Abstract
The cytotoxicity of many antineoplastic agents is due to their capacity to damage DNA and there is evidence indicating that DNA repair contributes to the cellular resistance to such agents. DNA strand breaks constitute a significant proportion of the lesions generated by a broad range of genotoxic agents, either directly, or during the course of DNA repair. Strand breaks that are caused by many agents including ionizing radiation, topoisomerase I inhibitors, and DNA repair glycosylases such as NEIL1 and NEIL2, often contain 5’-hydroxyl and/or 3’-phosphate termini. These ends must be converted to 5’-phosphate and 3’-hydroxyl termini in order to allow DNA polymerases and ligases to catalyze repair synthesis and strand rejoining. A key enzyme involved in this end-processing is polynucleotide kinase (PNK), which possesses two enzyme activities, a DNA 5’-kinase activity and a 3’-phosphatase activity. PNK participates in the single-strand break repair pathway and the non-homologous end joining pathway for double-strand break repair. RNAi-mediated down-regulation of PNK renders cells more sensitive to ionizing radiation and camptothecin, a topoisomerase I inhibitor. Structural analysis of PNK revealed the protein is composed of three domains, the kinase domain at the C-terminus, the phosphatase domain in the centre and a forkhead associated (FHA) domain at the N-terminus. The FHA domain plays a critical role in the binding of PNK to other DNA repair proteins. Thus each PNK domain may be a suitable target for small molecule inhibition to effectively reduce resistance to ionizing radiation and topoisomerase I inhibitors.
Keywords: DNA damage, DNA repair, small molecule inhibitors, polynucleotide kinase, FHA, kinase, phosphatase
Cancer therapy and DNA damage and repair
Cancer continues to be a major health problem. This year (2007) it is estimated that approximately 1.6 million individuals will be diagnosed with cancer in the USA and Canada (excluding basal and squamous cell skin cancer) and that approximately 630,000 will die from cancer (American Cancer Society and Canadian Cancer Society). Despite major advances in targeted chemotherapy, the major forms of therapy continue to be surgery, radiotherapy and conventional chemotherapy. The cellular target for radiotherapy and many conventional drugs is the DNA within the nuclei (and possibly mitochondria). Consequently, the levels of induced DNA damage and the repair of that damage would be expected to influence cell survival. Evidence that DNA repair affects clinical outcome has been reviewed by several authors [1–3]. As a result DNA repair systems are emerging as therapeutic targets to enhance the cytotoxicity of radiation and genotoxic drugs [4, 5]. Since human polynucleotide kinase (hPNK), the DNA repair enzyme that is the subject of this article, is involved in several pathways required to repair damage induced by many genotoxins, it is instructive to give a brief overview of the DNA lesions induced by a selection of well-known anti-cancer agents, the pathways responsible for their repair, and the role of hPNK within each pathway. Special attention will be paid to the DNA strand break termini generated either by the genotoxic agents directly or during the course of repair.
