Summary
In the two signal model of T cell activation, the outcome of antigen recognition is determined by the integration of multiple cues in the immune microenvironment. mTOR is an evolutionarily conserved PI3-kinase family member that plays a central role in integrating environmental cues in the form of amino acids, energy, and growth factors. Recently, an increasingly important role for mTOR in directing T cell activation and differentiation has become apparent. Here we review recent findings demonstrating the ability of mTOR to interpret signals in the immune microenvironment and program the generation of CD4+ effector versus regulatory T cells, the generation of CD8+ effector versus memory cells, T cell trafficking, and T cell activation versus anergy. The key theme to emerge from these studies is that the central role of mTOR provides a direct link between T cell metabolism and function.
Introduction
A central role for mTOR in regulating immune responses is emerging. mTOR has been implicated in neutrophil, monocyte, dendritic cell, B cell, gamma-delta and alpha-beta T cell function (Delgoffe and Powell, 2009; Mills and Jameson, 2009; Weichhart and Saemann, 2009). This review focuses on recent findings elucidating the ability of mTOR to regulate T cell differentiation and activation. Specifically, observations demonstrating the link between the unique metabolic requirements of T cells and the ability of mTOR to integrate environmental cues to direct T cell differentiation and function will be discussed.
The disassociation of recognition and function in the adaptive immune response
In the most primitive host defenses, recognition and function occur nearly simultaneously (Braun et al., 1994). For example, proteins produced by one strain of bacteria recognize receptors and kill other strains of bacteria. As organisms became more complex, the need for more sophisticated immune responses evolved. Recognition of Pathogen Associated Molecular Patterns (PAMPs) by different receptors led to differential signaling and responses (Janeway, 1989). For example in Drosophila PAMPs can lead to different responses depending whether the Toll or Immune Deficiency (IMD) pathway is activated. Signaling via Toll pathways leads to the elaboration of the antimicrobial protein attacin while IMD signaling leads to the expression of drosomycin (Tanji and Ip, 2005). That is, recognition of Toll leads to a distinct form of activation when compared to activation of IMD. Still, recognition and activation occur via a single signal or receptor.
The development of adaptive immune responses heralded the disassociation of recognition and response. Antigen receptors of adaptive responses are the products of stochastic mechanisms of generating diversity. Whereas such receptors are characterized by exquisite specificity, recognition of antigen does not impart information in terms of what type of response should ensue. Rather, this information is provided by accessory (“second” or “third”) signals (Curtsinger and Mescher; Sharpe, 2009). That is, when a T cell receptor recognizes its cognate peptide (for example ovalbumin peptide) there is nothing about the recognition of ovalbumin that tells the T cell whether to become a T helper 1 (Th1), Th2, Th17 or regulatory cell. Likewise, this recognition does not instruct a differentiated cell to become activated or anergic. Such instructions come from the presence or absence of signals derived from the environment that not only mediate early innate immunity but qualitatively modulate adaptive immunity. As such, with this increase in sophistication with respect to antigen recognition came the necessity to develop a mechanism to integrate environmental cues.
The mammalian Target of Rapamycin as an environmental sensor
Rapamycin was originally identified as an anti-fungal compound derived from Streptomyces hygroscopicus, found in soil samples collected from Easter Island (locally known as Rapa nui) (Dennis et al., 1999). Although rapamycin was found to be a poor antibiotic, it did turn out to have potent immunosuppressive activity. Initially, its immunosuppressive properties were ascribed to its ability to inhibit cell proliferation. This was in contrast to another macrolide antibiotic, Cyclosporine A (CSA), which suppresses immune responses by inhibiting calcineurin and thereby blocking TCR signaling (Powell and Zheng, 2006).
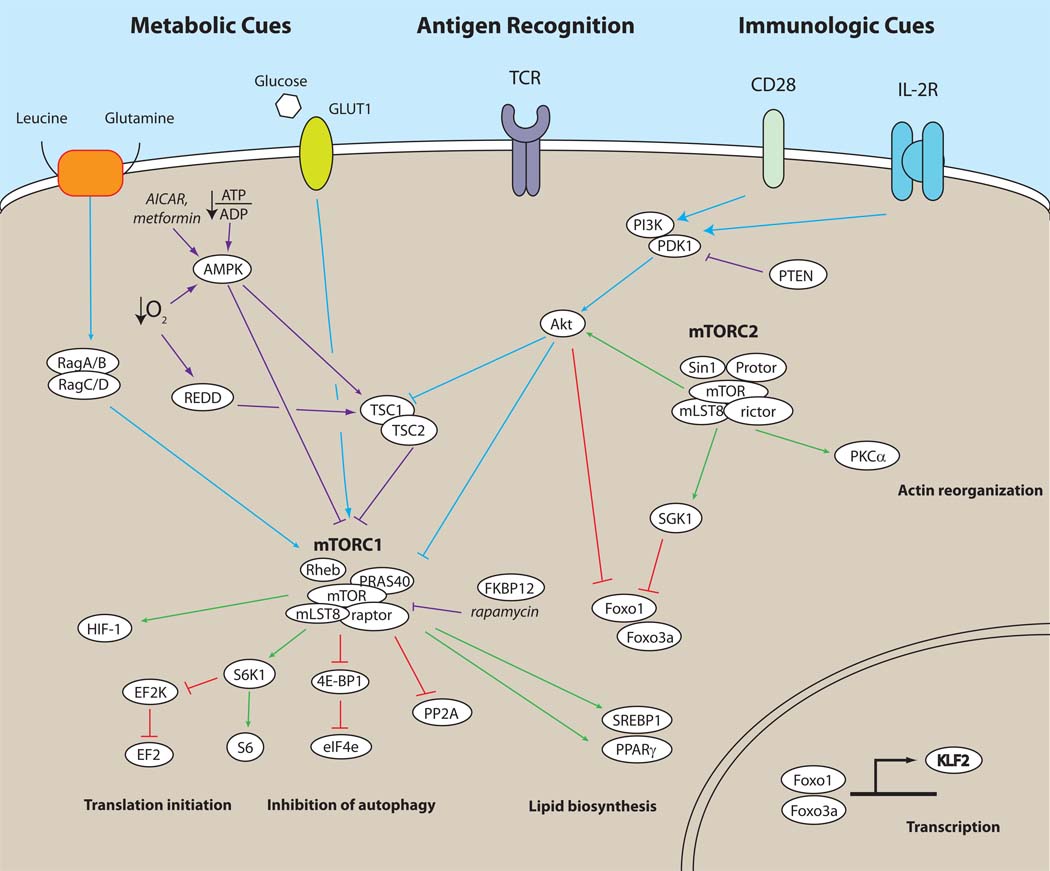
Efforts to identify the mechanism by which rapamycin exerted its effects led to the discovery of the Target of Rapamycin 1(TOR1) and TOR2 in yeast and subsequently the mammalian homolog, mTOR (also known as FKBP-rapamycin-associated protein(FRAP), rapamycin and FKBP12 targets (RAFT)) (Schmelzle and Hall, 2000). Genetic and biochemical analysis of mTOR revealed that it was a ~289 kDa protein that had substantial sequence homology with the phosphoinositide 3-kinase (PI3-kinase) family. However, mTOR is not a lipid kinase but rather a serine-threonine protein kinase. Interestingly, although in yeast there are two separate TOR genes (TOR1 and TOR2), in mammalian cells, mTOR is encoded as a single gene whose protein product signals via two distinct complexes (mTORC1 and mTORC2). mTOR complex 1 (mTORC1) contains the scaffolding protein regulatory-associated protein of mTOR (raptor) as well as mammalian lethal with Sec13 protein 8 (mLST8), the proline-rich Akt substrate 40 kDa (PRAS40) and DEP-domain-containing mTOR-interacting protein (Deptor) (see (Laplante and Sabatini, 2009b)for an up-to-date comprehensive review of mTOR signaling). The second mTOR signaling complex (mTORC2) consists of mLST8, the scaffolding protein raptor-independent companion of TOR (rictor), mSIN1 proteins, and the protein observed with rictor (Protor) (Laplante and Sabatini, 2009b).
