In this issue of Molecular Cell, Brickner and colleagues (Light et al., 2010) identify a DNA sequence which mediates transcriptional memory and retention of recently active INO1 at the nuclear pore complex.
In addition to their fundamental role of mediating macromolecular traffic between the nucleus and the cytoplasm, nuclear pore complexes (NPCs) also physically associate with certain actively transcribed genes, and this association affects transcriptional activity (Taddei, 2007). These interactions may be evolutionarily conserved, although their location is not: genes relocate from the nucleoplasm to the nuclear periphery where they associate with NPCs in the budding yeast, Saccharomyces cerevisiae, and nuclear pore subunits associate with active genes within the nucleoplasm in the fruit fly, Drosophila melanogaster (Capelson et al., 2010, Kalverda et al., 2010, Taddei, 2007, Vaquerizas et al., 2010).
Though the association between active genes and NPC components is well established in these two model organisms, questions remain concerning the mechanisms and physiological relevance of this phenomenon. Multiple factors have been implicated in the relocation of active genes to the NPC, including transcriptional activators, mRNA processing and export factors, and distinct NPC subunits (Taddei, 2007). As different loci were analyzed in the studies which identified these diverse factors, these results may indicate that distinct genes utilize different mechanisms for NPC association. Indeed, Brickner and colleagues recently identified two DNA elements, termed Gene Recruitment Sequences (GRS I and GRS II), in the budding yeast INO1 gene which direct its relocation to the NPC when transcriptionally active (Ahmed et al., 2010). However, other genes which associate with the NPC did not contain GRS sequences in their promoters, suggesting that there may be other as-yet unidentified GRS-like sequences which mediate relocation of these other loci to the NPC.
In addition to relocating to the NPC when transcriptionally active, some genes remain in contact with the NPC following transcriptional shutoff. This persistent association mediates a “transcriptional memory” which alters the gene’s re-induction kinetics (Brickner et al., 2007). The association of recently repressed genes with the NPC and their altered transcriptional kinetics require incorporation of the histone variant H2A.Z (Brickner et al., 2007), indicating that chromatin state plays a role in gene-NPC interactions. Another factor in transcriptional memory is gene looping (Lainé et al., 2009, Tan-Wong et al., 2009). Gene loops form immediately upon transcriptional induction as a result of physical interactions between the promoter and the 3′ end of the gene. Two recent studies provide evidence that gene loop maintenance and continued association with the NPC following transcriptional shutoff contribute to transcriptional memory (Lainé et al., 2009, Tan-Wong et al., 2009). Taken together, these results indicate that gene-NPC association can consist of at least two stages: initial relocation of active genes, followed by retention and transcriptional memory after transcriptional shutoff.
In this issue of Molecular Cell, Brickner and colleagues provide mechanistic insight into the retention and transcriptional memory step of gene-NPC association (Light et al., 2010). The authors identify a DNA sequence, dubbed a Memory Recruitment Sequence (MRS), in the promoter of the budding yeast INO1 gene, which mediates INO1 association with the NPC following transcriptional shutoff. This association, which can persist for multiple cell divisions, is required for INO1 transcriptional memory. Consistent with this result, the authors find that the MRS is required for incorporation of the histone variant H2A.Z, which is also necessary for INO1 transcriptional memory. Interestingly, the authors also find that recently repressed INO1 associates with the NPC even when the GRS sequence within the promoter has been mutated to prevent association when transcriptionally active, indicating that retention of recently repressed INO1 is mechanistically independent from association of active INO1 with the NPC. Finally, the authors provide evidence that recently repressed INO1 associates with the NPC via the MRS through different NPC subunits than those associated with active INO1, indicating distinct roles for different NPC subunits in different stages of gene-NPC association. Taken together, these results provide evidence for the molecular basis of the second, “memory” step of locus association with the NPC.
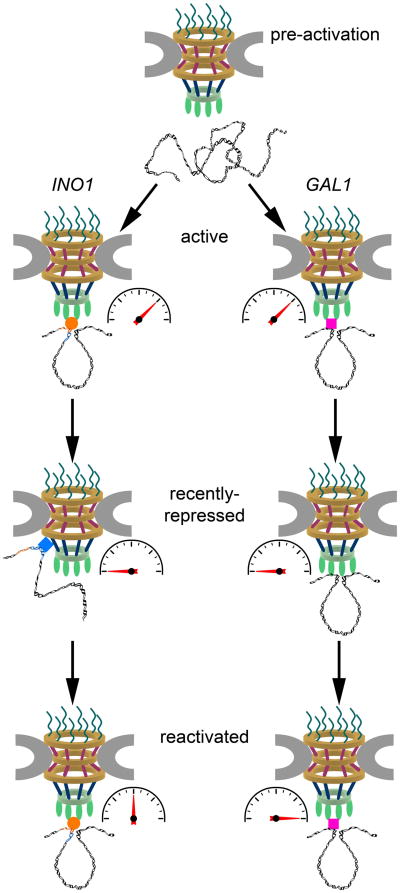
These findings suggest a two-step model for INO1 association with the NPC (Figure 1, INO1), where actively transcribed INO1 associates primarily with components of the NPC nuclear basket such as Nup60 and Mlp2 via GRS I and GRS II. Following transcriptional shutoff, the MRS mediates H2A.Z incorporation into the INO1 promoter and association of recently repressed INO1 with the Nup84 subcomplex and Nup100 in the NPC central core. This association persists for up to three cell divisions and correlates with faster re-activation dynamics for INO1 than occur without NPC-gene association.