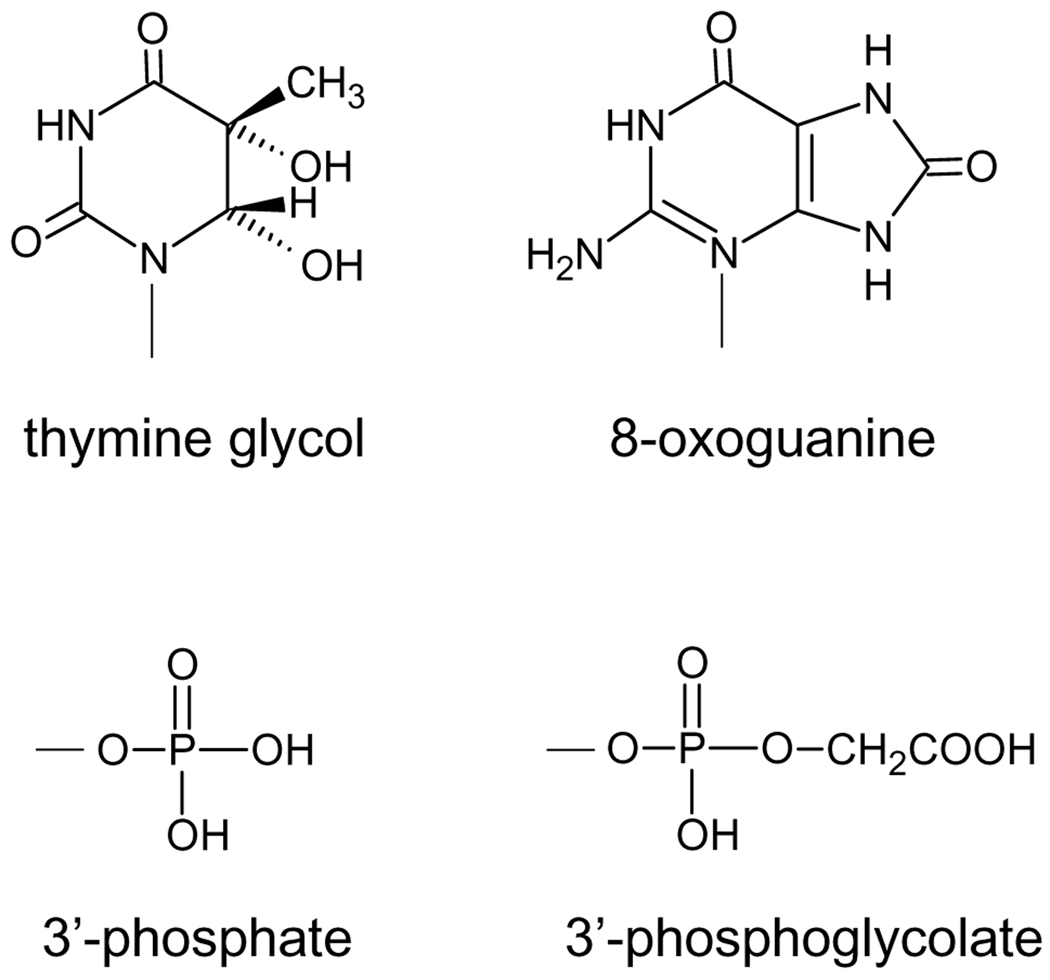
The form(s) of DNA damage are dependent on the anti-cancer agent used. Ionizing radiation, either directly or through the production of hydroxyl free radicals, generates a plethora of DNA lesions, involving all four bases and the sugar-phosphate backbone [6]. Examples of radiation-induced DNA base lesions include thymine glycol and 8-oxoguanine (Fig. 1). Radical reaction at the deoxyribose usually results in base loss and DNA strand cleavage [6]. Hydrogen abstraction from the C4’ carbon of the deoxyribose ultimately leads to strand breaks with either a 3’-phosphate or 3’-phosphoglycolate (Fig. 1), the two primary end-groups found at 3’-termini in irradiated DNA [6, 7]. Production of 3’-phosphoglycolate termini requires the presence of oxygen or a radiosensitizer such as misonidazole or hypoxic cell sensitizers such as tirapazamine [8, 9]. The predominant end group found at 5’-termini following irradiation are 5’-phosphate groups, but 5’-hydroxyl groups have also been observed in irradiated cellular DNA [10, 11]. A feature of ionizing radiation resulting from its mode of energy deposition, which results in the generation of a high local concentration of free radicals, is the production of complex DNA lesions, i.e. two or more lesions within 1–2 helical turns of DNA [12–14]. Such lesions comprise of damaged bases and/or strand breaks on the same or opposite strands and include double-strand breaks. (The other complex lesions may give rise to double-strand breaks during the course of DNA repair.)
Figure 1.
Common DNA lesions generated by exposure to ionizing radiation.
