create pTSJ3. To make pTSJ4, narK2 was removed from pTSJ1 with BamHI and SacI and cloned into the same sites in pBluescript SK+ (pSK).
RESULTS
Nitrate reductase activity in culture.
To determine the nitrate reductase activity of cultures, nitrite concentrations were assayed during growth. Measurements were made on both aerobic cultures and in microaerobic NRP-1 phase cultures. In addition to the controlled oxygen depletion model, we also used some tubes of vigorously agitated, loosely capped cultures that were incubated long enough to exhibit the late plateau of senescent stationary phase (SSP) (47). This phase has been used by some investigators as a model for dormancy, so nitrite concentrations were also determined after the growth of aerated cultures reached an optical density plateau. Since the nitrate reductase system has also been studied in M. bovis BCG (18, 53), both virulent and avirulent strains of this species were included for comparison purposes.
Aerobic shaken cultures of M. tuberculosis, virulent M. bovis, and the avirulent vaccine strain M. bovis BCG were started at a concentration of 2.5 × 106 cells/ml in DTA with 5 mM NaNO3, which allowed for approximately 140 h of logarithmic growth before the onset of SSP. After 112 h of aerobic growth, which represented mid-logarithmic phase (OD580 of ∼0.5), M. tuberculosis cultures had produced an average of 130 μM nitrite, while M. bovis and M. bovis BCG cultures produced only 13 and 7 μM, respectively (Table 2). After 5 days in SSP, the concentration of nitrite in the medium had increased to 2,600 and 180 μM for M. tuberculosis and M. bovis, respectively, while M. bovis BCG had produced only 11 μM.
TABLE 2.
Production of nitrite in whole-cell culture by M. tuberculosis and M. bovis
| Mycobacterium | Treatmenta | Nitrite concnb
|
||
|---|---|---|---|---|
| Aerobicc | NRP-1d | SSPe | ||
| M. tuberculosis | 130 ± 23 | 1,100 ± 80 | 2,600 ± 430 | |
| 400 μM Mo | 120 ± 6 | 780 ± 55 | ND | |
| 100 μM W | 9 ± 1 | 120 ± 9 | ND | |
| 400 μM Mo + 100 μM W | 23 ± 3 | 500 ± 5 | ND | |
| 40 μM azide | 3 ± 1 | 16 ± 7 | ND | |
| M. bovis | 13 ± 1 | 5 ± 2 | 180 ± 52 | |
| M. bovis BCG | 7 ± 1 | 2 ± 1 | 11 ± 5 | |
Treatment with the ions molybdate (Mo) and tungstate (W) and sodium azide.
Mean nitrite concentration (micromolar) ± standard deviation.
After 112 h of growth (OD580 of ∼0.4).
After 112 h, approximately 45 h in NRP-1 (OD580 of 0.1).
After 280 h of growth (OD580 > 1.0). ND, not done.
When the same number of cells was incubated in slowly stirred sealed tubes (0.5 HSR configuration) (49), they grew logarithmically for about 67 h before replication abruptly ceased due to hypoxia, and cells entered microaerobic NRP-1. At 112 h, after approximately 45 h in NRP-1, the nitrite concentration produced by M. tuberculosis was 1,100 μM, while the nitrite concentration produced by M. bovis was 5 μM and that produced by M. bovis BCG was 2 μM (Table 2).
After 112 h of incubation, aerobic cultures of M. tuberculosis produced eightfold-less nitrite than those in NRP-1, despite the fact that the aerobic cultures had a cell density approximately fourfold greater. Thus, nitrate reduction in M. tuberculosis showed the typical hypoxic induction seen in many other bacteria. Virulent M. bovis showed only weak activity, without any increase under hypoxia, while the avirulent M. bovis BCG showed only background levels of nitrite production. Cells began to clump and die as they entered SSP, so it was not possible to make a reliable assessment of the induction of nitrate reductase activity only on the basis of nitrite levels.
The effects of several ions on the production of nitrite in culture were also determined (Table 2). The nitrate reductase activity in M. tuberculosis was inhibited by the addition of 100 μM tungstate, which is a characteristic response of molybdenoproteins (41). This inhibition could be partially reversed by the addition of 400 μM molybdate in both aerobic and NRP-1 cultures. A low concentration of sodium azide also inhibited activity in M. tuberculosis; this is characteristic of the membrane-bound class of nitrate reductase enzymes (1, 2). Nitrate reductase enzymes also use chlorate as a substrate and reduce it to the toxic chlorite. M. tuberculosis was unable to grow in 5 mM sodium chlorate. This suggests that M. tuberculosis contains a membrane-bound molybdenum-containing nitrate reductase enzyme system.
Cell-free nitrate reductase assay.
