Abstract
Nascent transcripts of the phage HK022 put sites modify the transcription elongation complex so that it terminates less efficiently at intrinsic transcription terminators and accelerates through pause sites. We show here that the modification also suppresses termination in vivo at two factor-dependent terminators, one that depends on the bacterial Rho protein and a second that depends on the HK022-encoded Nun protein. Suppression was efficient when the termination factors were present at physiological levels, but an increase in the intracellular concentration of Nun increased termination both in the presence and absence of put. put-mediated antitermination thus shows no apparent terminator specificity, suggesting that put inhibits a step that is common to termination at the different types of terminator.
After initiating RNA synthesis, RNA polymerase (RNAP) continues to elongate the transcript until it reaches a termination site. At such sites, the enzyme has a high probability of dissociating from the transcript and the template (reviewed in references 26 and 32). Bacteria have two basic types of transcription termination signals, which differ in their requirements for halting elongation. Intrinsic terminators can stop transcription through the action of the nascent transcript. Formation of an RNA stem-loop immediately upstream of a U-rich stretch in nascent RNA disrupts RNA-DNA base pairs within the transcription elongation complex, and this destabilizes the complex (15, 20, 47). By contrast, factor-dependent terminators recruit a termination factor to the nascent transcript. Two termination factors have been well characterized: the bacterial Rho protein and the bacteriophage-encoded Nun protein. After binding to nascent transcripts, they both act on the nearby elongation complex. Rho has an ATP-driven RNA-DNA helicase activity, which is thought to destabilize the elongation complex (7, 30). Nun is transferred from its RNA binding site to the elongation complex, where it is thought to anchor RNAP to the DNA template within a few hundred nucleotides downstream of the binding site (16, 39, 43). Dissociation of Nun-arrested polymerase from the template and the transcript has not been observed in vitro and appears to require an additional factor or factors. Recent evidence suggests that the Escherichia coli Mfd protein can stimulate the dissociation of Nun-arrested complexes (42).
E. coli and its bacteriophages alter the efficiency of transcription termination in order to control the expression of genes located downstream of terminators (reviewed in reference 44). For example, the phage λ antitermination proteins N and Q modify RNAP so that it reads through intrinsic and rho-dependent terminators. Both N and Q recognize specific phage sequences (nut and qut, respectively) before they modify polymerase, and this limits antitermination to RNAP molecules that are transcribing phage DNA. Elongating RNAP that has been modified by interaction with either protein retains the modification as it translocates, as shown by its ability to read through multiple, sequential terminators. The action of both proteins is enhanced by host-encoded factors. Transcription of the rRNA operons in E. coli is also subject to antitermination control and, as in the case of λ N and Q, cis-acting sequences (boxA sites) located near rRNA promoters limit antitermination to polymerase molecules that are transcribing rRNA operons (6, 11). Ribosomal antitermination also requires trans-acting factors (41, 48).
Bacteriophage HK022 is a relative of λ that also antiterminates transcription in order to increase the expression of genes located downstream of termination sites (reviewed in reference 45). However, in contrast to the factor-dependent antitermination mechanisms outlined above, transcription of cis-acting, promoter-proximal phage sequences (put sites) is sufficient to convert RNAP into a termination-resistant form; no dedicated factors are absolutely required. We refer to this as intrinsic antitermination. put differs in sequence from the nut, qut, and ribosomal operon boxA sites. Computer modeling and enzymatic probing of RNAs synthesized in vitro suggest that the put transcripts fold into two stem-loops separated by an unpaired base (4). The stems are required for put function, since mutations that prevent base pairing reduce antitermination, and additional mutations that reestablish base pairing but not the original sequence restore antitermination (18). Nascent put RNA binds to the transcription elongation complex and remains associated with it through subsequent translocation. Stable binding is required for antitermination (35).
The distinction between intrinsic and factor-dependent antitermination is highlighted by the following observations. First, E. coli mutants that are defective in put-mediated antitermination supported N-mediated, Q-mediated, and ribosomal operon antitermination (10). Conversely, host mutants that are defective in λ N-mediated antitermination supported the growth of HK022 (3). The two types of mutations changed different host proteins: those defective in HK022 antitermination altered the β′-subunit of RNAP, and those defective in λ antitermination altered the E. coli Nus proteins or the β-subunit of RNAP (13, 24). Finally, purified wild-type polymerase efficiently read through multiple sequential intrinsic terminators that were fused to a wild-type put site. Efficient readthrough did not require additional protein factors but was prevented by a β′ mutation that is defective for antitermination in vivo (10, 18).