Chemotherapeutic agents also produce a wide variety of DNA lesions depending on their mechanisms of action. Alkylating agents, such as dacarbazine (DTIC), used to treat malignant melanoma, lymphoma and soft tissue sarcoma, and temozolomide (TMZ), used to treat brain cancers and tumors that have metastasized to the brain, act mainly via SN2 reactions to methylate the N7 and O6 of guanine and N3 of adenine [15, 16]. Bifunctional alkylating agents can cause DNA crosslinks. For example, 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU, carmustine) used in the treatment of brain tumors and certain lymphomas, generates a chlorethyl adduct at the O6-position of guanine that subsequently undergoes an intramolecular rearrangement and subsequently reacts with cytosine on the opposite DNA strand [17]. This reagent also reacts at alternative sites in DNA (N7 of guanine and N3 of adenine) and causes DNA strand breaks with 5’-hydroxyl termini, probably through alkylation of DNA internucleotide phosphates [18]. Other chemotherapeutic agents, such as bleomycin, which is used in the treatment of squamous cell carcinoma and non-Hodgkin’s lymphomas, attack the deoxyribose moiety of DNA, thereby inducing single- and double-strand cleavage. In the case of bleomycin, this reaction, which requires the presence of oxygen and a redox-active metal ion such as iron, is initiated by hydrogen abstraction from the C4’-position of the deoxyribose and generates 3’-phosphoglycolate termini [19, 20].
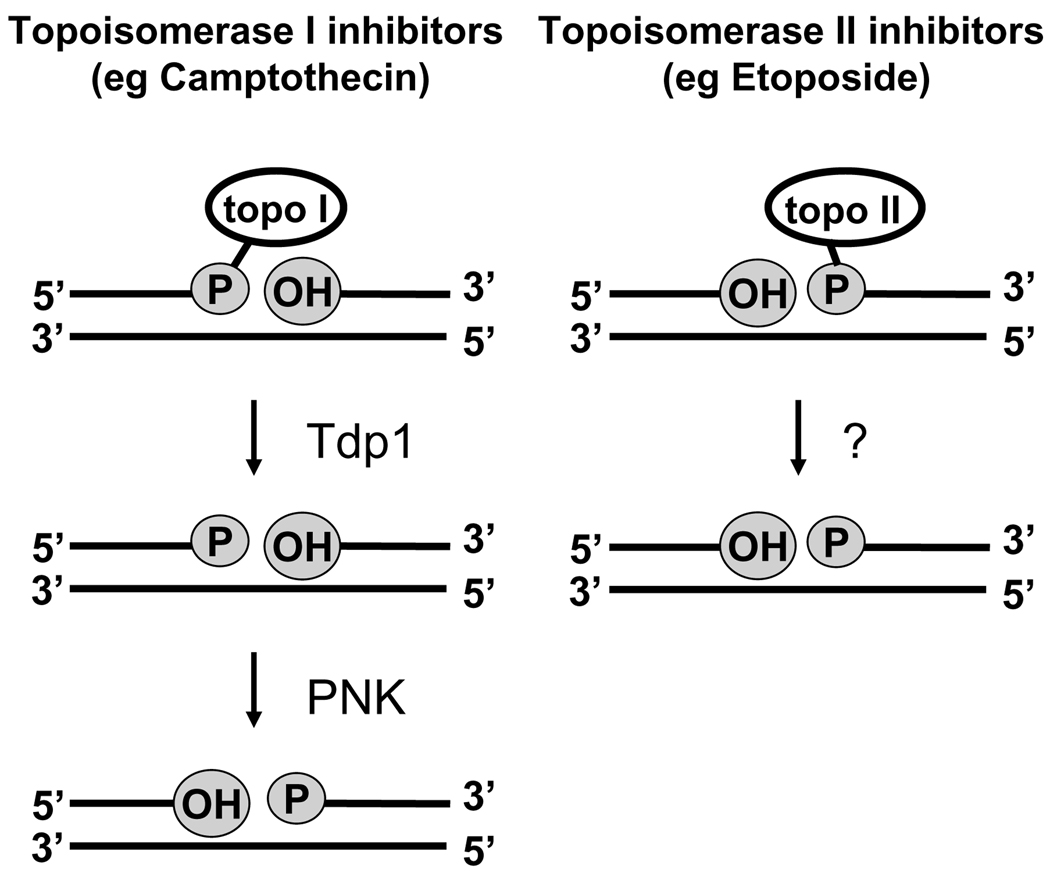
Topoisomerase inhibitors represent one further class of genotoxic chemotherapeutic agents. These compounds do not directly damage DNA, but block the action of topoisomerases at the stage in which the enzyme has cleaved the DNA and slow the reclosure step of the nicking-closing reaction [21, 22]. Synthetic derivatives of the plant alkaloid camptothecin, such as irinotecan (colorectal cancer) and topotecan (ovarian and small cell lung cancer), inhibit topoisomerase 1 by forming a “dead-end complex”, in which the topoisomerase is covalently bound to a 3’-phosphate at the site of the broken strand [23] (Fig. 2). On the other hand, topoisomerase II inhibitors, such as etoposide (used to treat many cancers), stabilize the covalent complex of topoisomerase II bound to the 5’-phosphate of the topoisomerase II-cleaved DNA [24].