A cell-free assay was developed to measure the levels of nitrate reductase enzyme. This system utilized cell sonicates rather than whole cells and allowed the measurement of enzyme activity in a measured amount of bacillary protein, independent of nitrate and nitrite transport. Cells of M. tuberculosis were grown without or with 5 or 20 mM nitrate and harvested from either aerobic, NRP-1, or SSP cultures (Table 3). There was no significant difference in the specific activity of cell-free sonic extracts from cultures that had been grown with and without nitrate, suggesting that the level of nitrate reductase enzyme in each extract is also equal, and therefore independent of the nitrate concentration in the environment. Furthermore, the activity from extracts of actively growing aerobic cultures and NRP-1 cultures were not significantly different from each other. Cell extracts of SSP cultures showed lower specific activity than extracts from growing aerobic cultures or NRP-1 cultures (Table 3). This could be due simply to a decrease in the number of viable cells in SSP. In summary, the nitrate reductase activity of cell extracts was constant under a variety of conditions, despite strong differences in the apparent activity shown by intact cells.
TABLE 3.
Nitrate reductase activity in cell-free sonicates
| Culture treatment | Nitrite reductase activitya
|
||
|---|---|---|---|
| Aerobicb | NRP-1c | SSPd | |
| No nitrate | 20 ± 1 | 22 ± 5 | 9 ± 1 |
| 5 mM NO3− | 21 ± 1 | 18 ± 3 | 6 ± 1 |
| 20 mM NO3− | 21 ± 2 | 22 ± 4 | ND |
Mean nitrite reductase activity (in nanomoles of NO2− per minute per milligram of protein) ± standard deviation.
After 112 h of growth (OD580 of ∼0.4).
After 112 h, approximately 45 h in NRP-1 (OD580 of 0.1).
After 280 h of growth (OD580 > 1.0). ND, not done.
Expression of nar genes.
A promoterless reporter plasmid containing lacZ was constructed from the integration vector pMH94 (23) and named pMP100. The narGHJI upstream promoter region was inserted into this plasmid to create pMP101-narG, and that of the narK2X region to make pMP102-narK2X (21). These plasmids were electroporated into M. tuberculosis at a site where the mycobacteriophage int gene and attachment site allowed integration of a single copy of the plasmid into the chromosome.
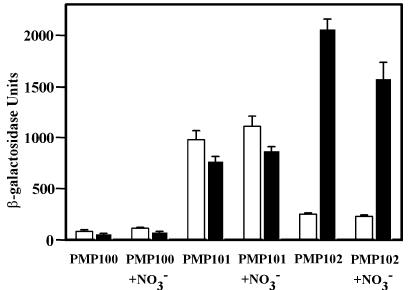
β-Galactosidase levels were determined from samples of each strain taken either during mid-logarithmic phase or after approximately 45 h in NRP-1. The narGHJI promoter of M. tuberculosis strain PMP101 in aerobic and NRP-1 cultures showed similar levels of activity (Fig. 2). In contrast, the narK2X promoter of M. tuberculosis strain PMP102 showed approximately eightfold induction under the controlled hypoxic conditions of NRP-1 in comparison to aerobic levels.
FIG. 2.
β-Galactosidase activity in M. tuberculosis containing narGHJI and narK2X promoter constructs. β-Galactosidase assays were performed with cell extracts from aerobic actively growing (open bars) or NRP-1 (filled bars) cultures. Each strain had lacZ controlled by the upstream region of narG (PMP101) or narK2X (PMP102) or no insert (PMP100). The means ± standard deviations (error bars) of three determinations are shown.
Neither promoter showed any response with the addition of 5 mM nitrate to the growth medium (Fig. 2). Since significant nitrite was also produced from nitrate in all cultures, this implies that the regulation of both the narGHJI and narK2X promoters are independent of both nitrate and nitrite.
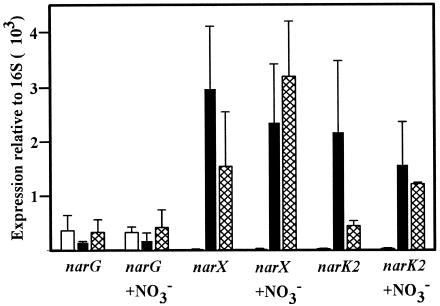
Real-time reverse transcriptase PCR was used to measure mRNA levels quantitatively. The levels of narG, narX, and narK2 were determined and compared to the level of the stable 16S rRNA (13, 37). Samples were taken from aerated cultures in mid-logarithmic phase, just after the onset of SSP, and from sealed cultures after approximately 45 h in NRP-1. The levels of narG transcripts were similar in aerobic, NRP, and SSP cultures (Fig. 3). In contrast, narX and narK2 mRNA levels were low in aerobic cultures but showed strong induction in both NRP and SSP cultures. Again, the three genes did not show a significant response to the addition of 5 mM nitrate to the growth medium (Fig. 3).
FIG. 3.
Quantitative real-time PCR of narG, narX, and narK2. RNA levels are expressed relative to the level of stable 16S rRNA (13, 37). The means ± standard deviations (error bars) of three determinations are shown.
