Abstract
The ability of archaea to salvage cobinamide has been under question because archaeal genomes lack orthologs to the bacterial nucleoside triphosphate:5′-deoxycobinamide kinase enzyme (cobU in Salmonella enterica). The latter activity is required for cobinamide salvaging in bacteria. This paper reports evidence that archaea salvage cobinamide from the environment by using a pathway different from the one used by bacteria. These studies demanded the functional characterization of two genes whose putative function had been annotated based solely on their homology to the bacterial genes encoding adenosylcobyric acid and adenosylcobinamide-phosphate synthases (cbiP and cbiB, respectively) of S. enterica. A cbiP mutant strain of the archaeon Halobacterium sp. strain NRC-1 was auxotrophic for adenosylcobyric acid, a known intermediate of the de novo cobamide biosynthesis pathway, but efficiently salvaged cobinamide from the environment, suggesting the existence of a salvaging pathway in this archaeon. A cbiB mutant strain of Halobacterium was auxotrophic for adenosylcobinamide-GDP, a known de novo intermediate, and did not salvage cobinamide. The results of the nutritional analyses of the cbiP and cbiB mutants suggested that the entry point for cobinamide salvaging is adenosylcobyric acid. The data are consistent with a salvaging pathway for cobinamide in which an amidohydrolase enzyme cleaves off the aminopropanol moiety of adenosylcobinamide to yield adenosylcobyric acid, which is converted by the adenosylcobinamide-phosphate synthase enzyme to adenosylcobinamide-phosphate, a known intermediate of the de novo biosynthetic pathway. The existence of an adenosylcobinamide amidohydrolase enzyme would explain the lack of an adenosylcobinamide kinase in archaea.
To date, de novo coenzyme B12 (Fig. 1) biosynthesis has only been reported to occur in prokaryotes (2, 13, 28, 30, 31, 38). This major biosynthetic pathway has mostly been studied in bacterial systems, with the majority of the work being focused on the anaerobic biosynthesis of the corrin ring in Salmonella enterica (11, 27), Propionibacterium freundenreichii subsp. shermanii (29), and Bacillus megaterium(6, 23, 24) and on aerobic biosynthesis of the corrin ring in Pseudomonas denitrificans (4). This large body of work has given considerable insight into the details of cobamide biosynthesis and has set the basis for comparisons with other organisms (26, 38).
FIG. 1.
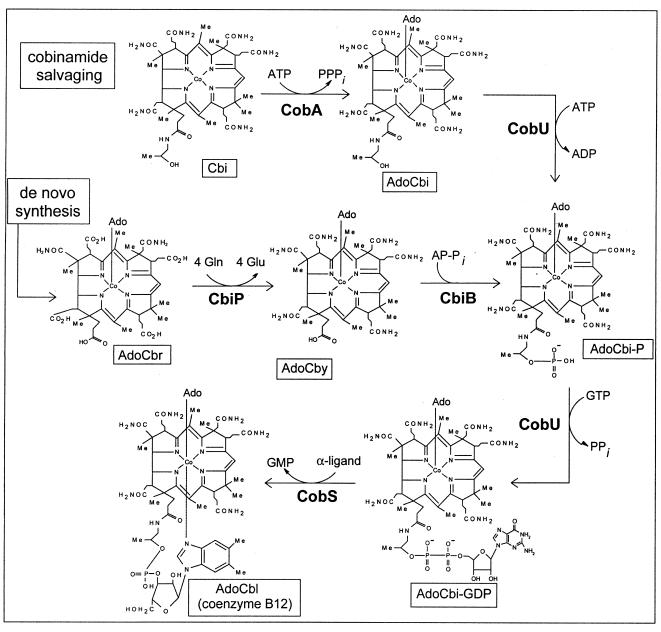
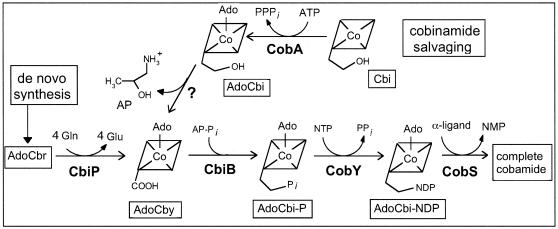
Late steps of cobamide biosynthesis in the bacterium S. enterica. Intermediates are boxed and indicated below structures. Abbreviations: AP-Pi, aminopropanol phosphate; AdoCbr, adenosylcobyrinic acid a,c-diamide; AdoCbl, adenosylcobalamin; CobS, cobalamin (5′-P) synthase.
At present, our knowledge of how archaea synthesize cobamides is very limited (7, 36, 39). It is clear that some archaea synthesize and require cobamides to live. For example, methanogenic archaea require cobamides for methanogenesis from H2 and CO2, acetate, or methanol (10). The extremely halophilic archaeon Halobacterium sp. NRC-1 has been shown to produce and require cobamides under certain growth conditions, but it is unclear why they are needed (39). Some archaea may possess cobamide-dependent ribonucleotide reductases that are required for DNA synthesis, as suggested by genome sequence analysis. In fact, cobamide-dependent ribonucleotide reductases have been isolated from Thermoplasma acidophilum and Pyrococcus furiosus (25, 34). The availability of several archaeal genome sequences has allowed researchers to predict which organisms may have complete de novo cobamide pathways and which may have only enough genetic information for precursor salvaging.
Analysis of the available archaeal genome sequences revealed the absence of an archaeal ortholog to the bacterial ATP:adenosylcobinamide (AdoCbi) kinase/GTP:adenosylcobinamide-phosphate (AdoCbi-P) guanylyltransferase (CobU in S. enterica). The transferase activity was shown to be required for de novo biosynthesis of cobamides and for the salvaging of unphosphorylated Cbi (19). The kinase activity, on the other hand, is only required for the salvaging of Cbi (8, 36) (Fig. 1). Recently, it was shown that the conserved archaeal cobY gene is the nonorthologous replacement of the S. enterica cobU gene. The CobY protein has the nucleoside triphosphate (NTP):AdoCbi-P nucleotidyltransferase activity required for de novo synthesis of cobamides but lacks the NTP:AdoCbi kinase activity necessary to salvage Cbi via the pathway used by bacteria (5, 36, 39).
The lack of an NTP:AdoCbi kinase ortholog in archaea raises three important questions. (i) Are archaea able to salvage Cbi? (ii) If they can, does an alternative, nonorthologous replacement of the bacterial NTP:AdoCbi kinase exist in these prokaryotes? (iii) If a nonorthologous replacement of the bacterial NTP:AdoCbi kinase does not exist in archaea, does an alternative, uncharacterized Cbi-salvaging pathway exist? Previous studies of Methanobacterium thermoautotrophicum strongly suggested that this archaeon can salvage Cbi (32). However, to the best of our knowledge, there are no reported studies of the pathway used by this or any other archaeon to salvage Cbi.