Evolutionarily conserved from yeast to man, mTOR integrates environmental cues as a means of regulating cellular size, growth, proliferation, survival and metabolism (Guertin and Sabatini, 2007). As such, it is not surprising that mTOR activation can be regulated by diverse stimuli such as amino acid availability, oxygen, energy status and growth factors. The upstream signaling events that lead to the activation of mTOR are complex and incompletely understood (Figure 1). Although various pathways leading to mTORC1 activation have been described, the precise upstream signals leading to mTORC2 activation remain basically undefined (Laplante and Sabatini, 2009b). In a general scheme, growth factor or cytokine signaling leads to the activation of PI-3kinase which in turn leads to the activation of phosphoinositide-dependent kinase 1 (PDK1) which then promotes the phosphorylation of Akt at threonine 308. The proximal, activation motif phosphorylation of Akt leads to the phosphorylation and inactivation of Tuberous Sclerosis Complex (TSC). TSC (which is comprised of TSC1 and TSC2) functions as a GTPase-activating protein (GAP) for the ras homologue enriched in brain (Rheb). Rheb is a small GTPase that has been shown to be a crucial regulator of mTORC1 signaling (Saucedo et al., 2003; Yamagata et al., 1994; Yee and Worley, 1997). When TSC is inhibited, the active, GTP-bound form of Rheb interacts with mTORC1 to stimulate its activity(Manning and Cantley, 2003). From the immunologist’s perspective notably, the PI-3K-Akt-mTORC1 axis is activated by CD28 engagement and IL-2 receptor signaling (Colombetti et al., 2006). In addition, IL-7 promotes mTOR activity, helping to prevent atrophy in T cells (Rathmell et al., 2001). IL-4-induced mTOR activity has been shown to promote proliferation and prevent apoptosis through the PI3K-Akt axis (Cardoso et al., 2009; Stephenson et al., 2005). Furthermore, IL-12 and IFN-γ have also been shown to promote the sustained activation of mTORC1 (Lekmine et al., 2004; Rao et al. 2010). Finally, evidence is accumulating demonstrating the ability of Wnt signaling to regulate mTORC1 activity in a glycogen synthase kinase 3 (GSK3)-dependent fashion (Inoki et al., 2006).
Figure 1. mTOR signaling for the immunologist.
Based on the review of Laplante and Sabatini, this figure is designed to be a ready reference for upstream and downstream signaling as well as the components of mTORC1 and mTORC2. Details of mTORC1 and mTORC2 signaling pathways are described in the main text. Although the figure is not meant to be exhaustive, we try to emphasize the connections between CD28 and IL-2R signaling with signals derived from leucine, oxygen and energy. Note, blue lines indicate signals upstream of mTOR that ultimately lead to its activation. Purple lines depict upstream signals that lead to the inhibition of mTOR. Green lines show downstream mTOR signaling that promotes a particular function while red lines depict downstream inhibitory signals.
mTORC1 activity is profoundly regulated by amino acids, especially leucine (Proud, 2007). When leucine is available from the environment, it is transported in a glutamine-dependent fashion into the cell (Laplante and Sabatini, 2009b). Recently it has been shown that in the presence of leucine (and other amino acids), Rag proteins (in this case a family of small GTPases, not recombinases) bind to raptor and promote the interaction between mTORC1 and Rheb (Sancak et al., 2008). During amino acid deprivation, even potent upstream activators of Rheb fail to activate mTOR. Mimicking leucine deficiency by using the leucine antagonist N-acetyl-leucine-amide (NALA), our group and others have been able to inhibit T cell function (Hidayat et al., 2003; Zheng et al., 2009).
Energy status as determined by the ratio of intracellular ATP/ADP also regulates mTORC1 activity. A paucity of ATP leads to the activation of AMP-activated protein kinase (AMP-kinase) that in turn inhibits mTORC1 activity by phosphorylating TSC2 and enhancing its GAP activity. As such, the AMP-kinase activators AICAR and the Type 2 diabetes drug metformin have both been shown to affect T cell function (Nath et al., 2005; Pearce et al., 2009).
Availability of oxygen can also regulate mTOR function. Under minimally hypoxic conditions, there is a decrease in ATP leading to AMP-kinase activity (Laplante and Sabatini, 2009b). Alternatively, hypoxia can inhibit mTORC1 activity by enhancing the expression of DNA damage response 1 (REDD1). REDD1 inhibits mTORC1 by causing TSC2 to disassociate from 14-3-3 proteins and enhancing its ability to block Rheb.
The diverse upstream activators of mTOR lead to equally diverse downstream consequences. For example, in a T cell, activating mTOR with insulin will not have the same functional consequences as activating mTOR via CD28 (Sabatini, 2006; Zheng et al., 2007). In this regard the list of downstream substrates and pathways for mTOR is ever increasing. Typically mTORC1 and mTORC2 activation is measured by phosphorylation of canonical substrates. mTORC1 activity can be monitored by the phosphorylation of p70 ribosomal S6 kinase 1(S6K1) or its downstream substrate S6 as well as by the phosphorylation of the translational inhibitor eukaryotic initiation factor 4E binding protein 1(4EBP-1)(Beugnet et al., 2003). A major role for mTORC1 is to regulate protein translation. In addition, mTORC1 activation also serves to inhibit autophagy, promote lipid metabolism and mitochondrial biogenesis (Laplante and Sabatini, 2009a). The net sum of these activities is to promote growth and differentiation.
The archetypical indicator of mTORC2 activity is the phosphorylation of Akt at its hydrophobic motif, serine 473 (Bhaskar and Hay, 2007). It is clear from this observation that Akt phosphorylation is both upstream and downstream of mTOR. Activation of mTORC1 by Akt (as indicated by phosphorylation of threonine 308) is not dependent upon mTORC2-mediated activation of Akt (as indicated by phosphorylation of serine 473). In general, mTORC2-dependent Akt activity positively regulates processes that promote proliferation and survival. In addition, Akt promotes the phosphorylation of the transcription factors Forkhead box O1(Foxo1) and Foxo3a leading to their exclusion from the nucleus and hence their inactivation (Laplante and Sabatini, 2009b). Another mTORC2 substrate, the serum and glucocorticoid-induced protein kinase 1(SGK1), is also involved in the regulation of the Foxo family of transcription factors (Huang et al., 2009). Finally, mTORC2 activation has been implicated in promoting cytoskeletal reorganization. This is believed to be mediated by the ability of mTORC2 to promote the phosphorylation and hence activation of protein kinase C alpha (PKC-α) (Ikenoue et al., 2008).
T cell responses are metabolically demanding
The metabolic demands of T cells are extraordinary, rivaling that of cancer cells (Fox et al., 2005). As a consequence, T cell metabolism is highly regulated. However, it is not that T cell activation leads to an upregulation in metabolism; rather, an increase in the metabolic machinery is an integral component of T cell activation (Jones and Thompson, 2007). Quiescence in T cells is an actively maintained state (Berger et al., 2010; Buckley et al., 2001; Modiano et al., 2008). A number of molecules including Schlafen-2 (Slfn2), transducer of ERB2 1(TOB1), Krüppel-like factor 2 or lung Krüppel-like factor (KLF2 or LKLF) and Foxos have all been implicated in actively suppressing T cell function by promoting the expression of inhibitors of activation. Both KLF2 (LKLF) and Foxo have also been implicated in regulating metabolism. The resting state is characterized metabolically by catabolism. Quiescent T cells employ autophagy to derive molecules necessary for energy and baseline protein synthesis.