Inactivation of narG and narX.
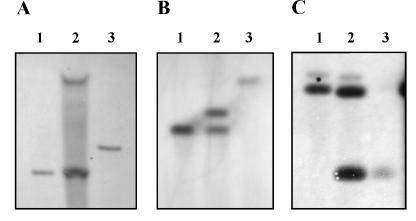
To help determine the role of narGHJI and narX of M. tuberculosis in the reduction of nitrate under different culture conditions, the selected genes were inactivated. A kanamycin resistance marker, aphI, was inserted into a cloned copy of narG or narX and recombined by single crossover into the chromosome of M. tuberculosis. Double crossovers were then isolated in which the chromosomal copy was replaced by the inactive cloned copy, and all plasmid sequences were lost (Fig. 4).
FIG. 4.
Southern blot analysis of narG and narX insertional mutants. Chromosomal DNA from strains used to produce the RVW1 (A), RVW2 (B), and RVW3 (C) mutants was isolated from the wild type (lanes 1), single crossovers containing two copies of the nar gene (lanes 2), and double crossovers containing aphI inserted into nar (lanes 3). DNA was cut with KpnI (A and B) or SmaI (C) and probed with a probe specific for either narG (A), narX (B), or narK2 (C).
There were no obvious differences in the growth curves (measured by optical density) between M. tuberculosis strain RVW1 narG::aphI, RVW2 narX::aphI, and wild type, whether the mycobacteria were grown in aerobic, NRP-1, or the truly anaerobic conditions of NRP-2 with or without 5 mM nitrate. To measure hypoxic survival after shiftdown, viable cell counts were determined by plating for a total of 12 weeks after inoculation. There were no obvious differences in survival with or without nitrate for the wild-type and mutant strains (C. D. Sohaskey, unpublished data). However, cultures of RVW1 narG::aphI did not reduce nitrate, while those of RVW2 narX::aphI showed wild-type levels of activity (Table 4). In cell-free assays, the narX knockout also showed enzyme levels similar to those of the wild type, but only background activity was seen in extracts from the narG knockout (Table 5). RVW1 narG::aphI was able to grow in 20 mM chlorate in contrast to RVW2 narX::aphI, which was as sensitive as the wild type. Thus, the insertion in narX had no effect on nitrate reductase activity, while that in narG eliminated the activity, indicating that only narGHJI was responsible for nitrate reductase activity during both aerobic and NRP-1 conditions.
TABLE 4.
Nitrate reductase activity of narG, narX, and narK2 mutants
| Strain | Genotype | Nitrite concna
|
||
|---|---|---|---|---|
| Aerobicb | NRP-1c | SSPd | ||
| Wild type | Wild type | 140 ± 10 | 1,400 ± 49 | 1,900 ± 410 |
| RVW1 | ΔnarG | 3 ± 1 | 2 ± 1 | 4 ± 2 |
| RVW1(pNarGHJI1)e | ΔnarG + narG | 160 ± 12 | 1,700 ± 72 | 1,000 ± 150 |
| RVW2 | ΔnarX | 150 ± 7 | 1,400 ± 34 | 2,600 ± 350 |
| RVW3 | ΔnarK2 | 110 ± 8 | 35 ± 5 | 910 ± 80 |
Nitrate reductase activity as measured by the nitrite concentration (micromolar). Values are means ± standard deviations.
After 112 h of growth (OD580 of ∼0.4).
After 112 h, approximately 45 h in NRP-1 (OD580 of 0.1).
After 280 h of growth (OD580 > 1.0).
Strain RVW1 (ΔnarG) with plasmid pNarGHJI1 (narG) regained Nar activity.
TABLE 5.
Nitrate reductase activity of narG, narX, and narK2 mutants in the cell-free assay
| Strain | Genotype | Nitrate reductase activitya
|
||
|---|---|---|---|---|
| Aerobicb | NRP-1c | SSPd | ||
| RVW1 | ΔnarG | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.02 |
| RVW2 | ΔnarX | 18 ± 2 | 21 ± 2 | 8 ± 2 |
| RVW3 | ΔnarK2 | 25 ± 4 | 23 ± 2 | 6 ± 1 |
Mean nitrite reductase activity (in nanomoles of NO2− per minute per milligram of protein) ± standard deviation.
After 112 h of growth (OD580 of ∼0.4).
After 112 h, approximately 45 h in NRP-1 (OD580 of 0.1).
After 280 h of growth (OD580 > 1.0).
Cloning of narGHJI and narX.