Figure 2.
Schematic representation of DNA strand breaks induced by topoisomerase inhibitors and the role of PNK in the pathways responsible for their repair. Topo I inhibitors, such as camptothecin, produce strand breaks with a 5’-hydroxyl group and the enzyme covalently attached to a 3’-phosphate. Hydrolysis of the protein-DNA bond by tyrosyl-DNA phosphodiesterase (Tdp1) leaves a 3’-phosphate group. Therefore, both the 3’ and 5’ termini need to be acted upon by PNK. In contrast topo II inhibitors, such as etoposide, generate strand breaks with 3’-hydroxyl groups and the enzyme covalently linked to a 5’-phosphate. Although the mechanism(s) for repairing these lesions has yet to be fully elucidated, it is unlikely that PNK is required.
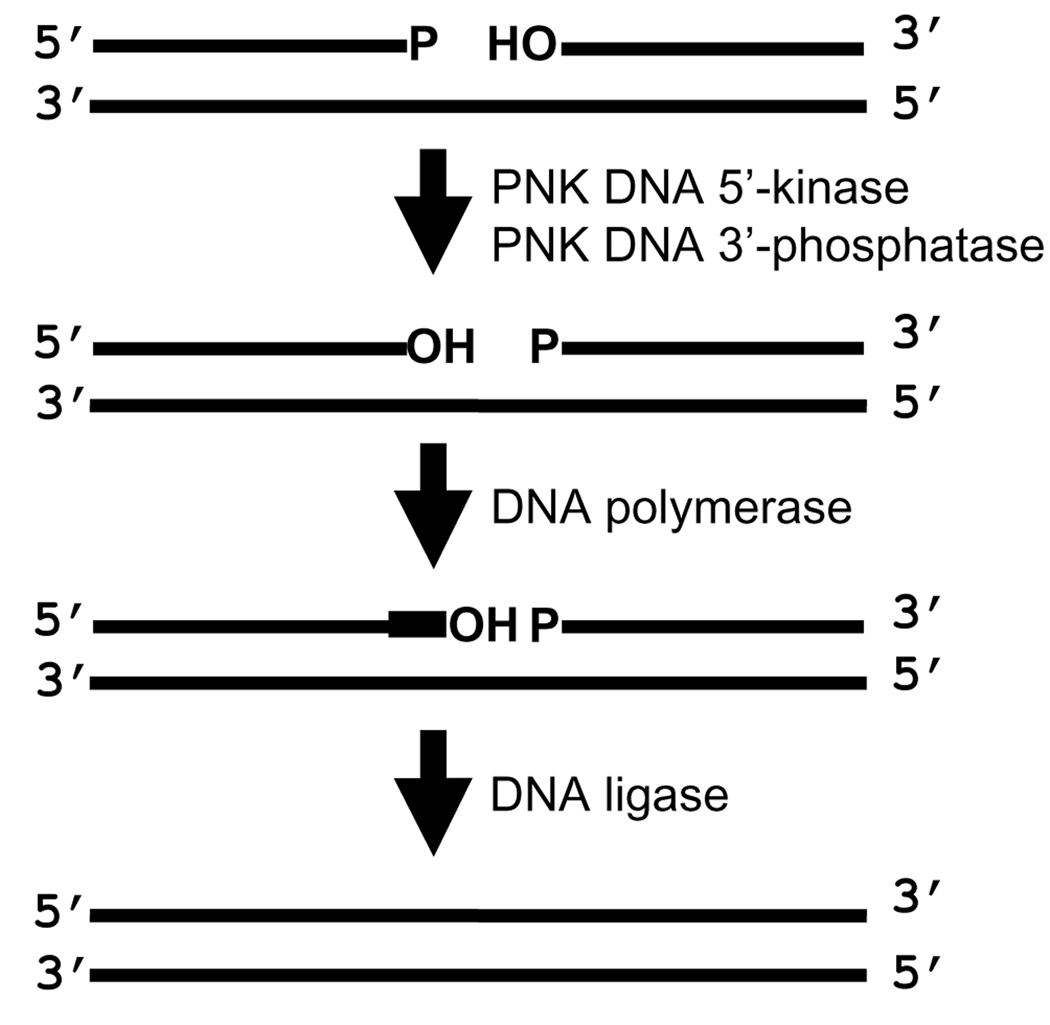
Aside from DNA repair pathways that directly reverse base modification (e.g. the removal of the methyl group from O6-methyl guanine by MGMT), all the repair pathways require DNA polymerases to replace missing nucleotides and DNA ligases to mediate strand rejoining. These enzymes have an absolute requirement for specific functional end groups on the deoxyribose at the strand break termini; DNA polymerases require 3’-OH groups and DNA ligases require 3’-OH groups and 5’-phosphate groups. Mammalian PNK is involved in the processing of strand break termini to ensure that they are suitable for DNA polymerases and ligases. PNK possesses both a 5’-kinase activity that catalyzes the transfer of phosphate from ATP to a 5’-hydroxyl (OH) terminus and also a 3’-phosphatase activity that converts 3’-phosphate termini to 3’-OH termini (Fig. 3) [25–27].