Upon activation there is a switch from catabolism to anabolism (Fox et al., 2005; Jones and Thompson, 2007). In spite of adequate amounts of oxygen in the environment, T cells employ glycolysis to generate energy. This state of oxidative glycolysis is known as the Warburg effect and is also employed by cancer cells. At first, it seems counterintuitive that cells that have markedly increased their demand for energy would employ a relatively inefficient means to generate ATP. To answer this question, Thompson has proposed that whereas glycolysis is less efficient in generating ATP, the glycolytic pathway provides substrates for the generation of amino acids, nucleotides and lipids (Vander Heiden et al., 2009). T cells can afford to inefficiently produce ATP because essentially ATP is not limiting. However, T cell activation demands adequate amounts of the essential components for protein, lipid, and DNA biosynthesis. mTOR plays a critical role in regulating these processes. It is within this context that an essential component of T cell activation is the upregulation of the metabolic machinery supporting these events. Whereas a critical function of CD28-mediated costimulation is to promote the generation of IL-2, an equally important aspect of costimulation is to promote the metabolic machinery necessary to support T cell activation (Frauwirth et al., 2002; Frauwirth and Thompson, 2004). Along these lines, Th1 cells rendered anergic (with TCR stimulation alone) fail to express IL-2 upon rechallenge with TCR and CD28 stimulation (Zheng et al., 2009). Likewise, anergic T cells cannot fully activate mTOR when rechallenged, and failing to fully upregulate the metabolic machinery required for glucose transport, protein and lipid synthesis. Based on these observations, one might speculate that the inability of anergic T cells to become metabolically active plays a role in maintaining the state of T cell anergy.
In light of the robust metabolic demands of T cell activation, it is not surprising that regulation of T cell activation can be achieved by limiting nutrient availability. The work of Mellor and Munn and subsequently others has shown the ability of tryptophan availability to inhibit immune responses (Mellor and Munn, 2004). The precise role if any of mTOR in tryptophan-mediated regulation of T cell function remains unknown. Recently, it has been shown that not only is glutamine required for T cell function, but glutamine uptake is increased with CD28 signaling (Carr et al., 2010). In addition, the ability of leucine to regulate T cell function is emerging. The leucine analogue NALA can inhibit T cell function, and TCR engagement in the presence of NALA promotes T cell anergy (Hidayat et al., 2003; Zheng et al., 2009). In as much as a lack of leucine inhibits mTORC1 activation, these latter findings are consistent with the observations that TCR engagement in the absence of mTOR activity promotes anergy (Powell et al., 1999). Similarly, glucose analogues which block metabolism have also been shown to block T cell function (Cham et al., 2008). As is the case with NALA, the glucose analogue 2-deoxyglucose (2DG) can inhibit mTOR function (most likely via the AMP-kinase pathway) and thus promote anergy. Finally, the AMP-kinase activator AICAR, by mimicking energy depletion, can acutely inhibit T cell function, mitigate Experimental Autoimmune Encephalomyelitis and promote anergy by inhibiting mTOR (Jhun et al., 2005) (Nath et al., 2005)(Zheng et al., 2009). More recently, it has been shown that another AMP-kinase activator, metformin, can promote CD8+ memory T cell development (discussed in more detail below) (Pearce et al., 2009).
The precise role that regulation of nutrient availability plays in controlling mTOR under natural circumstances remains to be determined. Modulation of T cell function by regulating tryptophan and cysteine concentrations has already been described (Angelini et al., 2002; Mellor and Munn, 2004). Recently, it has been shown that regulatory T cells can inhibit T cell function by expressing enzymes that deplete the environment of essential amino acids (Cobbold et al., 2009). The depletion of amino acids in turn leads to the inhibition of mTOR in target T cells that in turn leads to the upregulation of the transcription factor Foxp3. Likewise, one might predict that mechanisms regulating mTOR by controlling glutamine, leucine, glucose and energy availability will also be revealed.
Signal 1 in the absence of mTOR activation leads to anergy in Th1 cells
When CD4+ Th1 T cells recognize antigen through their TCR (Signal 1) in the absence of costimulation (Signal 2) they are rendered anergic (Schwartz, 2003). That is, such cells will fail to vigorously proliferate and produce IL-2 upon subsequent rechallenge with full (Signal 1 + 2) stimulation. [In this section we refer to Signal 2 as described by Lafferty and Cunningham(Lafferty and Cunningham, 1975). That is, a “second” APC derived signal that is required for the full activation of a T cell. Later on in this review we propose that the scope of Signal 2 be broadened with mTOR as a central integrator of its function]. Our laboratory and others demonstrated that anergy could be induced even in the presence of costimulation (for example, CD28 engagement) if the mTOR inhibitor rapamycin was present during TCR engagement (Powell et al., 1999; Vanasek et al., 2001). Originally, it was thought that rapamycin promoted anergy by inhibiting cell cycle progression, thus preventing the dilution of regulatory “anergic factors” (Jenkins, 1992). However, it became clear that it was not cell cycle progression that prevented anergy but rather the activation of mTOR itself (Allen et al., 2004; Colombetti et al., 2002; Zheng et al., 2007). For example, both rapamycin and sanglifehrin A inhibit T cell proliferation in G1. However, in spite of its ability to block proliferation, sanglifehrin A does not promote T cell anergy. For CD4+ Th1 cells, TCR engagement in the absence of mTOR activation leads to anergy. Such findings suggested that mTOR was a biochemical tranducer of Signal 2. In other words, just as mTOR plays a role in interrogating the environment for nutrient status, mTOR also senses the immune microenvironment for “danger signals” in T cells. Such signals, transmitted through the presence of costimulatory molecules, tell the T cell that antigen recognition should promote activation. In the absence of such signals (hence, a lack of mTOR activation), recognition leads to tolerance. As mentioned above, subsequent studies have revealed that, in addition to rapamycin, blocking metabolic pathways necessary for mTOR activation can also promote the induction of anergy (Zheng et al., 2009). Teleologically, the necessity of mTOR activation to promote Th1 effector responses makes sense; the metabolic demands of such a response require mTOR activation.
mTOR senses the immune microenvironment to direct CD4+ T cell differentiation
The ultimate fate of a naïve T cell is dictated by multiple cues from the immune microenvironment (Bettelli et al., 2007). Naïve CD4+ T cells activated in the presence of IFN-γ and IL-12 develop into Th1 effectors, wheras CD4+ T cells stimulated in the presence of IL-4 are fated to become Th2 effector cells. CD4+ T cells stimulated in the presence of IL-6 and TGF-β differentiate into IL-17 producing Th17 cells. Further, when naïve CD4+ T cells are stimulated in the presence of high concentrations of TGF-β they do not become effector cells but rather Foxp3+ regulatory T cells (Chen et al., 2003). In vitro, differentiation into these T cell subsets is easily achieved by culturing cells in high concentrations of cytokines and cytokine-neutralizing antibodies. However, in vivo, differentiation results from simultaneous integration of multiple, even opposing, signals.
Having previously demonstrated that mTOR played a role in promoting effector function in Th1 T cells, our group hypothesized that mTOR might also play a role in promoting the transition from naïve to effector CD4+ T cells (Delgoffe et al., 2009). To test this hypothesis we created mice in which mTOR was deleted in T cells. Indeed, the mTOR-deficient T cells failed to become Th1, Th2 or Th17 effector cells when stimulated under appropriate skewing conditions both in vitro and in vivo. Along these lines, it has recently been shown that rictor deficient T cells display an inability to become Th2 cells and a diminished capacity to become Th1 cells (Lee et al., 2010). By targeting Rheb in T cells, our lab has observed that Th1 and Th17 differentiation but not Th2 differentiation is mTORC1 dependent (unpublished findings). Likewise, we find that Rictor deficient T cells fail to develop into Th2 cells, however, in contrast to the work of Lee et al., we find no defect in Th1 differentiation in the rictor null T cells(Lee et al., 2010). At this point it is unclear whether mTORC1 and mTORC2 are differentially activated during Th1 and Th2 differentiation. For example, it may be that mTORC1 is equally activated under Th1 and Th2 conditions but under Th1 conditions specific Th1-promoting mTORC1-dependent substrates are expressed. Overall, given the activation of naïve CD4+ T cells is metabolically demanding, it seems logical that effector differentiation cannot occur in the absence of mTOR activation (Figure 2).