In this paper, we provide genetic evidence for the ability of the extremely halophilic archaeon Halobacterium sp. strain NRC-1 to efficiently salvage exogenous Cbi via an alternative pathway to the one used by bacteria. These studies demanded the functional characterization of two genes whose putative function had been annotated exclusively on the basis of their homology to the bacterial adenosylcobyric acid (AdoCby) and AdoCbi-P synthases (cbiP and cbiB, respectively) present in S. enterica (Fig. 1).
MATERIALS AND METHODS
Strains and plasmids.
The genotypes of the Halobacterium sp. strain NRC-1 and S. enterica strains and the plasmids used in this work are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Marker(s)a | Relevant genotype | Description | Reference or sourceb |
|---|---|---|---|---|
| Halobacterium strains | ||||
| MPK414 | Δura3 | Strain with de novo cobamide biosynthetic capability | 39 | |
| JE6738 | Δura3 ΔcbiP | Strain with in-frame deletion of cbiP | ||
| JE6791 | Δura3 ΔcbiB | Strain with in-frame deletion of cbiB | ||
| JE6930 | Δura3 ΔcbiB ura3::cbiB+ | Strain used to test for complementation of cbiB | ||
| JE7001 | Δura3 ΔcbiP ura3::cbiP+ | Strain used to test for complementation of cbiP | ||
| S. enterica strains | ||||
| TR6583 | metE | S. enterica wild type for this study | Laboratory collection | |
| JE588 | cbiP metE | S. enterica strain used for cbiP complementation studies | Laboratory collection | |
| JE6368 | cbiB metE | S. enterica strain used for cbiB complementation studies | Laboratory collection | |
| Plasmids | ||||
| pMPK428 | 5-FOAs, Mevr | ura3+ | Plasmid used to generate in-frame deletions of targeted genes | 22 |
| pMPK424 | 5-FOAs, Mevr | ura3+ | Plasmid contains flanking sequence to ura3 to allow recombination at the ura3 locus | 21 |
| pCBIP2 | 5-FOAs, Mevr | ura3+ ΔcbiP | Plasmid transformed into MPK414 to delete cbiP | |
| pCBIP7 | 5-FOAs, Mevr | ura3+cbiP+ | Plasmid used to recombine cbiP into ura3 locus | |
| pVng1578-2 | 5-FOAs, Mevr | ura3+ ΔcbiB | Plasmid transformed into MPK414 to delete cbiB | |
| pVng1578-3 | 5-FOAs, Mevr | ura3+cbiB+ | Plasmid used to recombine cbiB into ura3 locus | |
| pT7-7 | Apr | Cloning vector used for complementation studies in S. enterica | 33 | |
| pCBIP9 | Apr | cbiP+ | Plasmid used to provide S. enterica cbiP in trans | |
| pMmCBIP1 | Apr | cbiP+ | Plasmid used to provide M. mazei cbiP in trans | |
| pSeCBIB4 | Apr | cbiB+ | Plasmid used to provide S. enterica cbiB in trans | Laboratory collection |
| pMmCBIB1 | Apr | cbiB+ | Plasmid used to provide M. mazei cbiB in trans |
Abbreviations: Mevr, resistance to mevinolin; 5-FOAs, sensitivity to 5-fluoroorotic acid; Apr, resistance to ampicillin.
Unless otherwise stated, strains and plasmids were constructed during the course of this study.
Chemicals, culture media, and growth conditions.
All chemicals used in this work were commercially available, high-purity compounds. When corrinoids were added to the medium, they were used at concentrations of 100 pM for Halobacterium studies and 15 nM for S. enterica studies. All corrinoids were added in their cyano form. Cbi dicyanide was purchased from Sigma (St. Louis, Mo.). Cbi-GDP dicyanide was synthesized as previously described (36). Cobyric acid dicyanide [(CN)2Cby] was a gift from Paul Renz (Universität-Hohenheim, Stuttgart, Germany), 5-fluoroorotic acid (5-FOA) was purchased from Zymo Research (Orange, Calif.), and mevinolin was purchased from LKT Laboratories, Inc. (St. Paul, Minn.).
Halobacterium studies.
Strains were grown in liquid peptone (Oxoid, Hampshire, England) medium (18) lacking trace metals. Halobacterium cultures were grown to stationary phase at 37°C with shaking for 5 days. Cells used as inocula were harvested by centrifugation (10,000 × g for 2 min) with a Microfuge 18 centrifuge (Beckman-Coulter, Fullerton, Calif.) and washed once in a chemically defined medium (14). Cells were diluted 100-fold and used to inoculate the defined medium containing the appropriate corrinoid supplements. Cultures were grown at 37°C with shaking. Growth was monitored every 24 h by measuring the absorbance of the culture at 650 nm with a Spectronic 20D spectrophotometer (Milton Roy, Rochester, N.Y). In all cases, media were supplemented with uracil (450 μM).
S. enterica studies.
Plasmids were introduced into S. enterica by passing them first through a restriction-deficient strain (37).
Anaerobic growth studies.
Four independent colonies of each strain were patched onto Luria-Bertani-ampicillin (100 μg/ml) agar (6.6%), grown for 5 h at 37°C, and replica printed onto defined, no-carbon E medium (3) supplemented with glucose (11 mM), MgSO4 (1 mM), 1,2-propanediol (10 mM), CoCl (5 μM), ampicillin (25 μg/ml), and trace minerals (1). (CN)2Cby was added as indicated. Plates were incubated anaerobically in an ANA-PAK system (Scott Laboratories, Inc., Fiskeville, R.I.), with a BBL GasPak anaerobic system (Becton Dickinson, Cockeysville, Md). The growth of the strains after 24 h indicated de novo cobamide biosynthesis.
Aerobic growth studies.
S. enterica strains were grown to full density in nutrient broth (Difco) supplemented with ampicillin (100 μg/ml). Cells were diluted 100-fold and used to inoculate the defined no-carbon E medium supplemented with glucose (11 mM), MgSO4 (1 mM), 1,2-propanediol (10 mM), ampicillin (25 μg/ml), and trace minerals (1). Corrinoid supplements were added as indicated. Cultures were monitored while grown at 37°C with continuous shaking (19 Hz) in an EL808 Ultra Microplate Reader (Bio-Tek Instruments, Inc., Winooski, Vt).
Plasmid constructions.