FIGURE 1. Models for Transcriptional Memory at the NPC.
Prior to transcriptional activation, the INO1 and GAL1 genes are located in the nuclear interior. Upon transcriptional activation, both genes loop and relocate to the nuclear periphery where they associate with components of the nuclear basket of the NPC, such as Nup60, Mlp1, and Mlp2. GRS I and GRS II mediate these interactions between INO1 and the NPC through an as-yet unidentified factor (orange circle). Transcriptional activators including the SAGA complex mediate interactions between GAL1 and the NPC (pink square). Following transcriptional shutoff, INO1 loses its gene loop and associates with the Nup84 subcomplex and Nup100 within the NPC core. The MRS promotes these interactions and the incorporation of H2A.Z through an unknown MRS-binding factor (blue diamond). In contrast, GAL1 maintains both its gene loop and its association with Mlp1 upon transcriptional repression. Gene induction rates in each case are indicated by gauges. Re-induction kinetics for recently repressed INO1 are slower than initial INO1 activation, whereas re-induction of recently repressed GAL1 is more rapid than initial GAL1 activation.
While this study provides evidence to support distinct steps in gene-NPC association, it also raises many questions concerning the mechanics and the universality of this model. First, what are the DNA binding protein(s) or complexes which recognize the GRS and MRS sequences and mediate the activities attributed to these sequences? The identification of these proteins or complexes may reveal the molecular basis for association of active and recently repressed INO1 with distinct NPC subunits as well as the mechanism of H2A.Z incorporation into the recently repressed INO1 promoter.
Second, what regulates the switch between the association of active INO1 with the NPC subunits of the nuclear basket versus association of recently repressed INO1 with the central Nup84 subcomplex and Nup100? Under normal cellular conditions, this switching is presumably a hand-off between those NPC subunits which interact with active INO1 and those which mediate its recently repressed state. However, as these two steps can occur independently, they must also be mechanistically distinct, such that the “switch” between these NPC subunits is solely dependent upon the transcriptional status of the INO1 gene rather than current NPC association. This possibility, coupled with the fact that both active and recently repressed gene association appear to be regulated by distinct DNA elements, raises the question of whether some genes associate with the NPC only when active, while others do so only when recently repressed. Moreover, this question suggests the related question of how general the MRS sequence may be, and thus how many gene-specific MRS sequences may exist.
This study also presents results which are not entirely consistent with the two recent reports that identified a fundamental role for gene loops in mediating transcriptional memory (Lainé et al., 2009, Tan-Wong et al., 2009). These reports found that gene loop maintenance after transcriptional shutoff is required for transcriptional memory of the budding yeast GAL1 and HXK1 genes, and that the nuclear pore basket protein Mlp1 is required for both gene looping and transcriptional memory (Figure 1, GAL1). In contrast, INO1 does not retain a gene loop following transcriptional shutoff, and its transcriptional memory depends upon central core NPC components rather than nuclear basket components. In fact, the transcriptional memory of INO1 is fundamentally different from that of other loci examined thus far. GAL1 and other genes which demonstrate transcriptional memory have faster reactivation kinetics than initial activation (Brickner et al., 2007, Lainé et al., 2009, Tan-Wong et al., 2009). The reactivation of INO1, however, is slower than initial activation, but is slower still if INO1 is not retained at the NPC or if H2A.Z is not incorporated into the promoter (Brickner et al., 2007, Light et al., 2010). This difference between INO1 and, for example, GAL1 transcriptional memory may be linked to differences in gene looping and NPC association.
In the broader context, how general are these regulatory mechanisms across the genome? Can these models be applied to other genes, or do only a subset of genes associate with the NPC to modulate transcriptional activity and transcriptional memory? While many questions remain regarding this phenomenon, these studies suggest that transcription and transcriptional memory at the NPC may represent an additional level of gene regulation with the same degree of complexity as that imparted by classical transcription factors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. Nat Cell Biol. 2010;12:111. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. PLoS Biology. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Cell. 2010;140:372. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Cell. 2010;140:360. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. Genes & Development. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. Molecular Cell. 2010;39 doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A. Current Opinion in Cell Biology. 2007;19:305. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Genes & Development. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]