Figure 3.
Processing of DNA strand break termini by PNK. PNK catalyzes the phosphorylation of 5’-hydroxyl (OH) termini and dephosphorylation of 3’-phosphate (P) termini so that subsequent nucleotide insertion and strand rejoining can be mediated by DNA polymerases and ligases, respectively.
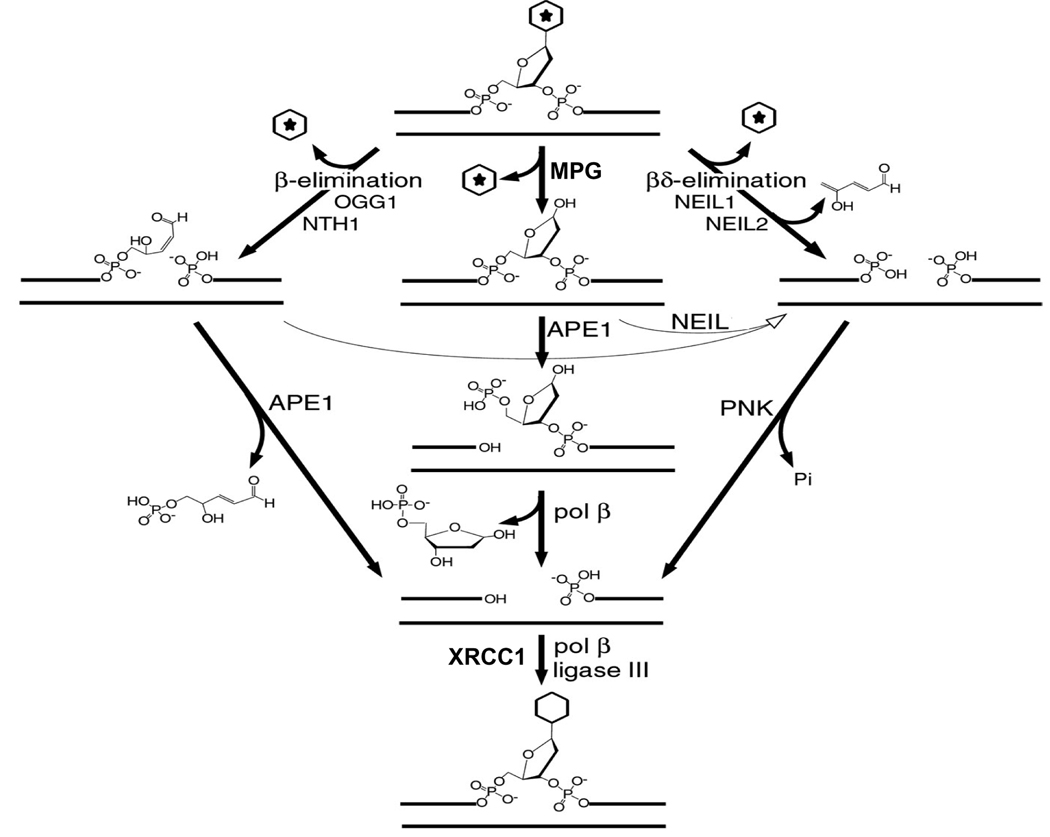
Most of the smaller modifications to bases, such as radiation-mediated oxidation or methylation of the bases (with the exception of O6-methylguanine), are handled by the base excision repair (BER) pathway [reviewed in [28]]. The first step (Fig. 4) entails release of the modified base by enzyme-catalyzed hydrolysis of the glycosyl bond to generate an abasic (AP) site. Several DNA glycosylases each deal with one or more modified bases and in some cases have overlapping functionality. For example, oxidized pyrimidines, such as thymine glycol, can be removed by NTH1 (human endonuclease III), or by NEIL1 or NEIL2; oxidized purines, such as 8-oxoguanine, are removed by OGG1 and NEIL1; and N3-methyladenine is released by MPG. The subsequent step in the process, cleavage of the DNA at the abasic site, depends on whether or not the glycosylase possesses an AP lyase activity, and if so, the type of AP lyase. If the DNA glycosylase does not possess an AP lyase activity (e.g. MPG), the abasic site is usually acted upon by APE1, an AP endonuclease that cleaves the DNA 5’ to the abasic site to generate strand break termini with 3’-hydroxyl and 