Plasmids were propagated in the Escherichia coli strain DH5α except where noted. In all cases, Halobacterium sp. strain NRC-1 genomic DNA for PCR was prepared as previously described (39). Methanosarcina mazei strain Goe1 DNA for PCR was a gift from Gerhard Gottschalk (Göttingen, Germany). All primers were purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). Underlined portions of the primer sequences (see below) indicate introduced restriction sites.
Halobacterium plasmids.
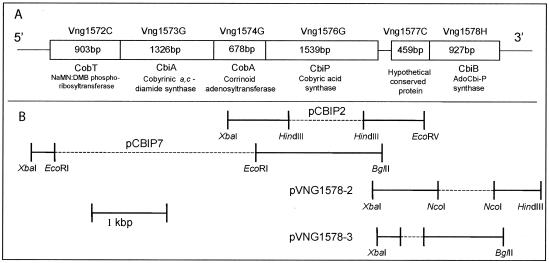
A diagram of the Halobacterium sp. strain NRC-1 DNA included in the most relevant plasmids is included in Fig. 2B.
FIG. 2.
Putative operons in Halobacterium sp. strain NRC-1 containing cbiP (Vng1576G) and cbiB (Vng1578H) and plasmid constructions. (A) The reported ORF designation is shown above each rectangle with our annotation below it. The reported length (base pairs) of each ORF is indicated within each box. (B) Brackets connected by solid lines indicate the regions of DNA that were included in plasmids pCBIP2, pCBIP7, pVNG1578-2, and pVNG1578-3. Dashed lines indicate regions that were not included in the plasmids. The DNA restriction enzyme sites used for cloning purposes are labeled below the brackets.
(i) Plasmid pCBIP1.
The 5′ primer cbiPΔHindIII5′#2 (GTTCGGGAAAAGCTTCGCACGCAG) and the 3′ reverse primer cbiPΔEcoRV3′ (CTGGAGTGGGATATCGGTGAGCAAC) were used to amplify an 804-bp PCR fragment from strain MPK414 genomic DNA. Amplified DNA was cut with HindIII/EcoRV restriction enzymes (unless otherwise noted, the underlined portion of the sequence is the restriction enzyme site), purified with a QIAquick gel extraction kit (QIAGEN; Valencia, Calif.), and cloned into the HindIII/SmaI restriction site of plasmid pMPK428, which contains the wild-type allele of the Halobacterium sp. ura3 gene and a mevinolin resistance determinant (22). The resulting plasmid is referred to as pCBIP1.
(ii) Plasmid pCBIP2.
Plasmid pCBIP2 (ΔcbiP ura3+) carries an in-frame deletion of the Halobacterium sp. strain NRC-1 cbiP gene and was constructed as follows. The 5′ primer cbiPΔXbaI5′ (GCACGTGGTCTAGATGATGAAAG) and reverse 3′ primer cbiPΔHindIII3′ (CACGACGAGTAAGCTTTCGGCGTC) were used to amplify an 807-bp fragment from MPK414 genomic DNA. The fragment was cut with XbaI/HindIII restriction enzymes, gel purified, and cloned into the XbaI/HindIII restriction site of plasmid pCBIP1 to create plasmid pCBIP2. The latter contained an in-frame deletion of cbiP that replaced bases 303 to 1376 with a 6-bp HindIII restriction site, thus deleting 358 of the 512 amino acids. Plasmid pCBIP2 also carries the mevinolin resistance determinant and a wild-type allele of the ura3 gene.
(iii) Plasmid pCBIP4.
The 5′ primer cbiPCompEcoRI5′ (TCTAGAGAATTCGAGCCGACGTTCGTGACCGAG) and reverse primer cbiPCompBglII3′ (AGATCTTAGATCTAAAAGCCGCGCCGGTTCAAACGACGTTGACACGGTAG) were used to amplify a 1,739-bp PCR product from strain MPK414 genomic DNA. The fragment was cloned into pGEM-T with the Promega pGEM-T cloning kit (Madison, Wis.) to yield the plasmid pCBIP4.
(iv) Plasmid pCBIP5.
The fragment carried on plasmid pCBIP4 was excised as a 1,721-bp fragment with an EcoRI/BglII digest, gel purified, and cloned into the EcoRI/BglII restriction site of pT7-7 (33) to yield plasmid pCBIP5.
(v) Plasmid pCBIP6.
The 5′ primer cbiPCompXbaI5′ (TCTAGATCTAGACCCAACTGTGGTTGCATACG) and reverse primer cbiPCompEcoRI3′ (GAATTCGAATTCGCCGTACGTCAGCAGTTCG) were used to amplify a 286-bp PCR product from strain MPK414 genomic DNA. The fragment was cut with XbaI/EcoRI restriction enzymes, gel purified, and cloned into the XbaI/EcoRI restriction site of plasmid pCBIP5 to yield plasmid pCBIP6.
(vi) Plasmid pCBIP7.
The 268-bp XbaI/EcoRI and 1,721-bp EcoRI/BglII fragments from plasmid pCBIP6 were excised as a single 1,989-bp fragment with XbaI/BglII restriction enzymes, gel purified, and cloned into the XbaI/BglII restriction site of plasmid pMPK424 (21), which was prepared from the dam mutant strain GM2163 (New England Biolabs, Manchester, Mass.) to yield plasmid pCBIP7 (ura3+ cbiP+). Plasmid pCBIP7 contained the 1,989-bp fragment flanked by a sequence that would allow recombination at the Halobacterium sp. strain NRC-1 ura3 locus. The resulting plasmid carried a wild-type copy of the cbiP gene, including 107 bases 5′ of the putative start codon and 218 bases upstream of the putative operon. To include these sequences, parts of the Vng1572C and Vng1574G open reading frames (ORFs) were also cloned, but the segments carried an in-frame fusion that fused amino acid residue 15 (of 300) of Vng1572C to residue 191 (of 225) of Vng1574G with Glu and Phe encoded by the introduced EcoRI site (Fig. 2B). Including these sequences should preserve the regulation of cbiP in its own operon without including other genes. Flanking the 3′ end was a 16-bp sequence derived from the bop transcription terminator sequence (9) to ensure termination of the cbiP mRNA transcript.
(vii) Plasmid pVNG1578-1.
The 5′ primer Vng1578NcoI5′ (CCATGGCCATGGGTCGTCTACGCCGGAGGTGG) and 3′ reverse primer Vng1578HindIII3′ (AAGCTTAAGCTTACCTCGAACAGCGGCTTCTCG) were used to amplify an 855-bp PCR fragment from strain MPK414 genomic DNA. The fragment was cut with NcoI/HindIII restriction enzymes, gel purified, and cloned into the NcoI/HindIII restriction site of plasmid pMPK428, which contains the wild-type allele of Halobacterium sp. strain NRC-1 ura3 and a mevinolin resistance determinant (22). The resulting plasmid is referred to as pVng1578-1.