The different factor and site requirements of the antitermination systems cited above could influence the spectrum of terminators that each is capable of suppressing. The N/nut and Q/qut pathways prevent termination at both intrinsic and Rho-dependent terminators, suggesting that they interfere with a step that is common to both types. The ribosomal boxA pathway promotes efficient readthrough of Rho-dependent terminators but is ineffective or less effective against intrinsic terminators (1). The put pathway is known to suppress several intrinsic terminators. Here we show that put also promotes readthrough of three factor-dependent terminators, one that requires Rho (λ TR1) and two that require Nun (λ nutL and λ nutR).
MATERIALS AND METHODS
Bacteria, phages, and plasmids.
The strains, phages, and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacteria, plasmids, and phage strainsa
| Strain | Relevant genotype and/or reference |
|---|---|
| Bacteria | |
| TAP114 | W3110 ΔM15[lacZ] lacIq; T. Patterson and D. Court, unpublished |
| RW3926 | TAP114 rpoC-Y75N btuB::Tn10(kan) |
| MC1000 | 37 |
| MOC23 | MC1000 rpoC-Y75N; reference 10 |
| Plasmids | |
| pJT6 | PTAC-putL-λTR1-lacZ fusion in pRS415 (Ampr); contains HK022 sequence from +2 to +174 relative to start of PL transcription |
| pML042 | pGB2ts (Cmr); M. Lobocka |
| pMOC170 | Expresses HK022 cI repressor (Spcr); reference 9 |
| pNUN | PLac-nun (Cmr); reference 5 |
| pNUNΔ | PLac (Cmr); reference 5 |
| pNL150 | PTACpsu+, Cmr, in pGZ119EH; reference 23 |
| pNL151 | PTAC Δpsu, Cmr, in pGZ119EH; reference 23 |
| pRAK31 | A pRS415 derivative that contains HK022 sequences from −144 to +150 relative to the start of PL transcription |
| pRAK122 | PTAC-putL-lacZ fusion in pRS415; contains HK022 sequence from +2 to +174 relative to start of PL transcription |
| pRAK161 | PTAC-ΔputL-lacZ fusion in pRS415; contains HK022 sequence from +2 to +21 relative to the start of PL transcription |
| pRAK166 | PTAC-ΔputL-λTR1-lacZ fusion in pRS415; pRAK161 with a phage λ DNA segment from bp 38042 to 38360 inserted between the HK022 sequences and lacZ |
| pRAK262 | HK022 cI inserted into pML042 |
| pRAK292 | PTAC-putLLS-λTR1-lacZ fusion in pRS415; contains HK022 sequence from +2 to +174 relative to start of PL transcription; the sequence contains linker scanning mutation G in putL (18) |
| pRAK296 | PL(HK022)-putL-λ nutL-lacZ fusion in pRS415; the λ nutL site (bp 35506 to 35558) was inserted into pRAK31 between putL and lacZ |
| pRAK381 | PL(HK022)-putL-λ nutR-TR1-lacZ fusion in pRS415; the λ nutR site (bp 38241 to 38292) was inserted into pRAK31 between putL and lacZ |
| pRS415 | Reference 38; lacZ transcriptional fusion vector; confers ampicillin resistance |
| pSB513 | λ nutL clone; reference 39 |
| Phage | |
| λ RS88 | 38 |
Many derivatives of TAP114 and RW3926 are not listed.
Bacterial growth, media, biochemicals, and antibiotics.
Cell cultures were grown in Luria-Bertani (LB) or tryptone broth (TB) (25). Antibiotics were added (when required) at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; spectinomycin, 25 μg/ml. Fusions that contained the PTAC or PLac promoters were induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM final concentration; purchased from Gold Biotechnologies). o-Nitrophenyl-β-d-galactopyranoside was purchased from Sigma. Restriction enzymes, Klenow, and ligase were purchased from New England Biolabs. Bacteriophage were grown and assayed as described previously (2). Oligonucleotides were purchased from BioServe (Laurel, Md.).
Cloning of the λTR1 terminator.
A fragment containing the λTR1 terminator was amplified by PCR from λ DNA using oligos RK76 (5′-CATCGGATCCTGGAACAACGCATAACCC-3′) and RK78 (5′-TGCAGGATCCCTATGTAAGTATTTCC-3′). RK78 primes downstream of the λTR1 near the cII translation start site. RK76 primes at the beginning of the cro coding sequence and changes the initiating codon to prevent cro translation. The incorporated BamHI restriction sites used for cloning are underlined. The amplified fragment was digested with BamHI and cloned into the reporter constructs shown in Table 1. All fusions made in this study were sequenced at the University of Maryland Biopolymer Laboratory. All fusions that contain the cro-TR1 region of lambda are signified by a TR1 notation.
Cloning the λ nutL and nutR sites.