The narGHJI operon was cloned from an M. tuberculosis cosmid into plasmid pPE207 (28) to produce pNarGHJI1, and narX was cloned to produce pNarX5. Both plasmids were electroporated into M. smegmatis. M. smegmatis strains carrying either plasmid were tested for their ability to produce nitrite from nitrate. During aerobic growth at mid-logarithmic phase, wild-type M. smegmatis produced only 0.7 μM nitrite. M. smegmatis complemented with pNarGHJI1 produced 27 μM at the same stage of growth, but with pNarX, it produced only 2 μM. In NRP-1, wild-type M. smegmatis produced 2 μM nitrite; when it was complemented with pNarGHJI1, it produced 14 μM, and when it was complemented with pNarX5, it produced 2 μM. Thus, M. tuberculosis narGHJI was able to introduce nitrate reductase activity to M. smegmatis but not the characteristic hypoxic increase in activity. When this plasmid was electroporated into M. tuberculosis RVW1 narG::aphI, the complementation restored the lost Nar activity (Table 4).
Ability to support anaerobic growth of E. coli.
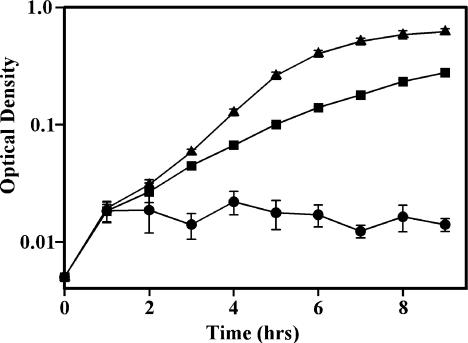
Anaerobically induced nitrate reductase enzymes support the anaerobic replication of many bacteria. Nevertheless, neither M. tuberculosis nor M. smegmatis with pNarGHJI1 could be induced to grow under anaerobic conditions either with or without nitrate. Therefore, the ability of the M. tuberculosis narGHJI operon to complement a nar E. coli mutant was investigated. E. coli JCB4023 lacks all three of the nitrate reductase enzymes of E. coli and consequently does not grow anaerobically with glycerol as the sole carbon source (31). JCB4023 that was complemented with pNarGHJI1 from M. tuberculosis acquired the ability to grow anaerobically but only in the presence of nitrate (Fig. 5). JCB4023 complemented with the vector pPE207 alone showed no anaerobic growth.
FIG. 5.
Anaerobic growth of an E. coli nar mutant with M. tuberculosis narGHJI1. E. coli was grown anaerobically in M9 medium containing glycerol and 20 mM NaNO3. Symbols: ▴, wild type; ▪, E. coli JCB4023 with pNarGHJI1 (complementation mutant); •, E. coli JCB4023 with pPE207 (mutant with only the vector).
Knockout of narK2.
Surprisingly, the transcription of narGHJI does not increase during hypoxia and enzyme levels do not change, despite a strong increase in nitrate reductase activity in whole cells. We hypothesized that this induction could be due to an increase in nitrate and nitrite transport under hypoxic conditions rather than an increase in nitrate reductase enzyme levels. The role of nitrate and nitrite transport in nitrate reduction was next addressed using an E. coli mutant. In E. coli, NarK and NarU are both proposed to be responsible for the transport of nitrate and nitrite (27, 34). In M. tuberculosis, four genes, narK1 to narK3 and narU are homologous to narK and narU (11). One of these genes, narK2, is located upstream of narX (Fig. 1) and is upregulated during hypoxia (Fig. 2 and 3).
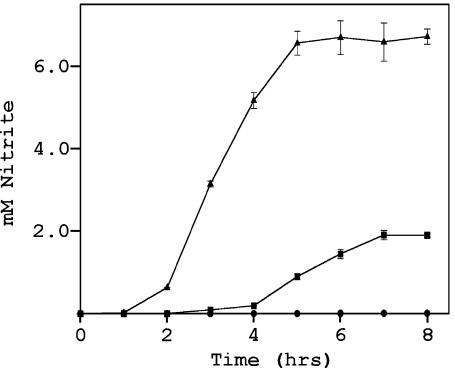
To determine whether narK2 of M. tuberculosis can function as a transporter of nitrate and nitrite, E. coli JCB4018 was used (9). In this mutant, the sole nitrate-reducing enzyme is located intracellularly and requires transport of nitrate into the cell for activity. This mutant also contains knockout mutations in both of the transporters genes (narK and narU) and therefore lacks nitrate reductase activity in culture despite functional NarGHJI enzyme (9). Under anaerobic conditions, JCB4018 with only vector pSK did not produce nitrite (Fig. 6). narK2 from M. tuberculosis was cloned into pSK to produce pTSJ4. JCB4018 with pTSJ4 was able to reduce nitrate, showing that narK2 can function in nitrate and nitrite transport. Interestingly, nitrite production from JCB4018 with narK2 from M. tuberculosis ceased at approximately 1.9 mM, while the wild-type strain with E. coli narK and narU genes continued to 6.5 mM. M. tuberculosis under hypoxic, but not aerobic, conditions ceases nitrite production at a level of approximately 2.5 mM (50). This suggests the possibility that this plateau in nitrite production may be due to narK2.
FIG. 6.