(viii) Plasmid pVNG1578-2.
Plasmid pVNG1578-2 (ΔcbiB ura3+) carried an in-frame deletion of the Halobacterium sp. strain NRC-1 cbiB gene and was constructed as follows. The 5′ primer Vng1578XbaI5′ (TCTAGATCTAGACGCGCACGTCGACCTCGACC) and reverse 3′ primer Vng1578NcoI3′ (CCATGGCCATGGCGTCCACGGTCGGTCGACG) were used to amplify an 841-bp fragment from MPK414 genomic DNA. The fragment was cut with XbaI/NcoI restriction enzymes, gel purified, and cloned into the XbaI/NcoI restriction site of plasmid pVNG1578-1 to create plasmid pVNG1578-2. The latter contained an in-frame deletion of cbiB that replaced bases 133 to 897 with a 6-bp NcoI restriction site, thus deleting 255 of the 308 amino acids. Plasmid pVNG1578-2 also carries the mevinolin resistance determinant and a wild-type allele of the ura3 gene.
(ix) Plasmid pVNG1578-3.
The plasmid pVng1578-3 (cbiB+ ura3+) carries a wild-type allele of the Halobacterium sp. strain NRC-1 cbiB gene and was constructed as follows. The 5′ primer cbiBCompXbaI5′ (GAATCCTCTAGATGACCGACCGATTCAAGTCC) and the reverse primer cbiBCompBglII3′ (GAATTCAGATCTAAAAGCCGCGCCGGTTGGTGATGAACGCCTCCCAG) were used to amplify a 1,398-bp PCR product from strain JE6693 (a derivative of MPK414) with an in-frame deletion on Vng1577, deleting bases 103 to 408 (J. C. Escalante-Semerena, laboratory collection) genomic DNA. The fragment was cut with Xba/BglII restriction enzymes, gel purified, and cloned into the Xba/BglII restriction site of plasmid pMPK424 (21) (prepared from the mutant strain GM2163 dam) (New England Biolabs, Manchester, Mass.) to yield plasmid pVNG1578-3 (ura3+ cbiB+). The latter contains the cloned fragment flanked by a sequence that would allow recombination at the ura3 locus of Halobacterium sp. strain NRC-1. The resulting plasmid carried a wild-type copy of the cbiB gene, including 47 bases upstream of the putative start codon and 200 bases upstream of the putative operon. To include these sequences, part of ORF Vng1577C was also cloned, but it carried an in-frame deletion spanning from residue 35 to residue 136 (of 152). Including theses sequences should preserve the regulation of cbiB in its own operon without including other genes. Flanking the 3′ end was a 16-bp sequence derived from the bop transcription terminator sequence (9) to ensure transcriptional termination of the cbiB mRNA transcript.
S. enterica plasmid pCBIP9.
The plasmid pCBIP9 contained a wild-type allele of S. enterica cbiP under the control of the lac promoter and ribosome-binding site and was constructed as follows. The fragment carried on plasmid pCBIP3 (Escalante-Semerena, laboratory collection) included only the S. enterica cbiP ORF and was excised as a 1,520-bp fragment with an NdeI/Xho1 digest, gel purified, and cloned into the NdeI/SalI restriction site of pT7-7 (33) to produce plasmid pCBIP9(cbiP+).
M. mazei plasmids. (i) Plasmid pMmCBIP1.
Plasmid pMmCBIP1 (cbiP+) contained a wild-type allele of M. mazei strain Goe1 cbiP (ORF Mma0093) under the control of the lac promoter and ribosome-binding site and was constructed as follows. The 5′ primer Mma0093-Blunt#1 (TGAATAATAAAAAGCCTGTTTGCGCAG) and the reverse primer Mma0093-Sal1-3′ (CGCGTGGTCGACTCAGACTCCTGC) were used to amplify a 1,512-bp PCR product from M. mazei genomic DNA. The fragment was treated with polynucleotide kinase, cut with SalI, gel purified, and cloned into the NdeI/SalI site of pT7-7 (prepared by cutting plasmid pT7-7 with NdeI, blunt ending with the MBI Fermentas [Amherst, N.Y.] DNA polymerase I large [Klenow] fragment, and digesting with SalI to produce the plasmid pMmCBIP1 [cbiP+]).
(ii) Plasmid pMmCBIB1.
Plasmid pMmCBIB1 (cbiB+) contained a wild-type allele of M. mazei strain Goe1 cbiB (ORF Mma2059) under the control of the lac promoter and ribosome-binding site and was constructed as follows. The 5′ primer MmcbiB-5′NdeI #2 (5′-AGCCTATCATATGATCATACCGGACAGC-3′) and the reverse primer MmcbiB-3′ SalI (5′-ATTGATCTGGAGTAAGTCGACTTTTCAGGG-3′) were used to amplify a 1,025-bp PCR product from M. mazei genomic DNA. The fragment was cut with NdeI/SalI restriction enzymes, gel purified, and cloned into the NdeI/SalI restriction site of plasmid pT7-7 to produce plasmid pMmCBIB1 (cbiB+).
Halobacterium strain constructions. (i) Construction of a ΔcbiP mutant strain.
An in-frame deletion of cbiP in the chromosome of strain MPK414 (Δura3) was generated by using previously described methodology (20). Briefly, strain JE6738 (Δura3 ΔcbiP) was constructed by transforming strain MPK414 with plasmid pCBIP2 as described previously (15). Flanking sequences of over 700 bases on each side of the deleted cbiP gene ensured efficient recombination of the fragment into the chromosome. Mevinolin-resistant transformants were selected as described previously (15) and replated on medium containing 5-FOA to select for the loss of the plasmid (20). Colonies resistant to 5-FOA were screened by PCR to identify the desired recombinant (ΔcbiP). DNA sequencing was used to confirm the in-frame deletion of the cbiP gene in the chromosome of strain JE6738.
(ii) Construction of a ΔcbiB mutant strain.
An in-frame deletion of cbiB in the chromosome of strain MPK414 was generated by using the same strategy as mentioned above. Strain JE6791 (Δura3 ΔcbiB) was constructed with strain MPK414 and plasmid pVNG1578-2. DNA sequencing was used to confirm the in-frame deletion of the cbiB gene in the chromosome of strain JE6791.
(iii) Construction of a cbiP complementation strain.
Complementation studies were performed with a single copy of the wild-type allele of the gene in question placed at the ura3 locus. For cbiP complementation studies, a wild-type allele of cbiP was placed at the chromosomal ura3 locus of strain JE6738. Plasmid pCBIP7 was transformed into strain JE6738, and strains carrying the cbiP+ allele at the chromosomal ura3 locus (strain JE7001 [Δura3 ΔcbiP ura3::cbiP+]) were isolated by using the same ura3-based gene replacement method for the isolation of deleted genes. PCR and DNA sequencing verified the presence of cbiP+ at the ura3 locus.