The λ nutL site was amplified from pSB513 with primers RK88 (5′-CAGCGAATTCTGAAGGTGACGCTCTTAAAAATT-3′) and SBS59 (5′-CGCCGGAGATCTCTGCAGTGGAGCGGGCAGCGGG-3′). The incorporated EcoRI site in RK88 is underlined. The purified PCR products were digested with EcoRI, and the ends of the resulting 64-bp nutL-containing fragment were filled with Klenow and cloned into the SmaI site of pRAK31 (18).
The λ nutR sequence was amplified from plasmid pJT6 with RK95 (5-TACGGATATCAATAACCCCGCTCTTAC-3′) and RK96 (5′-GCTGGATCCGTTTAATTTGATGCCCTTTTTC-3′). The EcoRV and BamHI sites used for the cloning are underlined. The purified products were digested with EcoRV and BamHI and cloned into SmaI-BamHI-digested pRAK31. All fusions that contain only the nutR-TR1 region are signified by a nutR-TR1 notation.
Crossing lacZ fusions onto λ.
The lacZ fusions in pRS415 were crossed from the plasmids in which they were constructed onto λ RS88 as described previously (18, 38). The copy number of λ prophage was determined as described elsewhere (31).
β-Galactosidase assays.
β-Galactosidase activities of cells carrying PTAC-lacZ fusions were assayed in microtiter plates as described previously (18). Overnight TB broth cultures were diluted into fresh TB containing antibiotics and incubated for approximately 2 h. The exponentially growing cultures were then diluted into TB alone or TB supplemented with 1 mM IPTG. The cultures were incubated for 1 h more and then assayed for β-galactosidase activity. β-Galactosidase activities of cells carrying PL(HK022)-lacZ fusions were determined as described elsewhere (25). Overnight cultures were diluted into LB broth with or without 1 mM IPTG and grown for 3 to 4 h to cell densities of 2 × 108 to 5 × 108/ml before assay.
Plasmid curing.
To measure steady-state levels of β-galactosidase in strains that carried single-copy HKPL-putL-nutL-lacZ (or nutR-lacZ) fusions, the plasmid that expresses HK022 repressor (pRAK262) was removed by growing cultures at 42°C in LB for several generations. The loss of pRAK262 was confirmed by screening for chloramphenicol sensitivity and for immunity to HK022.
RESULTS
put-mediated suppression of a Rho-dependent transcription terminator.
Lambda TR1 is a well-characterized Rho-dependent terminator (14, 21, 34). We measured the efficiency of termination at TR1 by comparing the activity of β-galactosidase produced after induction by cells containing a single copy of a PTAC-TR1-lacZ transcription fusion to that produced by cells containing a PTAC-lacZ transcription fusion. We measured put-mediated antitermination of TR1 in two ways. First, we compared β-galactosidase activity produced by a PTAC-putL-TR1-lacZ fusion to that produced by comparable fusions that lacked a functional putL site. Second, we compared β-galactosidase activity produced by a PTAC-putL-TR1-lacZ fusion in rpoC+ cells to that produced by the same fusion in rpoC-Y75N cells. This mutation, which alters the β′-subunit of RNAP, prevents put-mediated antitermination at intrinsic terminators (10, 18). The two methods gave similar results.
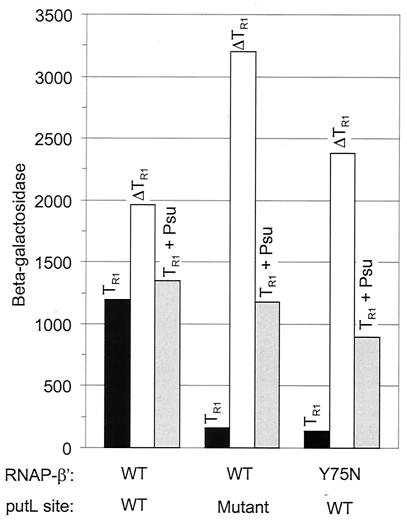
TR1 terminated transcription with an efficiency of 94 to 95% in the absence of put or when the cells contained the rpoC-Y75N mutation (Fig. 1 and Table 2). The presence of a functional putL site between the promoter and TR1 in rpoC+ cells reduced termination to 39%. We conclude that put partially suppresses termination at TR1 and that suppression is prevented by rpoC-Y75N. These conclusions are supported by measurements made with another fusion (see below).