Nitrite production by an E. coli mutant with M. tuberculosis narK2. E. coli was grown anaerobically in LB medium containing 20 mM NaNO3. Symbols: ▴, wild type; ▪, E. coli JCB4018 pTSJ4 (complementation mutant); •, E. coli JCB4018 with pSK (mutant with only the vector).
To determine the role of narK2 in the hypoxic induction of nitrate reductase activity in M. tuberculosis, this gene was inactivated. A kanamycin resistance marker was inserted into narK2 to create M. tuberculosis RVW3 narK2::aphI (Fig. 4). RVW3 produced essentially identical optical density growth curves to the wild type under aerobic and NRP conditions (Sohaskey, unpublished). This mutant showed wild-type levels of nitrite production in aerobic cultures (Table 4). However, there was no increase in activity in NRP-1 cultures, although low levels of nitrite were still produced. These levels were well above those of the narG knockout strain RVW1.
Induction of nitrate reductase activity in SSP.
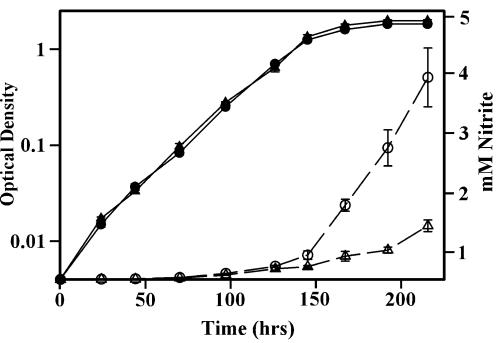
With the creation of the narK2 knockout mutant, the question of induction of nitrate reductase activity in SSP could again be addressed. Transcription of narGHJI is not induced in stationary phase, but narK2 is (Fig. 3). However, it was difficult to detect changes in nitrate reductase activity in the face of decreasing cell viability. If strain RVW3, in which the narK2 gene has been deleted, also lacks this induction, it should be easily detected by comparison to the wild type. The wild-type strain and RVW3 narK2::aphI were grown aerobically in parallel, and nitrite production was monitored (Fig. 7). The growth of both strains reached a plateau after 145 h of incubation, but the rate of nitrite production from the wild type continued to increase, while that of RVW3 decreased. This indicates that nitrate reducing activity is induced upon entry into SSP and that this upregulation is due to induction of narK2.
FIG. 7.
Induction of nitrate reductase levels in SSP. M. tuberculosis was grown aerobically in DTA medium with 5 mM NaNO3, and nitrite concentrations were determined at intervals. Growth (measured by OD580) (solid symbols) and nitrite levels (open symbols) of the wild-type strain (circles) and strain RVW3 narK2::aphI (triangles) are shown.
DISCUSSION
There are two sets of genes in M. tuberculosis, narGHJI and narX, that show homology with prokaryotic nitrate reductase genes, but only narGHJI is responsible for nitrate reducing activity in culture. Insertional inactivation of this locus eliminated the production of nitrite, and this activity could be restored by complementation with a plasmid-borne copy of the genes. Insertion in narX had no effect on the reduction of nitrate. The increase in nitrate reductase activity in hypoxic culture was due not to induction of narGHJI but to increased levels of the nitrate and nitrite transporter narK2.
Nitrate reductase activity of M. tuberculosis was sensitive to inhibition by both tungstate and azide, suggesting that this enzyme is a membrane-bound molybdenum-containing complex similar to the corresponding narGHJI of E. coli (41). It also appeared to be functionally similar to that of E. coli, having the ability to complement a defective strain of the latter in terms of both the ability to reduce nitrate and to support anaerobic growth. M. tuberculosis, unlike E. coli, does not increase either narGHJI mRNA (Fig. 3) or enzyme levels (Table 3) in response to hypoxia or stationary phase. In support, microarray analysis of transcription in M. tuberculosis has shown little induction of narG and narH after a shift from 20 to 0.2% oxygen, while under these same conditions, narK2X was induced (36). In M. bovis BCG, narK2X was induced during hypoxic shiftdown (20). narK2X, but not narGHJI, are members of the dosR-controlled NRP regulon. The dosR product induces a set of 48 genes in response to either hypoxia or NO (5, 45). Both of these conditions are thought to exist in granulomas and play a role in triggering nonreplicating persistence. Upstream of the transcription point of narK2X, there are two copies of a DosR binding sequence proposed to be important for this regulation (29). No such sequence is found upstream of narGHJI. Interestingly, there is a possible FNR box (39) upstream of narGHJI (TTGATnnnnATCCAAT [n is any nucleotide]) but not narK2X. In E. coli and many other bacteria, FNR regulates gene expression in response to hypoxia, but an FNR ortholog has not been identified in M. tuberculosis.
In SSP, narK2X but not narGHJI was induced in M. tuberculosis (Fig. 3). In aerobic shaking cultures, it is possible that a large number of respiring cells could deplete oxygen faster than it could dissolve, creating hypoxic conditions despite the apparent abundance of oxygen.