(iv) Construction of a cbiB complementation strain.
For cbiB complementation studies a wild-type allele of cbiB was placed at the chromosomal ura3 locus of strain JE6791. Plasmid pVNG1578-3 was transformed into strain JE6791, and a strain carrying the cbiB+ allele at the chromosomal ura3 locus (strain JE6930 [Δura3 ΔcbiB ura3::cbiB+]) was isolated. PCR and DNA sequencing verified the presence of cbiB+ at the ura3 locus.
RESULTS
Rationale used to probe into corrinoid salvaging in Halobacterium.
Because the growth of Halobacterium in defined medium requires cobamides, the growth of a corrinoid-deficient mutant in medium supplemented with incomplete cobamide precursors would be indicative of precursor salvaging. To block corrin ring biosynthesis in Halobacterium, in-frame deletions were introduced in the second-to-last step or in the last step of corrin ring biosynthesis. In S. enterica, these steps of the pathway are catalyzed by the AdoCby synthase (CbiP) enzyme and the AdoCbi-P synthase (CbiB) enzyme, respectively (38). It was hypothesized that a block in either one of these steps would render a strain dependent on exogenous Cby or Cbi precursors. The mutation in cbiP would block salvaging of cobyrinic acid a,c-diamide but should not interfere with Cby or Cbi salvaging. A mutation in cbiB would address the question of what the point of entry of Cbi is in the Halobacterium genome sequence. That is, if a cbiB mutation does not prevent Cbi salvaging, then an unidentified kinase may be responsible for the activation of Cbi to Cbi-P (the substrate of the CobY enzyme). Alternatively, the inability of a cbiB mutant to salvage Cbi would suggest the existence of a new pathway for the activation of Cbi in this archaeon.
Identification of the cbiP and cbiB genes of Halobacterium.
ORF Vng1576G (gene identification [gi] number 15790548) of the Halobacterium sp. strain NRC-1 genome sequence (17) was identified as the putative cbiP gene of this archaeon based on the 40% identity and 53% similarity of the predicted gene product to the CbiP protein of S. enterica. In the Halobacterium genome, the cbiP (ORF Vng1576G) gene is located at the 3′ end of a putative operon containing ORF Vng1574G and ORF Vng1573G, which encode the putative orthologs of the bacterial ATP:co(I)rrinoid adenosyltransferase (CobA in S. enterica) and the cobyrinic acid a,c-diamide synthase (CbiA in S. enterica), respectively (Fig. 2A). These two proteins are believed to modify the corrinoid immediately preceding the CbiP-catalyzed step (38).
ORF Vng1578H (gi number 15790550) of the Halobacterium genome sequence was identified as the putative cbiB gene of this archaeon based on the 30% identity and 43% similarity of the predicted gene product to the CbiB of S. enterica. In the Halobacterium genome, the cbiB gene is the promoter-distal gene in a putative operon containing one other ORF of unknown function (Fig. 2A).
cbiP (ORF Vng1576G) is a cobamide biosynthetic gene in Halobacterium.
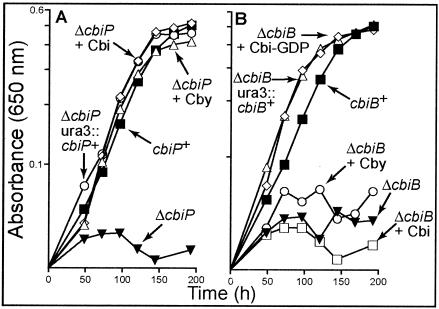
To determine if strain JE6738 (ΔcbiP) was deficient in cobamide biosynthesis, growth was assessed in defined medium where cobamides were essential for growth. Unlike strain MPK414 (cbiP+), strain JE6738 (ΔcbiP) failed to grow in the defined medium lacking corrinoids (Fig. 3A). To determine if the observed lack of growth of JE6738 was caused by the inability to synthesize cobamides de novo, the medium was supplemented with Cby (the nonadenosylated product of the CbiP-catalyzed reaction). The addition of Cby restored wild-type growth of JE6738 (Fig. 3A) but did not significantly enhance the growth of the wild-type strain (data not shown). The doubling times of strains MPK414 and JE6738 in medium supplemented with Cby were very similar (30 and 27 h, respectively), whereas doubling times could not be calculated for the strains that displayed extremely poor growth. These data strongly suggested that the absence of cbiP function correlated with the predicted phenotype of a strain lacking AdoCby synthase activity under conditions that demand de novo synthesis of cobamides. This finding led to the proposal that ORF Vng1576G was the archaeal ortholog of the CbiP.
FIG. 3.
Nutritional studies of Halobacterium sp. strain NRC-1 strains. Cobamide-dependent growth of Halobacterium sp. strain NRC-1 strains in the defined liquid medium at 37° is reported as absorbance at 650 nm as a function of time. The strains are indicated by their genotypes. The corrinoids added to the medium are indicated next to the genotypes. The strains used were MPK414 cbiP+ cbiB+, JE6738 ΔcbiP, JE7001 ΔcbiP ura3::cbiP+, JE6791 ΔcbiB, and JE6930 ΔcbiB ura3::cbiB+. Abbreviations: Cby, cobyric acid dicyanide; Cbi, cobinamide dicyanide; Cbi-GDP; cobinamide-GDP dicyanide. In all cases, corrinoids were used at concentrations of 100 pM.
Halobacterium can salvage Cbi.
Having a Halobacterium mutant blocked before the late steps of cobamide biosynthesis allowed us to test if this archaeon can salvage Cbi. In bacteria, AdoCbi is not an intermediate of the de novo pathway (8, 36, 39) (Fig. 1), and it is also not predicted to be an intermediate in archaea, based on the presence of CbiB. The salvaging of Cbi, therefore, would require additional enzymes or functions. The addition of Cbi to the medium allowed wild-type growth (i.e., 24-h doubling time) of strain JE6738 (ΔcbiP) (Fig. 3A) but did not significantly enhance the growth of the wild-type strain (data not shown). The ability of Halobacterium to salvage Cbi suggested the existence of an enzyme that can convert Cbi to a true intermediate of the de novo pathway. A mutation in the CbiB enzyme would block the pathway at a point that would allow us to ascertain whether the entry point for Cbi salvaging in archaea occurred via AdoCbi-P (as in bacteria) or via a new metabolic route.
cbiB (ORF Vng1578H) is a cobamide biosynthetic gene in Halobacterium.