5’-deoxyribose phosphate groups. The latter group is then removed by the deoxyribose phosphatase action of DNA polymerase β, leaving a 5’-phosphate terminus. Several DNA glycosylases, such as OGG1 and NTH1, possess an AP lyase activity that catalyses a β-elimination reaction disrupting the 3’-phosphodiester bond of the abasic site to generate a strand break with a 3’ unsaturated aldehydic α,β, 4-hydroxy-2-pentenal terminus and a 5’-phosphate terminus. APE1 can then remove the residual 4-hydroxy-2-pentenal group yielding a 3’-OH terminus. Other glycosylases, such as NEIL1 and NEIL2, possess AP lyases that catalyze a β,δ-elimination reaction that cleaves and removes the AP site leaving a strand break with 3’-phosphate and 5’-phosphate termini. The 3’-phosphate groups are hydrolyzed by hPNK because APE1 does not possess a potent 3’-phosphatase activity [29, 30]. However, it is important to note that the AP lyase activities of NEIL1 and NEIL2, can also act on the abasic sites generated by MPG and the 4-hydroxy-2-pentenal groups generated by OGG1 and NTH1, and this represents an alternative, hPNK-dependent pathway to the regular APE1-dependent BER pathway [30]. Although hPNK does not bind directly to NEIL1 or NEIL2, it can be isolated from human cell extracts together with these glycosylases in BER multi-protein complexes [30, 31]. The other proteins found in this complex include XRCC1, DNA polymerase β and DNA ligase III. The same three proteins together with hPNK form the complex required for single-strand break repair discussed below.