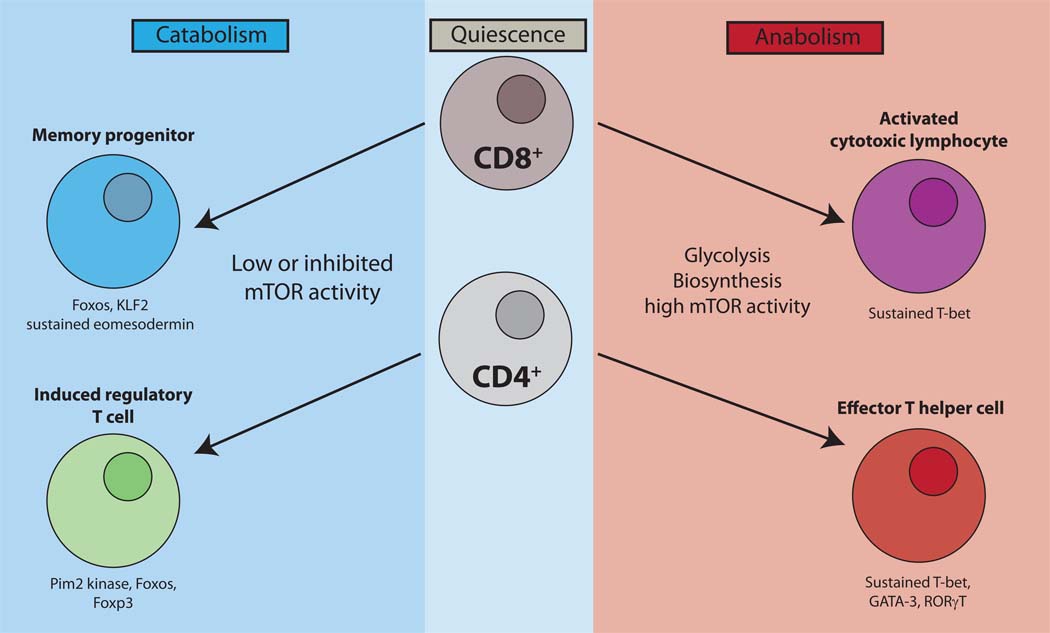
Figure 2. Linking metabolism and mTOR activity.
Quiescence in T cells is an active state that is maintained by Slfn2, Foxos, TOB and KLF2. Naïve T cells are catabolic and have low levels of mTOR activity. Activation and effector generation for both CD4+ and CD8+ T cells results in tremendous metabolic demands and a switch from catabolism to anabolism (right). By necessity mTOR activity is high which not only supports this increase in metabolism but also promotes both CD8+ and CD4+ effector T cell generation. Alternatively, inhibition of mTOR in CD8+ T cells promotes the generation of memory cells. Such cells are characterized by low metabolic demands, low mTOR activity, and an increase in the Foxos, KLF2 and eomesodermin. Likewise, activation in the context of mTOR inhibition promotes the generation of regulatory T cells. These cells, which are less metabolically demanding than their effector counterparts, display decreased mTOR activity but increased Pim2 kinase activity.
Mechanistically, the inability to become effector cells in the mTOR null T cell mice was associated with a failure to upregulate appropriate effector-specific transcription factors (Delgoffe et al., 2009). That is, the mTOR null T cells failed to upregulate sustained expression of T-bet in Th1 cells, GATA-3 in Th2 cells and RORγt in Th17 cells. This decrease in the upregulation of T cell-specific transcription factors was seen in the setting of decreased STAT activation in response to skewing cytokines. The precise role that mTOR plays in terms of regulating STAT activation in T cells still remains to be determined. However, links between mTOR and STAT signaling have been indentified in cancer cells, where for example, it has been shown that mTOR activation can promote STAT3 phosphorylation (Ma et al., 2010) Because STAT activation was not eliminated but rather diminished in the mTOR null T cells, it is likely that this effect contributes to but is not the sole mechanism accounting for the inability of such cells to differentiate into CD4+ effector cells. Along these lines recent findings suggest that mTORC2 activation of PKC-theta contributes to Th2 differentiation (Lee et al., 2010).
In the absence of mTOR naïve CD4+ T cells differentiate into Foxp3+ regulatory T cells (Delgoffe et al., 2009). That is, under normally effector skewing conditions, full T cell activation in the absence of mTOR leads to a default regulatory T cell pathway. Interestingly, whereas the generation of regulatory cells occurs in the absence of exogenous TGF-β, the mTOR null T cells displayed constitutively phosphorylated SMAD3 and neutralization of TGF-β diminishes the generation of Foxp3+ cells. These observations suggest that basal TGF-β signaling, unopposed by mTOR activation, can drive regulatory cell differentiation.
Consistent with the findings employing mTOR-deficient T cells are the observations that rapamycin can promote the generation of regulatory T cells both in vitro and in vivo (Battaglia et al., 2005; Kang et al., 2008; Kopf et al., 2007). Rapamcyin-induced regulatory T cell generation is associated with histone H3K4me2 and 3 methylation near the Foxp3 transcriptional start site (Sauer et al., 2008). Alternatively, activation of the Akt-mTOR axis by the expression of a constitutively active Akt impairs the generation of CD4+ Foxp3+ T cells in the thymus (Haxhinasto et al., 2008). In the periphery, it appears as if PD-L1 on the surface of antigen presenting cells can promote the development, maintenance and function of inducible regulatory T cells in part by inhibiting mTOR activation in T cells (Francisco et al., 2009).
Initially, it was thought that rapamycin specifically inhibited mTORC1 without affecting mTORC2 activity. As such, the ability of rapamycin to promote Treg generation was ascribed to its ability to inhibit mTORC1. However, studies in several different cell lines have demonstrated the ability of rapamycin to inhibit mTORC2 (Sarbassov et al., 2006). Our own group has noted that in primary T cells rapamycin at doses as low as 20nM can inhibit both mTORC1 and mTORC2 activity (unpublished findings). Along these lines, the genetic deletion of mTORC1 activity or mTORC2 activity alone has been shown to be insufficient to promote Treg differentiation (Delgoffe et al., 2009; Lee et al., 2010). Furthermore, more recently, focus has been placed on the role of Foxo1 and Foxo3a on regulating Foxp3 expression (Harada et al., 2010; Merkenschlager and von Boehmer, 2010; Ouyang et al., 2010). Recall that mTORC2-dependent phosphorylation of Foxo1 and Foxo3a leads to inactivation by promoting their sequestration in the cytoplasm (Laplante and Sabatini, 2009b). Both of these transcription factors have been shown to play a role in Foxp3 expression. Thus, in the presence of mTORC2 activation, Foxo1 and Foxo3a mediated Foxp3 expression is diminished.
When examining the link between T cell activation and metabolism, the observation that mTOR inhibition promotes regulatory T cell generation is quite logical (Figure 2). The metabolic demands of regulatory T cells appear to be much less than that of conventional CD4+ T cells. As a consequence, regulatory T cells are less dependent on mTOR activation for their function. Regulatory T cells thus have an advantage in terms of signaling, survival and proliferation in the presence of rapamycin when compared to conventional T cells (Strauss et al., 2009; Zeiser et al., 2008)(Whiteside and Negrin). It is thought that this relative resistance to rapamycin in regulatory T cells is imparted by an upregulation of Pim 2 kinase (Basu et al., 2008). Pim 2 is a serine-threonine kinase that shares many functions with Akt and is not inhibited by rapamycin. Rather, Pim 2 expression is increased when mTOR is inhibited. Further, the increased expression of Pim 2 appears to be mediated by Foxp3.