Unlike strain MPK414, strain JE6791 (ΔcbiB) cannot grow in the defined medium lacking corrinoids (Fig. 3B). To test if the lack of growth was due to the inability to synthesize cobamides, Cbi-GDP (a pathway intermediate downstream of the CbiB-catalyzed reaction) (Fig. 1) was added to the medium. Cbi-GDP restored the growth of strain JE6791 (30-h doubling time) (Fig. 3B) but did not significantly enhance growth of the wild-type strain MPK414 (data not shown). The addition of Cby (a pathway intermediate prior to the CbiB-catalyzed reaction), however, failed to restore growth of strain JE6791 (Fig. 3B). These results were consistent with a block in the synthesis of AdoCbi-P and led us to propose that ORF Vng1578H in Halobacterium encodes the archaeal ortholog of S. enterica CbiB enzyme.
CbiB activity is required for Cbi salvaging.
As mentioned above, strain JE6738 (ΔcbiP) can salvage Cbi; however, the addition of Cbi to the medium did not restore the growth of strain JE6791 (ΔcbiB) (Fig. 3B). These results confirmed that in Halobacterium Cbi must enter the de novo pathway at an entry point prior to the CbiB-catalyzed step. This finding is also consistent with the observation that Cbi and AdoCbi are not intermediates of the archaeal de novo pathway. If they were, strain JE6791 would be predicted to be able to salvage Cbi.
Complementation of cbiP and cbiB mutants of Halobacterium.
The observed AdoCby auxotrophy of JE6738 (ΔcbiP) and the AdoCbi-GDP auxotrophy of JE6791 (ΔcbiB) were corrected when the cbiP+ and cbiB+ alleles were reintroduced into the appropriate strains. Strain JE7001 (ΔcbiP ura3::cbiP+) and strain JE6930 (cbiB+ ura3::cbiB+) grew in the defined medium without any corrinoid supplementation (Fig. 3) with a doubling time of 26 and 34 h, respectively. The growth rate of these strains was similar to the rates of strains JE6738 (ΔcbiP) and JE6791 (ΔcbiB) growing on medium supplemented with the correct corrinoid supplements. These results showed that the cbiP+ or cbiB+ functions were necessary and sufficient to restore de novo cobamide synthesis in the mutant strains.
The archaeal cbiP and cbiB genes complement S. enterica cbiP and cbiB mutants.
To further support the conclusion that the archaeal orthologs of cbiP and cbiB do function as AdoCby and AdoCbi-P synthases in vivo, we tested the ability of archaeal cbiP and cbiB orthologs to complement S. enterica cbiP and cbiB mutants. To investigate this possibility, the cbiP and cbiB orthologs from the archaeal methanogen M. mazei strain Goe1 were cloned. Previous work in the laboratory has shown that Halobacterium genes do not express well in S. enterica, whereas genes from archaeal methanogens are well expressed (36). M. mazei ORF Mm0093 (gi number 21226195) showed 42% identity and 58% similarity to the Halobacterium cbiP gene, and ORF Mm2059 (gi number 21228161) showed 28% identity and 45% similarity to the cbiB gene of Halobacterium.
For this purpose, S. enterica strains carrying null alleles of metE and either cbiP or cbiB were used. The mutation in metE inactivates the cobamide-independent methionine synthase (MetE) enzyme, thus demanding cobamide-dependent methylation of homocysteine to yield methionine by the action of the MetH enzyme (35). An insertion in either cbiP or cbiB eliminated de novo cobamide synthesis.
For cbiP complementation, the positive control plasmid pCBIP9 (containing a wild-type allele of S. enterica cbiP+) or plasmid pMmCBIP1 (M. mazei cbiP+) was introduced into the S. enterica cbiP metE mutant strain JE588.
For cbiB complementation, a plasmid containing a wild-type allele of either S. enterica cbiB (the positive control plasmid pSeCBIB4) or M. mazei cbiB (plasmid pMmCBIB1) was introduced into the S. enterica cbiB metE mutant strain JE6368. Residual expression of the cbiP or cbiB genes in the absence of the T7 RNA polymerase allowed us to assess complementation. In both cases, plasmid pT7-7 was used as a vector-only negative control.
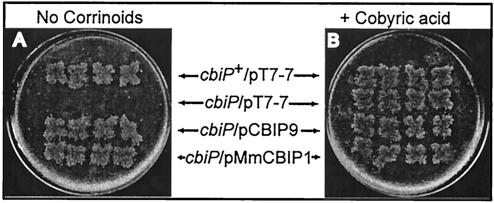
To test cbiP complementation, S. enterica was grown anaerobically, where the cells can synthesize cobamides de novo. Complementation of cobamide biosynthesis was observed when either S. enterica or M. mazei cbiP was provided in trans to JE588 but not with the control vector (Fig. 4A). Growth was similar for all strains when (CN)2Cby was added (Fig. 4B). These results were consistent with the archaeal CbiP enzyme having AdoCby synthase activity in vivo.
FIG. 4.
Nutritional studies of S. enterica cbiP mutants. Cobamide-dependent growth of S. enterica strains grown anaerobically in defined solid medium at 37°C without a corrinoid supplement (A) or with 15 nM (CN)2Cby (B). The strains are indicated by their genotypes. The strains used were TR6583 metE cbiP+ and JE588 metE cbiP. The plasmids used were pT7-7, vector-only control; pCBIP9, S. enterica cbiP+; and pMmCBIP1, M. mazei cbiP+.
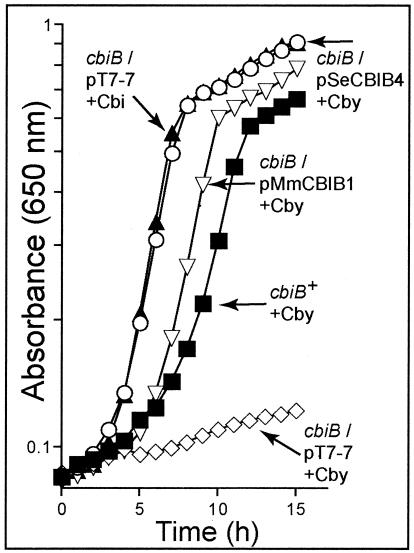
cbiB complementation was tested under aerobic conditions, where S. enterica must salvage cobamide precursors. In this case Cby was added to the medium. Cby salvaging requires a functional CbiB synthase enzyme (Fig. 1); hence, growth on this intermediate would indicate restoration of the de novo pathway of cbiB mutant strain JE6368. Complementation of Cby salvaging was observed when either S. enterica cbiB (pSeCBIB4) or M. mazei cbiB (pMmCBIB1) was provided in trans but not when the control vector was provided (Fig. 5). These data support the conclusion that the archaeal CbiB enzyme has AdoCbi-P synthase activity in vivo.