Unexpectedly, transcription of both narGHJI and narK2X were independent of nitrate and nitrite levels. Nitrate reductase activity in M. tuberculosis appears to be independent of the substrate concentration as determined by levels of mRNA (Fig. 3) and assays of cell extracts (Table 3). In E. coli, the transcription factor NarL regulates genes in response to nitrate in the environment. A possible NarL (Rv0844c) has been identified in M. tuberculosis. Both the upstream regions of narGHJI and narK2X lack good NarL binding sites (43).
It is especially interesting that despite the increase in activity of whole cells of M. tuberculosis exposed to hypoxic conditions, nitrate reductase does not appear to support actual anaerobic growth of this species. Instead, it shifts down to the state of nonreplicating persistence as microaerobic conditions develop. Shiftdown appears to be an orderly process, and the cessation of replication also appears to be part of the cell's adaptation to hypoxia rather than simply energy starvation (52). Since M. tuberculosis does not grow under anaerobic conditions, there may be no requirement for induction of narGHJI. The primary role for nitrate reductase in M. tuberculosis could be redox balancing, or it may serve only a temporary function to provide energy during shiftdown to NRP.
To be reduced, nitrate must enter the cell where the catalytic site of the enzyme is located. Subsequently, since M. tuberculosis is unable to reduce nitrite, which could accumulate to toxic levels, it must then be exported out of the cell. Early work in E. coli had suggested that narK was involved only in nitrite export (34), and so the homologous narK2 in M. tuberculosis was annotated as a “nitrite extrusion protein” (11). More recent work with an E. coli narK narU double mutant indicated that the two proteins could transport nitrate into and nitrite out of the cell (9). We show here that M. tuberculosis narK2 can complement this E. coli double mutant, supporting a role for narK2 in nitrate reduction by coding for a transporter of nitrate into and nitrite out of the cell.
M. tuberculosis RVW3 narK2::aphI, which lacks the nitrate and nitrite transporter, behaved like the wild-type strain in its nitrate reductase activity under aerobic conditions (Table 4). This low level of activity reflects the low rate of diffusion of nitrate into the cell, and this conclusion is supported by evidence that the rate of nitrate reduction by M. tuberculosis under aerobic, but not hypoxic, conditions is proportional to the nitrate concentration in the medium (50). During shiftdown to hypoxic NRP-1, nitrate reductase activity levels of RVW3 narK2::aphI lacked the strong induction seen in the wild type but instead continued at aerobic levels. This indicates that NarK2 is responsible for the hypoxic rise in activity by transporting nitrate into the cell.
In most bacteria in which transcription has been characterized, narGHJI and narK are induced by hypoxia (3, 26, 30, 54). This makes a determination of the role of each in the regulation of nitrate reductase activity difficult. In M. tuberculosis, narGHJI is not induced by hypoxia. Instead, narK2 is a major factor in the regulation of nitrate reductase activity. Nitrate reductase is regulated by control of the level of transcription of narK2, which controls the transport of nitrate into the cell. This is the first report of regulation of nitrate reduction solely by control of transcription of the nitrate transporter.
The gene narX has been found only in M. tuberculosis and M. bovis. It was designated a fused nitrate reductase, because it codes for a single protein that is homologous to sections of three proteins of the NarGHJI complex. The amino terminus from amino acids 1 to 256 is homologous to the same region of NarG, which is the catalytic subunit of the enzyme (10). The three cysteines and one histidine of NarG implicated in the binding of the [4Fe-4S] cluster (33) are present in NarX, but the amino acids thought to be responsible for the binding of the molybdopterin guanine dinucleotide cofactor are missing. Amino acids 257 to 413 of NarX are homologous to NarJ, which is not part of the nitrate reductase enzyme complex but is necessary for maturation of this complex (10). The carboxyl end of NarX from amino acids 417 to 652 is homologous to NarI, a b-type cytochrome. NarI is predicted to contain five membrane loops which are all present in NarX. The conserved histidine and glycines that are important for binding and packaging the two heme groups (4) are all found in NarX. Regulation of nitrate reductase activity does not appear to be associated with narX, and its function is still unknown. Transcripts of narX were detected in a low percentage of granulomas from the lungs of tuberculosis patients, showing that it is expressed in vivo (17).
Virulent M. bovis and avirulent BCG showed significantly less nitrate reductase activity than M. tuberculosis (Table 2), and this trait has been used to distinguish the two species (44, 48). M. bovis was not completely deficient in this activity, as can be seen by comparison to the narG knockout (Table 4), but it also did not show the hypoxic induction exhibited by M. tuberculosis. M. bovis BCG Pasteur lacked nitrate reductase activity in all stages of growth. M. bovis BCG Pasteur is reported to have a deletion of narH, which could explain this lack of activity (20). Virulent M. bovis has the complete narGHJI and narK2X operons (19). Analysis of the sequences of M. tuberculosis and M. bovis predicts two amino acid changes in NarG and one in NarI, while NarH, NarJ, and NarK2 are predicted to be identical in the two species. Transcription levels of these genes may also be different in the two species. This emphasizes the often overlooked variations between these species. M. bovis, for example, prefers a reduced oxygen tension and causes different disease manifestations (15, 51).