The ability of mTOR inhibition to inhibit CD4+ T cell effector differentiation and promote the generation of regulatory T cells has important clinical implications for transplantation and the treatment of autoimmune diseases. The ability to generate Foxp3+ regulatory cells from diabetes patients by ex vivo activation in the presence of rapamycin has been established (Battaglia et al., 2006; Putnam et al., 2009). Efforts are under way to generate ex vivo regulatory T cells to modulate graft versus host disease (Golovina et al., 2008). The Fowler lab has published a series of studies demonstrating the ability of rapamycin to promote Tc2 and Th2 cells ex vivo which when infused into mice undergoing bone marrow transplantation abrogate Graft Versus Host Disease (GVHD) (Foley et al., 2005; Jung et al., 2006). It is not precisely clear why their culture conditions favor the generation of Tc2 and Th2 cells in the presence of rapamycin. As previously mentioned, depending on the dose and duration of exposure in vitro, rapamycin can either specifically inhibit mTORC1 alone or inhibit both mTORC1 and mTORC2. As such it may be that these specific culture conditions favor a preferential inhibition of mTORC1 without inhibiting mTORC2.
The results of the use of rapamycin as an immunosuppressive agent in transplantation have not been as spectacular as one might have predicted based on the animal studies. This may be due to the fact that rapamycin does not acutely inhibit inflammation (Saemann et al., 2009). Clinically, often mTOR inhibitors had been employed in conjunction with calcineurin inhibitors (CSA and FK506) which will completely block the tolerance promoting abilities of mTOR inhibitors (Powell and Zheng, 2006). To avoid this pitfall, we employed the mTOR inhibitor sirolimus with CAMPATH and non-myeloablative doses of Total Body Irradiation in a matched sibling hematopoieitic stem cell transplant setting for the treatment of sickle cell disease. Using this regimen (which did not block TCR signaling with a calcineurin inhibitor), we were able to generate persistent mixed chimerism (Hsieh et al., 2009). In this regard, others have also employed mTOR inhibitors in the absence of calcineurin inhibitors in an effort to promote long lasting transplant tolerance (Swanson et al., 2002).
mTOR inhibition promotes the development of CD8+ T cell memory
Upon initial infection there is a tremendous increase in the frequency of antigen-specific effector cells (Araki et al., 2010). This expansion is followed after 2 weeks by a contraction phase and then the maintenance of a pool of antigen-specific memory cells poised to rapidly respond upon rechallenge. Overall this process is characterized by an increase in the cell surface expression of the IL-7 receptor (CD127), the re-expression of CD62L and an increase in the anti-apoptotic molecule Bcl2. The progressive increase in CD62L and CCR7, CD127 and Bcl-2 all seem to be associated with increased properties of self-renewing memory cells. Although the precise molecular details regulating the development of CD8+ T cell memory have yet to be determined, the transcription factors T-bet and Eomesodermin appear to play important roles (Intlekofer et al., 2005). During the initiation of the acute response, sustained T-bet expression, maintained by IL-12, drives the generation of effector cells (Joshi et al., 2007). IL-12 also serves to inhibit eomesodermin which is thought to promote the effector to memory transition (Takemoto et al., 2006). As inflammation subsides, IL-12 expression decreases leading to an increase in eomesodermin thus supporting memory cell development. From a metabolic perspective, the massive expansion of antigen-specific effector cells from the naïve T cell pool requires much energy and is characterized by increased, protein, lipid and nucleotide synthesis (Pearce)(Figure 2). As with CD4+ T cells, mTOR plays a central role in facilitating the anabolic processes that facilitate CD8+ effector generation. Alternatively, as antigen is cleared and there is a switch to the development of long lived memory cells, metabolism switches back to catabolism.
Recently there has been a series of reports describing the ability of rapamycin to enhance the generation of memory T cells. Araki et al. infected mice with Lymphocytic Choriomeningitis Virus (LCMV) and found that treatment with rapamycin led to an increase in antigen-specific CD8+ T memory cells (Araki et al., 2009). Specifically, treatment of mice during the expansion phase (days 0–8) led to an increase in memory cells. This was reported to be due to a decrease in the contraction of the antigen-specific cells. Additionally, treatment of mice during the contraction phase (days 8–30) seemed to accelerate the effector to memory cell transition and led to the development of more robust memory cells. Importantly, the dose of rapamycin used in these studies was suboptimal with regard to mTOR inhibition; higher doses of rapamycin resulted in suppression of CD8+ T cell expansion. Nonetheless, the effect appeared to be T cell-intrinsic and dependent on mTORC1 signaling. Gene targeting raptor in the antigen-specific CD8+ T cells reproduced the results obtained with low dose rapamycin treatment. Further, gene targeting the binding partner of rapamycin FK506 binding protein 12 (FKBP12) in the antigen-specific CD8+ T cells abrogated the ability of rapamycin to promote memory cell development.
In a separate report, Rao et al. also demonstrated the ability of rapamycin to enhance the generation of CD8+ memory cells. In their model, IL-12 led to the sustained activation of mTOR and the upregulation of T-bet (Rao et al., 2010). This promoted the generation of effector T cells. The presence of rapamycin inhibited mTOR activity leading to a decrease in T-bet and a subsequent increase in eomesodermin that in turn promoted the generation of memory T cells. A third study has also demonstrated the ability of mTOR inhibition as a means of enhancing the generation of CD8+ memory T cells (Pearce et al., 2009). In this report it was noted that TNF receptor associated factor 6 (TRAF6)-deficient T cells became effector cells but failed to generate memory T cells. This defect was associated with a failure to switch to catabolism with regard to fatty acid oxidation (FAO). AMP-kinase is an important regulator of FAO. It was noted that the TRAF6-deficient CD8+ T cells had lower amounts of AMP-kinase activity upon growth factor withdrawal. By treating such cells with the AMP-kinase activator metformin, this defect could be overcome and the ability of such cells to become memory cells restored. Not only did the metformin treatment lead to mTOR inhibition in such cells, but, as was the case with the previous two studies, the addition of rapamycin could also enhance the induction of memory cells. Our own unpublished studies examining naïve CD8+ T cells lacking mTORC1 signaling demonstrate that in vitro such cells proliferate more slowly, have decreased expression of effector cell markers but more readily upregulate memory markers. Thus, for both naïve CD4+ and CD8+ T cells mTOR plays a role in regulating effector function. Inhibition of mTOR in naïve CD4+ T cells promotes the generation of regulatory T cells while inhibition of mTOR in naïve CD8+ T cells promotes the generation of memory T cells. While functionally regulatory T cells and memory cells are quite diverse, metabolically, their demands seem similar.
In as much as rapamycin is an immunosuppressive agent, at first glance it seems somewhat paradoxical that the inhibition of mTOR would promote immunity in the form of T cell memory. However, in the context of the metabolic demands of T cell effector function such observations make more biological sense (Figure 2). Furthermore, these observations have potential clinical implications. Overall these findings suggest that the strategic inhibition of mTOR might enhance the development of memory cells in response to vaccines. Testing their findings in non human primates, Araki et al. were able to demonstrate that rapamycin treatment enhanced memory responses to Vaccinia virus (Araki et al., 2009). Likewise, others have shown that rapamycin and metformin can enhance anti-tumor responses in mouse models (Pearce et al., 2009; Rao et al., 2010).