FIG. 5.
Nutritional studies of S. enterica cbiB mutants. Cobamide-dependent growth of S. enterica strains grown aerobically in defined liquid medium at 37°C is reported as absorbance at 650 nm as a function of time. The strains are indicated by their genotypes. The corrinoids added to the medium are indicated next to the genotypes. The strains used were TR6583 metE cbiP+ and JE6368 metE cbiB. The plasmids used were pT7-7, vector control; pSeCBIB4, S. enterica cbiB+; and pMmCBIB1, M. mazei cbiB+. Abbreviations: Cby, cobyric acid dicyanide; Cbi, cobinamide dicyanide. In all cases, corrinoids were used at concentrations of 15 nM.
DISCUSSION
The contributions of this work are twofold. First, the functions encoded by two putative ORFs in two archaea are supported by in vivo evidence. Second, evidence for the existence of the pathway for salvaging the cobamide precursor Cbi in archaea has been obtained. The latter pathway is distinct from the one used by bacteria.
Biochemical roles of two archaeal genes in cobamide biosynthesis.
The results of the nutritional analysis of mutants of the extremely halophilic archaeon Halobacterium sp. strain NRC-1 showed that ORFs Vng1576G and Vng1578H were necessary for de novo cobamide biosynthesis and that ORF Vng1578H was necessary for salvaging cobyric acid from the environment. The conclusions drawn from these analyses were fully supported by complementation analyses of bona fide S. enterica mutants lacking either CbiP or CbiB activities by M. mazei strain Goe1 genes. On the basis of this work, we propose that Halobacterium ORF Vng1578H be annotated as encoding the AdoCbi-P synthase enzyme and that the putative annotation of Vng1576G as encoding the AdoCby synthase enzyme is correct. ORF Vng1578H should be renamed as cbiB to reflect its involvement in cobamide biosynthesis in archaea. This nomenclature should be extended to the ORFs Mm0093 (cbiP) and Mm2059 (cbiB) of M. mazei strain Goe1.
In this study, corrinoid intermediates have been assumed to be adenosylated in vivo. Although this fact has been established in bacteria (12), it is unknown if the corrinoids are adenosylated in archaea. Because archaea possess a putative ortholog of CobA and archaeal genes can complement S. enterica cob mutants, it is assumed that the corrinoid substrates for the archaeal enzymes are adenosylated.
The archaeal pathway for salvaging Cbi is different from the bacterial pathway.
The requirement for CbiB enzyme activity for the salvaging of Cbi by Halobacterium is key to the proposal that the archaeal pathway for salvaging this precursor is different from the one that operates in bacteria (Fig. 1 and 6). In bacteria, CbiB is not required for Cbi salvaging because the NTP:AdoCbi kinase activity of CobU directly converts AdoCbi to AdoCbi-P, the product of the CbiB enzyme (Fig. 1). The kinase activity of CobU effectively bypasses the need for CbiB. The tight block in Cbi salvaging observed in Halobacterium cbiB mutants strongly suggests that the point of entry of Cbi salvaging in this archaeon is AdoCby, which can then be converted by the action of CbiB to AdoCbi-P, the substrate for the next enzyme of the archaeal pathway, i.e., CobY (Fig. 6). It is unlikely that the point of entry is prior to AdoCby, because Halobacterium cbiP mutants can readily salvage Cbi. We propose that, in archaea, AdoCbi is the substrate for an unidentified amidohydrolase enzyme that cleaves off the (R)-1-amino-2-propanol moiety of AdoCbi to yield AdoCby, the substrate of CbiB (Fig. 6). We favor this hypothesis on the basis of preliminary data obtained in our laboratory, which show that this AdoCbi amidohydrolase activity is present in cell extracts of E. coli overexpressing a single gene of M. mazei (J. D. Woodson and J. C. Escalante-Semerena, unpublished results). The requirement of an adenosylated substrate is speculative, and it is possible that the corrin ring is adenosylated after entering the de novo pathway. The identification of the gene encoding the amidohydrolase activity and the isolation and characterization of this new cobamide biosynthetic enzyme will be reported elsewhere.
FIG. 6.
A new model for the late steps of cobamide biosynthesis in archaea. Intermediates are boxed and indicated below structures. The adenosylation of archaeal intermediates is putative. The putative archaeal orthologs of the bacterial CobA and CobS (16) proteins are indicated. Abbreviations: AP-Pi, aminopropanol phosphate; AP, aminopropanol; AdoCbr, adenosylcobyrinic acid a,c-diamide; AdoCby, adenosylcobyric acid; AdoCbi, adenosylcobinamide; CobS, cobalamin (5′-P) synthase; CobY, NTP:AdoCbi-P nucleotidyltransferase.
Acknowledgments
This work was supported by a grant GM40313 from the National Institutes of Health (NIH) to J.C.E.-S.; J.D.W. was supported in part by the Ira L. Baldwin Predoctoral Fellowship; C.L.Z. was supported in part by NIH Minority Access to Research Careers Predoctoral Fellowship F31-GM64009.
We thank P. Renz for his gift of (CN)2Cby and G. Gottschalk for his gift of M. mazei chromosomal DNA.
REFERENCES
- 1.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battersby, A. R. 2000. Tetrapyrroles: the pigments of life. Nat. Prod. Rep. 17:507-526. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanche, F., B. Cameron, J. Crouzet, L. Debussche, D. Thibaut, M. Vuilhorgne, F. J. Leeper, and A. R. Battersby. 1995. Vitamin B12: how the problem of its biosynthesis was solved. Angew. Chem. Int. Ed. Engl. 34:383-411. [Google Scholar]
- 5.Blanche, F., L. Debussche, A. Famechon, D. Thibaut, B. Cameron, and J. Crouzet. 1991. A bifunctional protein from Pseudomonas denitrificans carries cobinamide kinase and cobinamide phosphate guanylyltransferase activities. J. Bacteriol. 173:6052-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brey, R. N., C. D. B. Banner, and J. B. Wolf. 1986. Cloning of multiple genes involved with cobalamin (vitamin B12) biosynthesis in Bacillus megaterium. J. Bacteriol. 167:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brindley, A. A., E. Raux, H. K. Leech, H. L. Schubert, and M. J. Warren. 2003. A story of chelatase evolution: identification and characterization of a small 13-15-kDa “ancestral” cobaltochelatase (CbiXS) in the archaea. J. Biol. Chem. 278:22388-22395. [DOI] [PubMed] [Google Scholar]
- 8.Brushaber, K. R., G. A. O'Toole, and J. C. Escalante-Semerena. 1998. CobD, a novel enzyme with l-threonine-O-3-phosphate decarboxylase activity is responsible for the synthesis of (R)-1-amino-propanol O-2-phosphate, a proposed new intermediate in cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 273:2684-2691. [DOI] [PubMed] [Google Scholar]
- 9.DasSarma, S., U. L. RajBhandary, and H. G. Khorana. 1984. Bacterio-opsin mRNA in wild-type and bacterio-opsin-deficient Halobacterium halobium strains. Proc. Natl. Acad. Sci. USA 81:125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiMarco, A. A., T. A. Bobik, and R. S. Wolfe. 1990. Unusual coenzymes of methanogenesis. Annu. Rev. Biochem. 59:355-394. [DOI] [PubMed] [Google Scholar]
- 11.Escalante-Semerena, J. C. 1999. Regulation of adenosylcobalamin biosynthesis in Salmonella typhimurium, p. 577-594. In R. Banerjee (ed.), Chemistry and biochemistry of B12. Academic Press, Inc., New York, N.Y.