The role of nitrate reductase in the virulence of M. tuberculosis has not been investigated. However, immunodeficient SCID mice infected with an M. bovis BCG narG mutant showed smaller granulomas with fewer bacteria than those infected with the wild-type strain (53). The mutant produced tissue damage in the lungs of immunocompetent mice but was cleared from many organs, unlike the wild-type strain (18). The 50% lethal dose of a Salmonella enterica serovar Typhimurium nitrate reductase-deficient mutant was increased in mice relative to the wild-type strain, but its virulence was not completely attenuated (12). The M. tuberculosis equivalent, RVW1, may also be attenuated and might be a candidate for a safe and more effective live vaccine against tuberculosis.
It has been proposed that hypoxia may be partly responsible for the plateau in bacillus counts seen after primary infection with M. tuberculosis, which results in a latent infection (52). Differences between the avirulent and virulent forms of M. tuberculosis have been attributed in the past to enhanced ability of the former to grow at lower O2 concentrations (51). Hypoxic conditions within phagosomes of macrophages are probably sufficient to induce narK2X. Indeed, activated macrophages produce an oxygen gradient between the phagosomes and the extracellular space (22). The reduction of nitrate in the absence of oxygen may serve to provide an alternative energy source to the cell, as it adapts to decreasing oxygen levels. This adaptation allows tubercle bacilli to survive microaerobic conditions that may exist in granulomas or macrophages.
Acknowledgments
We thank Jeff Cole, William Jacobs, and Frank Collins for providing bacterial strains and cell lines. We thank Stewart Cole, Julian Davies, Graham Hatfull, Nadine Honoré, Michael Hynes, and Sunny Twelker for providing plasmids. We thank Sandra Sudberg for technical assistance.
This study was supported in part by the Medical Research Services of the U.S. Department of Veterans Affairs.
REFERENCES
- 1.Bedzyk, L., T. Want, and R. W. Ye. 1999. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J. Bacteriol. 181:2802-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, L. C., D. J. Richardson, and S. J. Ferguson. 1990. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. FEBS Lett. 265:85-87. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. Richardson. 1995. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 4.Berks, B. C., M. D. Page, D. J. Richardson, A. Reilly, A. Cavill, F. Quten, and S. J. Ferguson. 1995. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol. Microbiol. 15:319-331. [DOI] [PubMed] [Google Scholar]
- 5.Boon, C., and T. Dick. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito, B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 66:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canetti, G. 1955. The tubercle bacillus in the pulmonary lesions of man, p. 87-90. In The histobacteriogenesis of tuberculosis lesions: experimental studies. Springer Publishing Company, Inc., New York, N.Y.
- 8.Chang, L., L. I. C. Wei, J. P. Audia, R. A. Morton, and H. E. Schelhorn. 1999. Expression of the Escherichia coli NRZ nitrate reductase is highly growth phase dependent and is controlled by RpoS, the alternative vegetative sigma factor. Mol. Microbiol. 34:756-766. [DOI] [PubMed] [Google Scholar]
- 9.Clegg, S., F. Yu, L. Griffiths, and J. A. Cole. 2002. The roles of the polytopic membrane proteins of NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol. Microbiol. 44:143-155. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. 1996. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Gernier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. I. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Contreras, I., C. S. Toro, G. Troncoson, and G. C. Mora. 1997. Salmonella typhi mutants defective in anaerobic respiration are impaired in their ability to replicate within epithelial cells. Microbiology 143:2665-2672. [DOI] [PubMed] [Google Scholar]
- 13.DesJardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the alpha-crystallin chaperone protein homologues (hspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 183:5311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolin, P. J., M. C. Raviglione, and A. Kochi. 1994. Global tuberculosis incidence and mortality during 1990-2000. Bull W. H. O. 72:213-220. [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn, P. L., and R. J. North. 1995. Virulence ranking of some Mycobacterium tuberculosis and Mycobacterium bovis strains according to their ability to multiply in the lungs, induce lung pathology, and cause mortality in mice. Infect. Immun. 63:3428-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dye, C., S. Cheele, P. Olin, P. Ikram, and M. C. Aviglioen. 1999. Global burden of tuberculosis. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 17.Fenhalls, G., L. Stevens, L. Moses, J. Bezuidenhout, J. C. Betts, P. van Helden, P. T. Lukey, and K. Duncan. 2002. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect. Immun. 70:6330-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz, C., S. Maass, A. Kreft, and F.-C. Bange. 2002. Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect. Immun. 70:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnier, T., K. Eiglmeier, J.-C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutter, B., and T. Dick. 1999. Up-regulation of narX, encoding a putative ′fused nitrate reductase' in anaerobic dormant Mycobacterium bovis BCG. FEMS Microbiol. Lett. 178:63-69. [DOI] [PubMed] [Google Scholar]