The role of mTOR in regulating T cell trafficking
Trafficking of T cells to secondary lymphoid organs and tissues is intimately related to the functional state of the T cell. For CD8+ T cells, the cell surface markers that are employed to identify naïve, effector and memory T cells such as CD44, CD62L, and CCR7 are involved in controlling T cell migration (Finlay and Cantrell, 2010). Naïve T cells express CD62L and CCR7 which facilitates their trafficking between secondary lymphoid organs. Upon T cell activation, these receptors are down modulated and other receptors are upregulated facilitating their re-direction to inflamed tissues. From a metabolic perspective, catabolic naive cells down regulate CD62L and CCR7 as they become anabolic. This process is regulated by the PI-3-kinase-mTOR axis. Deletion of PTEN in CD8+ T cells which leads to increased PI3-kinase activity results in the loss of expression of CD62L and CCR7 (Sinclair et al., 2008). Alternatively, treatment with rapamycin prevents the downregulation of these molecules upon activation. The transcription factor KLF2 promotes the transcription of both CD62L and CCR7. KLF2 expression is promoted by Foxo1 (Fabre et al., 2008; Kerdiles et al., 2009). Thus, upon mTOR activation, Foxo1 is sequestered in the cytoplasm, leading to decreased KLF2 expression and subsequent decreased transcription of CD62L and CCR7. As an infection is cleared and T cell activation (and metabolic demands) decreases, mTOR activity decreases leading to re-expression of these cell surface markers. As such these newly formed memory cells display cell surface markers which promote their trafficking throughout secondary lymphoid tissues.
The G protein-coupled receptor Sphingosine 1-phosphate receptor 1 (S1P1) is also regulated in this fashion (Carlson et al., 2006). S1P1 promotes egress from lymph nodes through ligation of its natural receptor sphingosine 1-phosphate (Matloubian et al., 2004). S1P1 null T cells fail to leave the thymus while S1P1 transgenic T cells fail to traffic efficiently to lymphoid organs. Recently a role for S1P1 in modulating regulatory T cell development and function has emerged (Liu et al., 2009). Transgenic expression of S1P1 inhibits the development of regulatory T cells as well as impairs the ability of Foxp3+ cells to suppress. Interestingly, this ability to block T reg development and function was associated with the induction of mTOR activity. That is, rapamycin prevented the ability of S1P1 to mitigate T reg function. Thus, inhibition of mTOR promotes the generation of regulatory T cells and regulatory T cells display decreased mTOR activity. The S1P1 is upregulated by mTOR inhibition but signaling by S1P leads to increased mTOR activation and consequently decreased regulatory T cell function. From the perspective of the naïve T cell, antigen activation greatly increases the metabolic demands of the cell. In part, this demand can be facilitated by increased mTOR activation induced by S1P as the cell begins its egress from the lymph node. However, this activation in turn down modulates S1P1 (as well as CD62L and CCR7) allowing for the cells to traffic to the sites of inflammation.
Redefining Signal 2
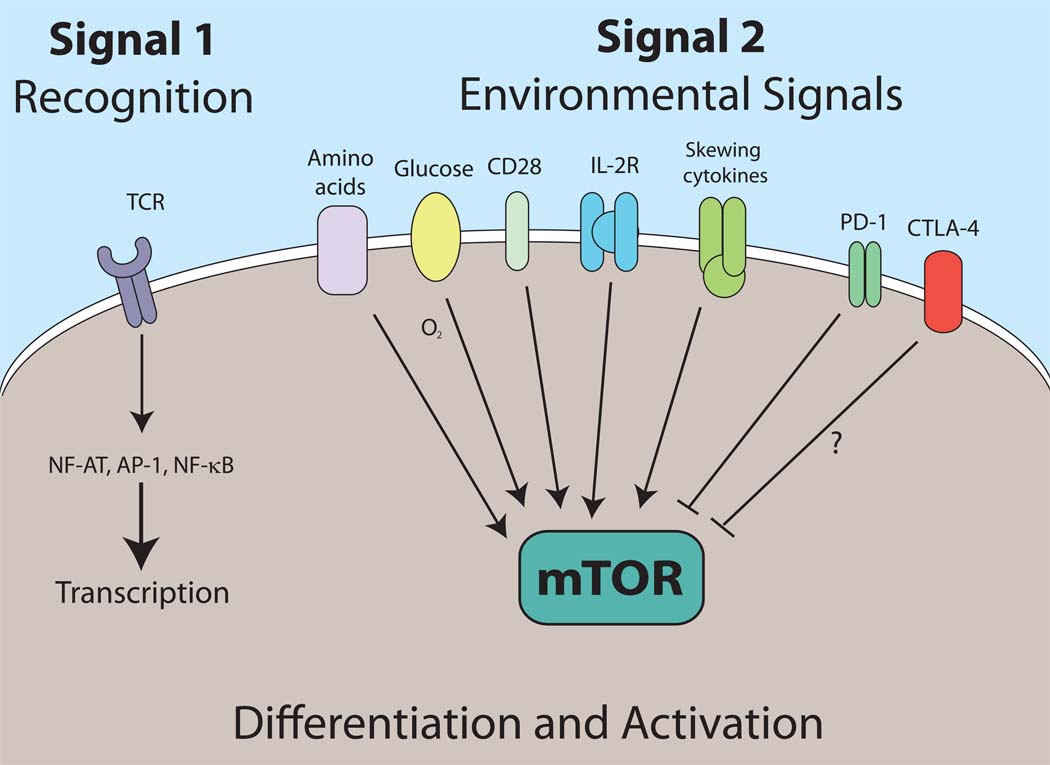
The adaptive immune response is characterized by the remarkable antigen receptor diversity (and thus, specificity). It is also characterized by ability to develop robust initial responses and rapid recall responses. A consequence of these functions is the fact that T cell activation encumbers prodigious metabolic demands. The stochastic generation of antigen receptors necessitates accessory signals derived from the environment to guide lymphocyte activation. The two signal model provides a framework for understanding how TCR recognition leads to appropriate T cell activation or tolerance. Antigen recognition through the TCR (Signal 1) in the presence of “danger” or “infectious non-self” signals (Signal 2) promotes activation while Signal 1 alone leads to tolerance (Fuchs and Matzinger, 1996; Medzhitov and Janeway, 2000). However, the consequences of antigen recognition are not simply activation versus tolerance. Rather, for both CD4+ and CD8+ T cells, TCR engagement in the context of specific environmental cues can lead to the generation of a diverse array of effector, regulatory and memory cells. While the term Signal 3 has been used to refer to signaling derived from inflammatory cytokines, one might also think of Signal 2 as the net sum of both activating and inhibitory signals derived from the inflammatory milieu. We propose to expand the role of Signal 2 to not only dictate activation versus tolerance but also to direct differentiation. In our model Signal 1 heralds antigen recognition whereas Signal 2 refers to the integrated sum of environmental cues. We propose that mTOR serves to integrate costimulatory, cytokine, nutritional and energetic environmental cues to dictate the course of T cell differentiation and function upon antigen recognition (Figure 3). TCR signaling (recognition) leads to activation of multiple signaling pathways (for example, AP-1, NF-AT and NF-kB) which in turn leads to the transcription of a large number of genes (Riley et al., 2002). TCR engagement leads to the expression of numerous molecules associated with activation (for example cytokines and chemokines such as TNF and MIP-1α) but also negative regulatory molecules such as Cbl-b. Likewise, even under specific effector skewing conditions upon initial stimulation, naïve T cells might express multiple T cell specific transcription factors. For example in response to activation in the presence of TGF-β naïve T cells can co-express Foxp3 and ROR-γt (Lee et al., 2009). Signal 2, which we define as the net sum of environmental cues will dictate the ultimate outcome of recognition. mTOR plays a central role in integrating these environmental cues to direct T cell activation and differentiation. For example, during inflammation under Th17 skewing conditions, Foxp3 expression will become extinguished and the cells will differentiate into Th17 effector cells. Alternatively, strong PD-1 engagement might tip the balance in favor of regulatory T cell development by inhibiting mTOR. Likewise, although a precise connection between mTOR and CTLA-4 has not been defined, CTLA-4 has been shown to mediate its effects in part by activating the phosphatase PP2A which has been shown to be inhibited by mTOR (Chuang et al., 2000; Peterson et al., 1999).