FIG. 1.
put-mediated antitermination of the Rho-dependent terminator, λTR1. The strains were rpoC+ or rpoC-Y75N derivatives of TAP114 and contained single copies of a PTAC-putL-TR1-lacZ or a PTAC-putL-lacZ fusion, as indicated. The mutant putL site has a multiple base substitution that prevents antitermination of Rho-independent terminators (linker scanning mutation G [18]). β-Galactosidase activities (in arbitrary units) were measured 1 h after addition of IPTG to growing cultures and are the means of at least four independent assays. The open bars (ΔTR1) report the activities from fusions lacking TR1, the shaded bars (TR1 plus Psu) report the activities from those carrying TR1 in cells that contained a plasmid with a PTAC-psu fusion (pNL150), and the filled bars (TR1) report the activities from fusions containing TR1 in cells that contained a PTAC-Δpsu fusion (pNL151). The standard error of the mean was less than 20%.
TABLE 2.
Rho-mediated termination and put-mediated antitermination at λTR1a
| putL | RNAP-β′ | Termination (%) |
|---|---|---|
| WTb | WT | 39 |
| Mutant | WT | 96 |
| WT | Y75N | 95 |
| Δ | WT | 96 |
| Δ | Y75N | 95 |
The putL site, when present, was located between the promoter and lacZ and TR1, when present, was located between putL and lacZ. Termination = [1 − (β-galactosidase activity in a fusion containing TR1)/(β-galactosidase activity in a comparable fusion lacking TR1)] × 100. β-Galactosidase activities are from Fig. 1 except for those of the ΔputL fusions with and without TR1, which were 268 and 6,688 U, respectively, in the rpoC+ strain and 296 and 6,252 U, respectively, in the rpoC-Y75N strain (see Fig. 1 legend for more details).
WT, wild type.
The apparent termination efficiency of TR1 was severalfold higher in our fusions than in phage λ (12). It has previously been observed that the activity of TR1 depends on its context (14, 29; D. Court, personal communication), and this might explain the difference. Nevertheless, to confirm that our fusions indicate Rho-dependent termination rather than termination at an uncharacterized intrinsic terminator, we measured β-galactosidase activity in the presence of phage P4 Psu, a protein that antagonizes Rho activity (22, 23). We found that Psu significantly increased β-galactosidase activity if the reporter fusion carried an inactive put site or if the strain carried the rpoC-Y75N mutation (Fig. 1). Therefore, the TR1-containing reporter fusions do, indeed, report Rho-dependent termination. Psu had no such effect on a put+ fusion in rpoC+ cells, as expected if put suppresses Rho-dependent termination (Fig. 1). Psu did not increase lacZ expression as much as did deletion of TR1, perhaps because Psu did not completely prevent Rho termination, or not enough time was allowed to reach the steady-state level of β-galactosidase after Rho action was blocked. The psu gene was derepressed for only 1 h before β-galactosidase was measured, because continuous high-level expression is lethal (23). Another possibility, discussed more fully below, is that deletion of TR1 increases the stability or translation efficiency of the lacZ message.
We note that a deletion and a base substitution mutation of put increased the accumulation of β-galactosidase 1.5- to 3.3-fold in fusions that lack TR1 (Fig. 1). (We consider possible explanations below.) The increased activity did not significantly change our estimate of TR1 termination, which was 95 to 96% regardless of whether antitermination was prevented by a put mutation or by rpoC-Y75N (Table 2).
put suppresses Nun-dependent termination.
The phage HK022-encoded Nun protein terminates transcription after binding to a nascent transcript of the λ nutL or nutR sites (see introduction). To measure Nun-dependent termination, we compared the steady-state levels of β-galactosidase produced by cells containing a single copy of a PL(HK022)-putL-nutL-lacZ transcription fusion in the presence and absence of Nun. In this fusion, the putL site is in its natural location, immediately downstream of the HK022 PL promoter. To see if put suppresses Nun-dependent termination, we inactivated putL by mutation or prevented put action with the rpoC-Y75N mutation. A plasmid that contains a PLac-nun fusion (pNUN) provided Nun at either low or high concentration according to whether the culture was grown in the absence or presence, respectively, of the lac operon inducer IPTG. The low concentration was similar to that found in a single-copy HK022 lysogen, and the high concentration was about 100 times greater (reference 19 and other data not shown).
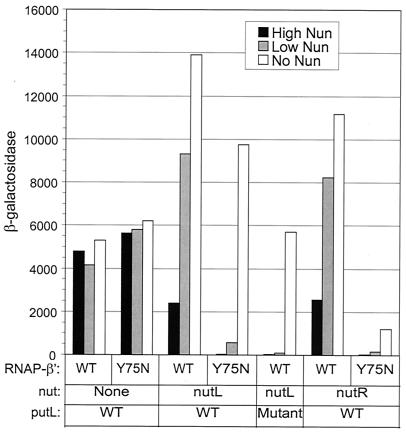
The efficiency of termination at a low Nun concentration was 33%, and mutation of rpoC or putL increased this efficiency to 94 or 83%, respectively (Fig. 2 and Table 3). The efficiency of termination at a high Nun concentration was 83%, and mutation of rpoC or putL increased this efficiency to 99.7 or 99.5%, respectively. We conclude that put antiterminates Nun-dependent termination at nutL, and that increasing the Nun concentration increases termination in both the presence and absence of put-mediated antitermination.