- 21.Hutter, B., and T. Dick. 2000. Analysis of the dormancy-inducible narK2 promoter in Mycobacterium bovis BCG. FEMS Microbiol. Lett. 188:141-146. [DOI] [PubMed] [Google Scholar]
- 22.James, P. E., O. Y. Grinberg, G. Michaels, and H. M. Swartz. 1995. Intraphagosomal oxygen in stimulated macrophages. J. Cell. Physiol. 163:241-247. [DOI] [PubMed] [Google Scholar]
- 23.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim, A., M. Eleuterio, B. Hutter, B. Murugasu-Oei, and T. Dick. 1999. Oxygen depletion-induced dormancy in Mycobacterium bovis BCG. J. Bacteriol. 181:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monahan, I. M., J. A. Mangan, and P. D. Butcher. 2001. Extraction of RNA from intracellular Mycobacterium tuberculosis. Methods Mol. Med. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 26.Moreno-Vivián, C., P. Cabello, M. Martínez-Luque, R. Blasco, and F. Castillo. 1999. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181:6573-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noji, S., T. Nohno, T. Saito, and S. Taniguchi. 1989. The narK gene product participates in nitrate transport induced in Escherichia coli nitrate-respiring cells. FEBS Lett. 252:139-143. [DOI] [PubMed] [Google Scholar]
- 28.Paget, E., and J. Davies. 1996. Apramycin resistance as a selective marker for gene transfer in mycobacteria. J. Bacteriol. 178:6357-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, H.-D., K. M. Guinn, M. I. Harrell, R. Liao, M. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of M. tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippot, L., and O. Højberg. 1999. Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1446:1-23. [DOI] [PubMed] [Google Scholar]
- 31.Potter, L. C., P. Millington, L. Griffiths, G. H. Thomas, and J. A. Cole. 1999. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem. J. 344:77-84. [PMC free article] [PubMed] [Google Scholar]
- 32.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 33.Ramírez-Arcos, S., L. A. Fernández-Herrero, and J. Berenguer. 1998. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta 1396:215-227. [DOI] [PubMed] [Google Scholar]
- 34.Rowe, J. J., T. Ubbink-Kok, D. Molenaar, W. N. Konings, and A. J. M. Driessen. 1994. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol. Microbiol. 12:579-586. [DOI] [PubMed] [Google Scholar]
- 35.Saunders, B. M., A. A. Frank, and I. M. Orme. 1999. Granuloma formation is required to contain bacillus growth and delay mortality in mice chronically infected with Mycobacterium tuberculosis. Immunology 98:324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, L., Y.-J. Jung, S. Tyagi, M. L. Gennaro, and R. North. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA 100:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 39.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 75:399-428. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, V. 1982. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J. Bacteriol. 151:1320-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, V. 1988. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol. Rev. 52:190-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timm, J., M. Lim Eng, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyson, K. L., A. I. Bell, J. A. Cole, and S. J. Busby. 1993. Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol. Microbiol. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 44.Virtanen, S. 1960. A study of nitrate reduction by mycobacteria. Acta Tuberc. Scand. Suppl. 47:1-119. [PubMed] [Google Scholar]
- 45.Voskuil, M. I., D. Schnappinger, M. I. Harrell, K. C. Visconti, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2002. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis persistence program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wayne, L. G. 2001. In vitro model of hypoxically induced nonreplicating persistence of Mycobacterium tuberculosis. Methods Mol. Med. 54:247-269. [DOI] [PubMed] [Google Scholar]
- 47.Wayne, L. G. 2003. Senescent stationary phase nomenclature. ASM News 69:108. [Google Scholar]
- 48.Wayne, L. G., and J. R. Doubek. 1965. Classification and identification of mycobacteria. II. Tests employing nitrate and nitrite as substrate. Am. Rev. Respir. Dis. 91:738-745. [DOI] [PubMed] [Google Scholar]
- 49.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wayne, L. G., and L. G. Hayes. 1999. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber. Lung Dis. 79:127-132. [DOI] [PubMed] [Google Scholar]
- 51.Wayne, L. G., and G. P. Kubica. 1986. The mycobacteria, p. 1435-1457. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 52.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 53.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F.-C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 54.Wood, N. J., T. Alizadeh, S. Bennett, J. Pearce, S. J. Ferguson, D. J. Richardson, and J. W. B. Moir. 2001. Maximal expression of membrane-bound nitrate reductase in Paracoccus is induced by nitrate via a third FNR-like regulator named NarR. J. Bacteriol. 183:3603-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]