Figure 3. Redefining Signal 2.
Signal 1 refers to antigen recognition through the TCR and is characterized by the transcription of many genes. The outcome of this recognition is dictated by the net sum of environmental signals (Signal 2). We propose that mTOR acts to integrate all of these signals which in turn direct the outcome of T cell differentiation and activation.
Concluding remarks
The ability of mTOR activation to enhance or inhibit T cell differentiation and function is intimately linked to the metabolic demands of the T cell. Future studies will seek to determine the precise pathways leading from mTOR to T cell function. In this regard, the ability of mTOR signaling to regulate the Foxo family of transcription factors has provided several incisive links (Merkenschlager and von Boehmer, 2010). Identification of additional downstream substrates and pathways of mTORC1 and mTORC2 should provide important insight in terms of how mTOR regulates specific effector, memory or regulatory T cell differentiation. Further, elucidating additional roles for mTOR signaling in T cells will enhance our understanding and ability to manipulate T cell responses to infection, in autoimmunity and post transplantation.
Acknowledgements
This work was supported in part by NIH grant R01AI077610-01A2. We thank Drs. Drew Pardoll and Ronald Schwartz for their suggestions as well as the members of the Powell Lab.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Allen A, Zheng Y, Gardner L, Safford M, Horton MR, Powell JD. The novel cyclophilin binding compound, sanglifehrin A, disassociates G1 cell cycle arrest from tolerance induction. J Immunol. 2004;172:4797–4803. doi: 10.4049/jimmunol.172.8.4797. [DOI] [PubMed] [Google Scholar]
- Angelini G, Gardella S, Ardy M, Ciriolo MR, Filomeni G, Di Trapani G, Clarke F, Sitia R, Rubartelli A. Antigen-presenting dendritic cells provide the reducing extracellular microenvironment required for T lymphocyte activation. Proc Natl Acad Sci U S A. 2002;99:1491–1496. doi: 10.1073/pnas.022630299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Youngblood B, Ahmed R. The role of mTOR in memory CD8 T-cell differentiation. Immunol Rev. 235:234–243. doi: 10.1111/j.0105-2896.2010.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Golovina T, Mikheeva T, June CH, Riley JL. Cutting edge: Foxp3-mediated induction of pim 2 allows human T regulatory cells to preferentially expand in rapamycin. J Immunol. 2008;180:5794–5798. doi: 10.4049/jimmunol.180.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Stabilini A, Migliavacca B, Horejs-Hoeck J, Kaupper T, Roncarolo MG. Rapamycin promotes expansion of functional CD4+CD25+FOXP3+ regulatory T cells of both healthy subjects and type 1 diabetic patients. J Immunol. 2006;177:8338–8347. doi: 10.4049/jimmunol.177.12.8338. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- Berger M, Krebs P, Crozat K, Li X, Croker BA, Siggs OM, Popkin D, Du X, Lawson BR, Theofilopoulos AN, et al. An Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence. Nat Immunol. 11:335–343. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Korn T, Kuchroo VK. Th17: the third member of the effector T cell trilogy. Curr Opin Immunol. 2007 doi: 10.1016/j.coi.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003;372:555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Braun V, Pilsl H, Gross P. Colicins: structures, modes of action, transfer through membranes, and evolution. Arch Microbiol. 1994;161:199–206. doi: 10.1007/BF00248693. [DOI] [PubMed] [Google Scholar]
- Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc--dependent pathway. Nat Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- Cardoso CR, Provinciatto PR, Godoi DF, Ferreira BR, Teixeira G, Rossi MA, Cunha FQ, Silva JS. IL-4 regulates susceptibility to intestinal inflammation in murine food allergy. Am J Physiol Gastrointest Liver Physiol. 2009;296:G593–G600. doi: 10.1152/ajpgi.90431.2008. [DOI] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cham CM, Driessens G, O'Keefe JP, Gajewski TF. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8(+) T cells. Eur J Immunol. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang E, Fisher TS, Morgan RW, Robbins MD, Duerr JM, Vander Heiden MG, Gardner JP, Hambor JE, Neveu MJ, Thompson CB. The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity. 2000;13:313–322. doi: 10.1016/s1074-7613(00)00031-5. [DOI] [PubMed] [Google Scholar]
- Cobbold SP, Adams E, Farquhar CA, Nolan KF, Howie D, Lui KO, Fairchild PJ, Mellor AL, Ron D, Waldmann H. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci U S A. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombetti S, Basso V, Mueller DL, Mondino A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J Immunol. 2006;176:2730–2738. doi: 10.4049/jimmunol.176.5.2730. [DOI] [PubMed] [Google Scholar]
- Colombetti S, Benigni F, Basso V, Mondino A. Clonal anergy is maintained independently of T cell proliferation. J Immunol. 2002;169:6178–6186. doi: 10.4049/jimmunol.169.11.6178. [DOI] [PubMed] [Google Scholar]
- Curtsinger JM, Mescher MF. Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol. 22:333–340. doi: 10.1016/j.coi.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Powell JD. mTOR: taking cues from the immune microenvironment. Immunology. 2009;127:459–465. doi: 10.1111/j.1365-2567.2009.03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- Fabre S, Carrette F, Chen J, Lang V, Semichon M, Denoyelle C, Lazar V, Cagnard N, Dubart-Kupperschmitt A, Mangeney M, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- Finlay D, Cantrell D. Phosphoinositide 3-kinase and the mammalian target of rapamycin pathways control T cell migration. Ann N Y Acad Sci. 1183:149–157. doi: 10.1111/j.1749-6632.2009.05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JE, Jung U, Miera A, Borenstein T, Mariotti J, Eckhaus M, Bierer BE, Fowler DH. Ex vivo rapamycin generates donor Th2 cells that potently inhibit graft-versus-host disease and graft-versus-tumor effects via an IL-4-dependent mechanism. J Immunol. 2005;175:5732–5743. doi: 10.4049/jimmunol.175.9.5732. [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Thompson CB. Regulation of T lymphocyte metabolism. J Immunol. 2004;172:4661–4665. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- Fuchs EJ, Matzinger P. Is cancer dangerous to the immune system? Semin Immunol. 1996;8:271–280. doi: 10.1006/smim.1996.0035. [DOI] [PubMed] [Google Scholar]
- Golovina TN, Mikheeva T, Suhoski MM, Aqui NA, Tai VC, Shan X, Liu R, Balcarcel RR, Fisher N, Levine BL, et al. CD28 costimulation is essential for human T regulatory expansion and function. J Immunol. 2008;181:2855–2868. doi: 10.4049/jimmunol.181.4.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Harada Y, Elly C, Ying G, Paik JH, DePinho RA, Liu YC. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. 2010;207:1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidayat S, Yoshino K, Tokunaga C, Hara K, Matsuo M, Yonezawa K. Inhibition of amino acid-mTOR signaling by a leucine derivative induces G1 arrest in Jurkat cells. Biochem Biophys Res Commun. 2003;301:417–423. doi: 10.1016/s0006-291x(02)03052-8. [DOI] [PubMed] [Google Scholar]
- Hsieh MM, Kang EM, Fitzhugh CD, Link MB, Bolan CD, Kurlander R, Childs RW, Rodgers GP, Powell JD, Tisdale JF. Allogeneic hematopoietic stem-cell transplantation for sickle cell disease. N Engl J Med. 2009;361:2309–2317. doi: 10.1056/NEJMoa0904971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Wu CL, Manning BD. Signaling events downstream of mammalian target of rapamycin complex 2 are attenuated in cells and tumors deficient for the tuberous sclerosis complex tumor suppressors. Cancer Res. 2009;69:6107–6114. doi: 10.1158/0008-5472.CAN-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. Embo J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Jenkins MK. The role of cell division in the induction of clonal anergy. Immunol Today. 1992;13:69–73. doi: 10.1016/0167-5699(92)90137-V. [DOI] [PubMed] [Google Scholar]