FIG. 2.
Antitermination of Nun-dependent terminators. The strains were derivatives of TAP114 carrying a single copy of a PL(HK022)-putL-lacZ fusion. Different fusions contained nutL, nutR, or neither nut site between putL and lacZ, and Nun was provided by a plasmid containing a PLac-nun fusion (pNUN). The shaded and filled bars show enzyme activity from mid-log cultures carrying pNUN grown in the absence and presence, respectively, of 1 mM IPTG. The open bars show enzyme activity from mid-log cultures carrying the vector plasmid (pNUNΔ) grown for 3 to 4 h in the presence of 1 mM IPTG. The mutant putL site (linker scanning mutation G [18]) has a multiple base substitution that prevents antitermination of Rho-independent terminators. Activities are averages of assays of at least two independent cultures at two different times during exponential growth. The standard errors of the mean ranged from 3 to 33%.
TABLE 3.
Nun-mediated termination and put-mediated antitermination at the λ nutL sitea
| nut | putL | RNAP-β′ | Nun termination (%)
|
|
|---|---|---|---|---|
| High Nun | Low Nun | |||
| nutL | WT | WT | 83 ± 3 | 33 ± 4 |
| nutL | WT | Y75N | 99.7 ± 0.1 | 93.9 ± 0.2 |
| nutL | Mutant | WT | 99.5 ± 0.1 | 98.3 ± 0.1 |
| None | WT | WT | 9 ± 9 | 22 ± 3 |
| None | WT | Y75N | 9 ± 20 | 6 ± 10 |
The putL site was adjacent to the promoter and the nutL site, when present, was between putL and the lacZ coding sequence. Nun termination = [1 − (β-galactosidase activity with Nun)/(β-galactosidase activity without Nun)] × 100, and the errors are standard errors of the mean. β-Galactosidase activities are from Fig. 2 (see Fig. 2 legend for more details). WT, wild type.
The effect of Nun on lacZ expression was, as expected, site specific: Nun, even when present at a high concentration, reduced the activity of a reporter fusion that lacked a nut site by only a small amount, and the rpoC-Y75N mutation did not further reduce β-galactosidase production (Fig. 2 and Table 3). Unexpectedly, β-galactosidase activity in the fusion lacking nutL was 0.3 to 0.4 that of the nutL+ fusion in the absence of Nun (Fig. 2). Therefore, we did not attempt to estimate the efficiency of Nun-dependent termination by comparing the activities of these two fusions in the presence of Nun (see below).
We used a PL(HK022)-putL-nutR-TR1-lacZ fusion to measure put suppression of Nun-dependent termination at the λ nutR site. The nutR region also contains the Rho-dependent λTR1 terminator; the rutA and rutB sequences, which are required for Rho action, lie immediately upstream and downstream, respectively, of the boxB element of nutR (14). nutL lacks rut sequences but is otherwise very similar to nutR. We hoped that comparison of the nutL- and nutR-containing fusions would tell us if Rho affects termination or antitermination at the nutR site. We note that the fusions used for these experiments have a different promoter, a different transcription start, and less transcribed DNA between putL and nutR than the PTAC-putL-TR1-lacZ fusion used to measure Rho-dependent termination in the experiments of Fig. 1 (see Materials and Methods). These differences could, in principle, alter termination and antitermination efficiencies.
We estimated Nun-dependent termination and put-mediated antitermination by measuring β-galactosidase activities in the presence or absence of Nun in rpoC+ or rpoC-Y75N cells, as described earlier for nutL. The efficiencies of Nun-dependent termination at nutR at low and high Nun concentrations were similar to those observed at nutL, and put suppressed Nun action at both sites to approximately the same extent (Fig. 2 and Table 4). Therefore, the presence of rut sites and their interaction with Rho do not appreciably alter Nun termination or put antitermination at nut (see also reference 33).
TABLE 4.
put-mediated antitermination of interdigitated Nun- and Rho-dependent terminatorsa
| RNAP-β′ | Nun termination (%)
|
Rho termination (%) | |
|---|---|---|---|
| High Nun | Low Nun | ||
| WT | 77 ± 3 | 26 ± 4 | <1b |
| Y75N | 98 ± 0.5 | 86 ± 2 | 80c |
All of the fusions contained putL immediately downstream of the promoter and either contained or lacked the nutR-TR1 region upstream of lacZ. The efficiency of Nun-dependent termination = [1 − (β-galactosidase activity with Nun)/(β-galactosidase activity without Nun)] × 100, and the errors are standard errors of the means. The efficiency of Rho-dependent termination, measured in cells lacking Nun, = [1 − (β-galactosidase activity in a fusion containing TR1)/(β-galactosidase activity in a comparable fusion lacking TR1)] × 100. β-Galactosidase activities are from Fig. 2. WT, wild type.