Figure 4.
Base excision repair, showing the importance of the AP lyase activities of the DNA glycosylases that act on a variety of base lesions. Under normal conditions, glycosylases that lack an AP lyase activity (e.g. MPG) or possess a β-elimination lyase activity (e.g. OGG1 and NTH1) depend on APE1 for the subsequent processing of the abasic sites. Glycosylases with a lyase acting by β.δ-elimination, such as NEIL1 and NEIL2, rely on PNK to remove the 3’-phosphate group. However, the lyase activity of NEIL1 and NEIL2 can also act on the intermediates generated by the other classes of DNA glycosylase, and therefore provide the basis for an alternative APE-independent repair pathway for these glycosylases. (Adapted from reference [30]).
Human PNK also processes many of the strand breaks induced directly by genotoxic agents such as ionizing radiation [32–37]. Of the two major 3’-end groups generated by ionizing radiation at single-strand breaks (SSBs), hPNK removes phosphate groups while APE1 is responsible for removal of phosphoglycolate groups [38]. In the case of double-strand breaks the situation is more complicated. APE1 can efficiently remove phosphoglycolate groups from recessed 3’-termini, but not from blunt-ended or overhanging 3’-termini [39, 40]. Evidence indicates that these APE1-refractory phosphoglycolate groups are initially acted upon by another enzyme, tyrosyl-DNA phosphodiesterase (Tdp1), which converts the phosphoglycolate to phosphate and the phosphate is then removed by hPNK [41]. 5’-hydroxyl termini found at both single and double-strand breaks are suitable substrates for mammalian PNK [27, 36]. The strand breaks introduced by topoisomerase I inhibitors, such as camptothecin, are handled by the same set of enzymes (Fig. 2) [42]. First, the phosphotyrosyl bond linking the stalled topoisomerase I to the 3’-terminus is incised by Tdp1 and then the 3’-phosphate and 5’-OH termini are reversed by PNK and the strand rejoined by DNA ligase III. The process is enhanced by the presence of XRCC1. In contrast, the lesions generated by topoisomerase II inhibitors do not require hPNK since the nick resulting from hydrolysis of the protein-DNA bond already possesses 3’-OH and 5’-phosphate termini (Fig. 2).
As mentioned above, for SSB repair (and BER) hPNK is part of a protein complex that also contains XRCC1, DNA polymerase β and DNA ligase III [35, 43]. XRCC1 is considered to be a scaffold protein required for the assembly and orchestration of the other proteins at the sites of DNA damage within the cell [35]. It can stimulate both the 3’-phosphatase and 5’-kinase activities of hPNK [35] and it has been determined that phosphorylated XRCC1 binds to hPNK through the forkhead associated (FHA) domain of hPNK [43]. It is still not entirely clear how XRCC1 stimulates hPNK, but there is evidence to suggest that XRCC1 may displace hPNK from the product of hPNK reaction with its substrates, thus increasing protein turnover ([44], Mani et al unpublished data).