- 12.Escalante-Semerena, J. C., S.-J. Suh, and J. R. Roth. 1990. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J. Bacteriol. 172:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedmann, H. C., and R. K. Thauer (ed.). 1992. Macrocyclic tetrapyrrole biosynthesis in bacteria, vol. 3. Academic Press, Inc., New York, N.Y.
- 14.Grey, V. L., and P. S. Fitt. 1976. An improved synthetic growth medium for Halobacterium cutirubrum. Can. J. Microbiol. 22:440-442. [DOI] [PubMed] [Google Scholar]
- 15.Krebs, M. P., R. Mollaaghababa, and H. G. Khorana. 1993. Gene replacement in Halobacterium halobium and expression of bacteriorhodopsin mutants. Proc. Natl. Acad. Sci. USA 90:1987-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maggio-Hall, L. A. 2001. Synthesis and incorporation of the lower ligand base of cobalamin. Ph.D. dissertation. University of Wisconsin, Madison.
- 17.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oesterhelt, D., and W. Stoeckenius. 1974. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 31:667-678. [DOI] [PubMed] [Google Scholar]
- 19.O'Toole, G. A., and J. C. Escalante-Semerena. 1995. Purification and characterization of the bifunctional CobU enzyme of Salmonella typhimurium LT2: evidence for a CobU-GMP intermediate. J. Biol. Chem. 270:23560-23569. [DOI] [PubMed] [Google Scholar]
- 20.Peck, R. F., S. Dassarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 21.Peck, R. F., C. Echavarri-Erasun, E. A. Johnson, W. V. Ng, S. P. Kennedy, L. Hood, S. DasSarma, and M. P. Krebs. 2001. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J. Biol. Chem. 276:5739-5744. [DOI] [PubMed] [Google Scholar]
- 22.Peck, R. F., E. A. Johnson, and M. P. Krebs. 2002. Identification of a lycopene beta-cyclase required for bacteriorhodopsin biogenesis in the archaeon Halobacterium salinarum. J. Bacteriol. 184:2889-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raux, E., A. Lanois, A. Rambach, M. J. Warren, and C. Thermes. 1998. Cobalamin (vitamin B12) biosynthesis: functional characterization of the Bacillus megaterium cbi genes required to convert uroporphyrinogen III into cobyrinic acid a, c-diamide. Biochem. J. 335:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raux, E., A. Lanois, M. J. Warren, A. Rambach, and C. Thermes. 1998. Cobalamin (vitamin B12) biosynthesis: identification and characterization of a Bacillus megaterium cobI operon. Biochem. J. 335:159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riera, J., F. T. Robb, R. Weiss, and M. Fontecave. 1997. Ribonucleotide reductase in the archaeon Pyrococcus furiosus: a critical enzyme in the evolution of DNA genomes? Proc. Natl. Acad. Sci. USA 94:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roessner, C. A., P. J. Santander, and A. I. Scott. 2001. Multiple biosynthetic pathways for vitamin B12: variations on a central theme. Vitam. Horm. 61:267-297. [DOI] [PubMed] [Google Scholar]
- 27.Rondon, M. R., J. R. Trzebiatowski, and J. C. Escalante-Semerena. 1997. Biochemistry and molecular genetics of cobalamin biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 56:347-384. [DOI] [PubMed] [Google Scholar]
- 28.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 29.Santander, P. J., C. A. Roessner, N. Stolowich, M. T. Holderman, and A. I. Scott. 1997. How corrinoids are synthesized without oxygen: nature's first pathway to vitamin B12. Chem. Biol. 4:659-666. [DOI] [PubMed] [Google Scholar]
- 30.Scott, A. I. 2003. Discovering nature's diverse pathways to vitamin B12: a 35-year odyssey. J. Org. Chem. 68:2529-2539. [DOI] [PubMed] [Google Scholar]
- 31.Scott, A. I., and C. A. Roessner. 2002. Biosynthesis of cobalamin (vitamin B12). Biochem. Soc. Trans. 30:613-620. [DOI] [PubMed] [Google Scholar]
- 32.Stupperich, E., I. Steiner, and H. J. Eisinger. 1987. Substitution of Coα-(5-hydroxybenzimidazolyl)cobamide (factor III) by vitamin B12 in Methanobacterium thermoautotrophicum. J. Bacteriol. 169:3076-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabor, S. 1990. Expression using the T7 RNA polymerase/promoter system, p. 16.2.1.-16.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y.
- 34.Tauer, A., and S. A. Benner. 1997. The B12-dependent ribonucleotide reductase from the archaebacterium Thermoplasma acidophila: an evolutionary solution to the ribonucleotide reductase conundrum. Proc. Natl. Acad. Sci. USA 94:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, R. T., and H. Weissbach. 1973. N5-methylenetetrahydrofolate-homocysteine methyltransferases, p. 121-165. In P. D. Boyer (ed.), The enzymes, vol. 9. Academic Press, Inc., New York, N.Y.
- 36.Thomas, M. G., and J. C. Escalante-Semerena. 2000. Identification of an alternative nucleoside triphosphate: 5′-deoxyadenosylcobinamide phosphate nucleotidyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 182:4227-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, S. P., R. J. Hartin, and J. Ryu. 1989. Transformation in restriction-deficient Salmonella typhimurium LT2. J. Gen. Microbiol. 135:2561-2567. [DOI] [PubMed] [Google Scholar]
- 38.Warren, M. J., E. Raux, H. L. Schubert, and J. C. Escalante-Semerena. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19:390-412. [DOI] [PubMed] [Google Scholar]
- 39.Woodson, J. D., R. F. Peck, M. P. Krebs, and J. C. Escalante-Semerena. 2003. The cobY gene of the archaeon Halobacterium sp. strain NRC-1 is required for de novo cobamide synthesis. J. Bacteriol. 185:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]