The level of β-galactosidase was 2.1-fold greater in the fusion containing nutR than in the fusion lacking nutR (Fig. 2).
This could be an underestimate of the termination efficiency (see text).
We estimated the efficiency of Rho-dependent termination and put-mediated antitermination at TR1 by measuring β-galactosidase produced by fusions with or without the nutR-TR1 region in rpoC+ or rpoC-Y75N cells. Since these cells did not contain Nun, there was no Nun-dependent termination at nutR. The estimated efficiency of termination in the rpoC mutant host was 80%, and termination was completely suppressed in the wild-type host (Table 4). These estimates are somewhat lower than those presented in Table 2, in which different fusions were used. The differences might be the result of differences in the fusions, but there are also uncertainties in our estimates of termination efficiency, as discussed below. Nun caused an additional reduction in β-galactosidase activity produced by fusions containing the nutR-TR1 region (Fig. 2), but the experimental uncertainties preclude quantitative estimates of any effect of Nun on Rho-dependent termination.
We wish to call attention to several unexpected observations. Mutation or deletion of putL increased the specific activity of β-galactosidase 1.5- to 3-fold in certain fusions (Fig. 1 and Table 2 footnote [ΔTR1 fusions]). In other fusions, deletion of nutL or nutR-TR1 decreased the specific activity of β-galactosidase to 0.3 to 0.4 (Fig. 2, No Nun). We speculate that these changes increased the stability and/or the translation efficiency of the lacZ message, but we have no independent evidence for this hypothesis. Because of this, estimates of termination and antitermination that are based on comparison of fusions that either contain or lack the terminator or antiterminator sites, respectively, should be considered as approximations. In many cases we estimated efficiencies of termination and antitermination by comparing identical fusions in the presence and absence of a trans-acting protein, such as Nun, Psu, or wild-type RNAP, and these estimates are probably more reliable. For example, our estimate that unsuppressed Rho-mediated termination efficiency at TR1 is 80% was based on comparison of different fusions (Fig. 2 and Table 4). Comparison of the amount of β-galactosidase produced by the fusion containing TR1 in rpoC+ and rpoC-Y75N cells (Fig. 2) suggested that the true termination efficiency in this fusion could be 90% or more.
DISCUSSION
We have shown that put suppresses Nun- and Rho-dependent transcription terminators in vivo. Suppression was efficient when the termination factors were present at physiological levels, but increasing the concentration of Nun increased termination (on templates containing a nut site) both in the presence and absence of a functional put site. We previously demonstrated efficient suppression of several strong intrinsic terminators in vivo and in vitro. We also showed that the HK022 put sites could replace the function of the λ N gene and nut sites in λ-HK022 hybrid phages (10, 18, 28). The hybrid phages showed no obvious defect in lytic growth, lysogenization, or lysogenic induction. It therefore appears that the put sites suppress the numerous λ terminators in the early operons to the extent required for normal phage growth. put-mediated antitermination thus shows no demonstrable terminator specificity. The λ N/nut and Q/qut antitermination pathways also suppress intrinsic and Rho-dependent terminators, and N suppresses Nun-dependent termination by competing for binding to nut RNA (5, 8, 16, 33, 46). Q has not yet been tested on a Nun-dependent terminator. This apparent lack of terminator specificity suggests that put inhibits a step that is common to termination at the different types of terminator. What might this step be?
A short region of RNA-DNA hybrid at the 3′ end of the nascent RNA chain is believed to play a critical role in stabilizing the transcript elongation complex (27, 36). Current models propose that intrinsic and Rho-dependent termination are a consequence of hybrid disruption (15, 20, 40, 47). It is, therefore, tempting to suggest that put RNA acts by increasing the stability of the hybrid or by preventing a step that follows hybrid disruption. However, this suggestion fails to account for put suppression of Nun-dependent terminators. Nun binds to nascent nut RNA and is then delivered to the nearby elongation complex, where it arrests translocation. Arrest is believed to be the result of a Nun-DNA interaction that anchors the elongation complex to the template so that it can no longer translocate (17, 43). Since the arrested complex is stable and catalytically active in vitro, it is not obvious how further stabilization by put RNA could prevent arrest or restart translocation once arrest had occurred or, if it did, how increasing the concentration of Nun would overcome this effect.