- Jhun BS, Oh YT, Lee JY, Kong Y, Yoon KS, Kim SS, Baik HH, Ha J, Kang I. AICAR suppresses IL-2 expression through inhibition of GSK-3 phosphorylation and NF-AT activation in Jurkat T cells. Biochem Biophys Res Commun. 2005;332:339–346. doi: 10.1016/j.bbrc.2005.04.126. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung U, Foley JE, Erdmann AA, Toda Y, Borenstein T, Mariotti J, Fowler DH. Ex vivo rapamycin generates Th1/Tc1 or Th2/Tc2 Effector T cells with enhanced in vivo function and differential sensitivity to post-transplant rapamycin therapy. Biol Blood Marrow Transplant. 2006;12:905–918. doi: 10.1016/j.bbmt.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Kang J, Huddleston SJ, Fraser JM, Khoruts A. De novo induction of antigen-specific CD4+CD25+Foxp3+ regulatory T cells in vivo following systemic antigen administration accompanied by blockade of mTOR. J Leukoc Biol. 2008;83:1230–1239. doi: 10.1189/jlb.1207851. [DOI] [PubMed] [Google Scholar]
- Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf H, de la Rosa GM, Howard OM, Chen X. Rapamycin inhibits differentiation of Th17 cells and promotes generation of FoxP3+ T regulatory cells. Int Immunopharmacol. 2007;7:1819–1824. doi: 10.1016/j.intimp.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty KJ, Cunningham AJ. A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci. 1975;53:27–42. doi: 10.1038/icb.1975.3. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009a;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009b;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian Target of Rapamycin Protein Complex 2 Regulates Differentiation of Th1 and Th2 Cell Subsets via Distinct Signaling Pathways. Immunity. 32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21:274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Lekmine F, Sassano A, Uddin S, Smith J, Majchrzak B, Brachmann SM, Hay N, Fish EN, Platanias LC. Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp Cell Res. 2004;295:173–182. doi: 10.1016/j.yexcr.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, Chi H. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10:769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q, Zha X, Wang F, Wang Y, Jing Y, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120:103–114. doi: 10.1172/JCI37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr How does the immune system distinguish self from nonself? Semin Immunol. 2000;12:185–188. doi: 10.1006/smim.2000.0230. discussion 257–344. [DOI] [PubMed] [Google Scholar]
- Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med. 2010;207:1347–1350. doi: 10.1084/jem.20101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Jameson JM. T cell dependence on mTOR signaling. Cell Cycle. 2009;8:545–548. doi: 10.4161/cc.8.4.7625. [DOI] [PubMed] [Google Scholar]
- Modiano JF, Johnson LD, Bellgrau D. Negative regulators in homeostasis of naive peripheral T cells. Immunol Res. 2008;41:137–153. doi: 10.1007/s12026-008-8017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N, Giri S, Prasad R, Salem ML, Singh AK, Singh I. 5-aminoimidazole-4-carboxamide ribonucleoside: a novel immunomodulator with therapeutic efficacy in experimental autoimmune encephalomyelitis. J Immunol. 2005;175:566–574. doi: 10.4049/jimmunol.175.1.566. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol. 22:314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci U S A. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J Immunol. 1999;162:2775–2784. [PubMed] [Google Scholar]
- Powell JD, Zheng Y. Dissecting the mechanism of T-cell anergy with immunophilin ligands. Curr Opin Investig Drugs. 2006;7:1002–1007. [PubMed] [Google Scholar]
- Proud CG. Amino acids and mTOR signalling in anabolic function. Biochem Soc Trans. 2007;35:1187–1190. doi: 10.1042/BST0351187. [DOI] [PubMed] [Google Scholar]
- Putnam AL, Brusko TM, Lee MR, Liu W, Szot GL, Ghosh T, Atkinson MA, Bluestone JA. Expansion of human regulatory T-cells from patients with type 1 diabetes. Diabetes. 2009;58:652–662. doi: 10.2337/db08-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167:6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- Riley JL, Mao M, Kobayashi S, Biery M, Burchard J, Cavet G, Gregson BP, June CH, Linsley PS. Modulation of TCR-induced transcriptional profiles by ligation of CD28, ICOS, and CTLA-4 receptors. Proc Natl Acad Sci U S A. 2002;99:11790–11795. doi: 10.1073/pnas.162359999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Saemann MD, Haidinger M, Hecking M, Horl WH, Weichhart T. The multifunctional role of mTOR in innate immunity: implications for transplant immunity. Am J Transplant. 2009;9:2655–2661. doi: 10.1111/j.1600-6143.2009.02832.x. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O'Connor E, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- Sharpe AH. Mechanisms of costimulation. Immunol Rev. 2009;229:5–11. doi: 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair LV, Finlay D, Feijoo C, Cornish GH, Gray A, Ager A, Okkenhaug K, Hagenbeek TJ, Spits H, Cantrell DA. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson LM, Park DS, Mora AL, Goenka S, Boothby M. Sequence motifs in IL-4R alpha mediating cell-cycle progression of primary lymphocytes. J Immunol. 2005;175:5178–5185. doi: 10.4049/jimmunol.175.8.5178. [DOI] [PubMed] [Google Scholar]
- Strauss L, Czystowska M, Szajnik M, Mandapathil M, Whiteside TL. Differential responses of human regulatory T cells (Treg) and effector T cells to rapamycin. PLoS One. 2009;4:e5994. doi: 10.1371/journal.pone.0005994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson SJ, Hale DA, Mannon RB, Kleiner DE, Cendales LC, Chamberlain CE, Polly SM, Harlan DM, Kirk AD. Kidney transplantation with rabbit antithymocyte globulin induction and sirolimus monotherapy. Lancet. 2002;360:1662–1664. doi: 10.1016/S0140-6736(02)11606-0. [DOI] [PubMed] [Google Scholar]
- Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- Tanji T, Ip YT. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005;26:193–198. doi: 10.1016/j.it.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Vanasek TL, Khoruts A, Zell T, Mueller DL. Antagonistic roles for CTLA-4 and the mammalian target of rapamycin in the regulation of clonal anergy: enhanced cell cycle progression promotes recall antigen responsiveness. J Immunol. 2001;167:5636–5644. doi: 10.4049/jimmunol.167.10.5636. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Saemann MD. The multiple facets of mTOR in immunity. Trends Immunol. 2009;30:218–226. doi: 10.1016/j.it.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- Yee WM, Worley PF. Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol Cell Biol. 1997;17:921–933. doi: 10.1128/mcb.17.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser R, Leveson-Gower DB, Zambricki EA, Kambham N, Beilhack A, Loh J, Hou JZ, Negrin RS. Differential impact of mammalian target of rapamycin inhibition on CD4+CD25+Foxp3+ regulatory T cells compared with conventional CD4+ T cells. Blood. 2008;111:453–462. doi: 10.1182/blood-2007-06-094482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Collins SL, Lutz MA, Allen AN, Kole TP, Zarek PE, Powell JD. A Role for Mammalian Target of Rapamycin in Regulating T Cell Activation versus Anergy. J Immunol. 2007;178:2163–2170. doi: 10.4049/jimmunol.178.4.2163. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol. 2009;183:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]