We offer two models to explain suppression of Nun-dependent termination. First, the efficiency of Nun arrest in vitro is decreased by conditions that increase the rate of translocation (16). It is possible that rapid translocation of RNAP away from the Nun binding site decreases the efficiency of transfer of Nun to the elongation complex. If so, put-mediated acceleration of the elongation complex through pause sites (18) could have the same effect on Nun transfer. Elevating the Nun concentration should quicken its binding to nut RNA and thus increase the probability of transfer. In the second model, put RNA impedes or delays binding of Nun to nut RNA. We have shown that nascent put RNA binds to elongating polymerase and that this complex persists as polymerase translocates (35). If the nascent put transcript binds close to the product RNA exit channel in RNAP, it might delay the binding of Nun to the nascent nut transcript until the elongation complex is too distant for efficient transfer of Nun to RNAP. This effect would be mitigated by increasing the Nun concentration. Either of these models can be adapted to explain the suppression of other classes of terminators by put RNA. However, it is possible that put suppresses different types of terminators in different ways and that no single mechanism suffices to account for its action.
Acknowledgments
We are grateful to Max Gottesman and Don Court for their comments on the manuscript. We also acknowledge Jennie Tang for constructing plasmid pJT6.
REFERENCES
- 1.Albrechtsen, B., C. L. Squires, S. Li, and C. Squires. 1990. Antitermination of characterized transcriptional terminators by the Escherichia coli rrnG leader region. J. Mol. Biol. 213:123-134. [DOI] [PubMed] [Google Scholar]
- 2.Arber, W. 1983. A beginner's guide to lambda biology, p. 381-394. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Atkinson, B. L., and M. E. Gottesman. 1992. The Escherichia coli rpoB60 mutation blocks antitermination by coliphage HK022 Q-function. J. Mol. Biol. 227:29-37. [DOI] [PubMed] [Google Scholar]
- 4.Banik-Maiti, S., R. A. King, and R. A. Weisberg. 1997. The antiterminator RNA of phage HK022. J. Mol. Biol. 272:677-687. [DOI] [PubMed] [Google Scholar]
- 5.Baron, J., and R. A. Weisberg. 1992. Mutations of the phage lambda nutL region that prevent the action of Nun, a site-specific transcription termination factor. J. Bacteriol. 174:1983-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, K. L., C. Squires, and C. L. Squires. 1989. Ribosomal RNA operon anti-termination. Function of leader and spacer region box B-box A sequences and their conservation in diverse micro-organisms. J. Mol. Biol. 209:345-358. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, C. A., A. J. Dombroski, and T. Platt. 1987. Transcription termination factor rho is an RNA-DNA helicase. Cell 48:945-952. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay, S., S. C. Hung, A. C. Stuart, A. G. Palmer III, J. Garcia-Mena, A. Das, and M. E. Gottesman. 1995. Interaction between the phage HK022 Nun protein and the nut RNA of phage lambda. Proc. Natl. Acad. Sci. USA 92:12131-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerget, M. 1991. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 3:780-788. [PubMed] [Google Scholar]
- 10.Clerget, M., D. J. Jin, and R. A. Weisberg. 1995. A zinc binding region in the β′ subunit of RNA polymerase is involved in antitermination of early transcription of phage HK022. J. Mol. Biol. 248:768-780. [DOI] [PubMed] [Google Scholar]
- 11.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Court, D., C. Brady, M. Rosenberg, D. L. Wulff, M. Behr, M. Mahoney, and S. U. Izumi. 1980. Control of transcription termination: a rho-dependent termination site in bacteriophage lambda. J. Mol. Biol. 138:231-254. [DOI] [PubMed] [Google Scholar]
- 13.Ghysen, A., and M. Pironio. 1972. Relationship between the N function of bacteriophage lambda and host RNA polymerase. J. Mol. Biol. 65:259-272. [DOI] [PubMed] [Google Scholar]
- 14.Graham, J. E., and J. P. Richardson. 1998. rut sites in the nascent transcript mediate Rho-dependent transcription termination in vivo. J. Biol. Chem. 273:20764-20769. [DOI] [PubMed] [Google Scholar]
- 15.Gusarov, I., and E. Nudler. 1999. The mechanism of intrinsic transcription termination. Mol. Cell 3:495-504. [DOI] [PubMed] [Google Scholar]
- 16.Hung, S. C., and M. E. Gottesman. 1995. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J. Mol. Biol. 247:428-442. [DOI] [PubMed] [Google Scholar]
- 17.Hung, S. C., and M. E. Gottesman. 1997. The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities. Genes Dev. 11:2670-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King, R. A., S. Banik-Maiti, D. J. Jin, and R. A. Weisberg. 1996. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell 87:893-903. [DOI] [PubMed] [Google Scholar]
- 19.King, R. A., P. L. Madsen, and R. A. Weisberg. 2000. Constitutive expression of a transcription termination factor by a repressed prophage: promoters for transcribing the phage HK022 nun gene. J. Bacteriol. 182:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komissarova, N., J. Becker, S. Solter, M. Kireeva, and M. Kashlev. 2002. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell 10:1151-1162. [DOI] [PubMed] [Google Scholar]
- 21.Lau, L. F., J. W. Roberts, and R. Wu. 1982. Transcription terminates at lambda tR1 in three clusters. Proc. Natl. Acad. Sci. USA 79:6171-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linderoth, N. A., and R. L. Calendar. 1991. The Psu protein of bacteriophage P4 is an antitermination factor for rho-dependent transcription termination. J. Bacteriol. 173:6722-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linderoth, N. A., G. Tang, and R. Calendar. 1997. In vivo and in vitro evidence for an anti-Rho activity induced by the phage P4 polarity suppressor protein Psu. Virology 227:131-141. [DOI] [PubMed] [Google Scholar]
- 24.Mason, S. W., and J. Greenblatt. 1991. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5:1504-1512. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Nudler, E., and M. E. Gottesman. 2002. Transcription termination and anti-termination in E. coli. Genes Cells 7:755-768. [DOI] [PubMed] [Google Scholar]
- 27.Nudler, E., A. Mustaev, E. Lukhtanov, and A. Goldfarb. 1997. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89:33-41. [DOI] [PubMed] [Google Scholar]
- 28.Oberto, J., M. Clerget, M. Ditto, K. Cam, and R. A. Weisberg. 1993. Antitermination of early transcription in phage HK022. Absence of a phage-encoded antitermination factor. J. Mol. Biol. 229:368-381. [DOI] [PubMed] [Google Scholar]
- 29.Patterson, T. A., Z. Zhang, T. Baker, L. L. Johnson, D. I. Friedman, and D. L. Court. 1994. Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J. Mol. Biol. 236:217-228. [DOI] [PubMed] [Google Scholar]
- 30.Platt, T. 1994. Rho and RNA: models for recognition and response. Mol. Microbiol. 11:983-990. [DOI] [PubMed] [Google Scholar]
- 31.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. (Erratum, 23:1278, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson, J. P., and J. Greenblatt. 1996. Control of RNA chain elongation and termination, p. 822-848. In F. C. Neidhardt, R. C. Curtiss III, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 33.Robert, J., S. B. Sloan, R. A. Weisberg, M. E. Gottesman, R. Robledo, and D. Harbrecht. 1987. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell 51:483-492. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg, M., D. Court, H. Shimatake, C. Brady, and D. L. Wulff. 1978. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature 272:414-423. [DOI] [PubMed] [Google Scholar]
- 35.Sen, R., R. A. King, and R. A. Weisberg. 2001. Modification of the properties of elongating RNA polymerase by persistent association with nascent antiterminator RNA. Mol. Cell 7:993-1001. [DOI] [PubMed] [Google Scholar]
- 36.Sidorenkov, I., N. Komissarova, and M. Kashlev. 1998. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell 2:55-64. [DOI] [PubMed] [Google Scholar]
- 37.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 39.Sloan, S. B., and R. A. Weisberg. 1993. Use of a gene encoding a suppressor tRNA as a reporter of transcription: analyzing the action of the Nun protein of bacteriophage HK022. Proc. Natl. Acad. Sci. USA 90:9842-9846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz, E. J., and T. Platt. 1994. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc. Natl. Acad. Sci. USA 91:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres, M., C. Condon, J. M. Balada, C. Squires, and C. L. Squires. 2001. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 20:3811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Washburn, R. S., Y. Wang, and M. E. Gottesman. 2003. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 329:655-662. [DOI] [PubMed] [Google Scholar]
- 43.Watnick, R. S., and M. E. Gottesman. 1999. Binding of transcription termination protein nun to nascent RNA and template DNA. Science 286:2337-2339. [DOI] [PubMed] [Google Scholar]
- 44.Weisberg, R. A., and M. E. Gottesman. 1999. Processive antitermination. J. Bacteriol. 181:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisberg, R. A., M. E. Gottesman, R. W. Hendrix, and J. W. Little. 1999. Family values in the age of genomics: comparative analyses of temperate bacteriophage HK022. Annu. Rev. Genet. 33:565-602. [DOI] [PubMed] [Google Scholar]
- 46.Yang, X. J., and J. W. Roberts. 1989. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc. Natl. Acad. Sci. USA 86:5301-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]
- 48.Zellars, M., and C. L. Squires. 1999. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol. Microbiol. 32:1296-1304. [DOI] [PubMed] [Google Scholar]