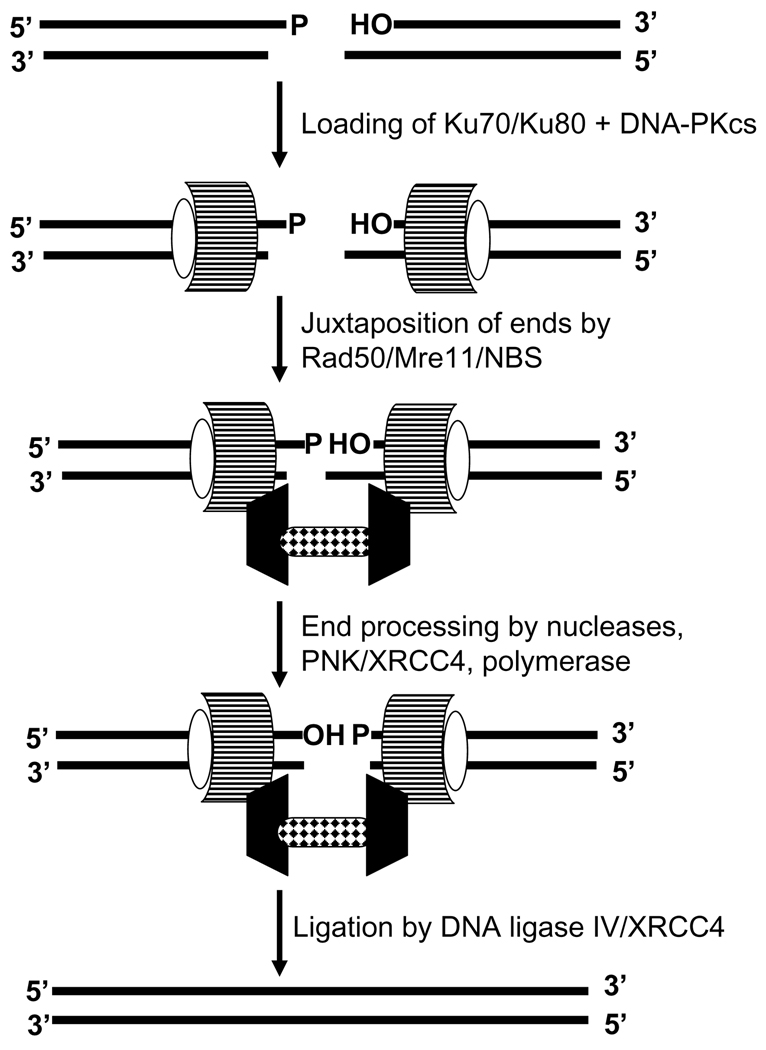
In the case of double-strand break (DSB) repair, hPNK participates in the nonhomologous end-joining pathway (NHEJ) [36, 37, 45]. The observation that a marked reduction in hPNK expression failed to influence the level of radiation-induced sister-chromatid exchange suggests that hPNK is not involved in homologous recombination, the other major DSB repair pathway[45]. There is still considerable uncertainty regarding the temporal progression of the NHEJ pathway (Fig. 5) [46, 47], but it is clear that the early steps require the binding of the Ku70/80 heterodimer to each end, followed by the binding of DNA-dependent protein kinase catalytic subunit (DNA-PKcs). At this point it is proposed that an assortment of proteins, including the Artemis nuclease and hPNK together with XRCC4, is recruited in order to process the termini to a form suitable for ligation by DNA ligase IV. The exact role of XRCC4 remains undefined but it may function in an analogous role to XRCC1 in SSB repair by localizing and orchestrating other proteins to the break site. Like XRCC1, phosphorylated XRCC4 can bind to the FHA domain of PNK [37, 48]. Recently, an alternative XRCC1/DNA ligase III-dependent DSB repair pathway has been described [49] that utilizes hPNK in the processing of termini [50].
Figure 5.
Outline of the basic steps in the nonhomologous end-joining pathway for DNA double-strand break repair. PNK is required to process the strand break termini. In this pathway it interacts directly with phosphorylated XRCC4.
The importance of PNK in the response of cells to genotoxic agents can be gauged through the use of cells with depleted levels of the enzyme. PNK was initially knocked out in Schizosaccharomyces pombe [51]. The fission yeast PNK has reasonable homology to the hPNK (34% sequence identity) and contains both a kinase and a phosphatase domain, but lacks an FHA domain. The PNK knockout was found to have elevated sensitivity to both ionizing radiation and camptothecin in comparison to the wild type. The level of hPNK expression has been stably downregulated by expressing an siRNA sequence in A549 cells, a human lung adenocarcinoma cell line [52]. These cells were approximately two-fold more sensitive to ionizing radiation across the full dose range tested (0–8 Gy). They also showed an elevated sensitivity to camptothecin (at doses > 1 µM), methyl methanesulfonate (a model alkylating agent) and hydrogen peroxide, but not etoposide or cisplatin [45, 52].
Structure and key interactions of mammalian PNK
Overview of the structure of PNK
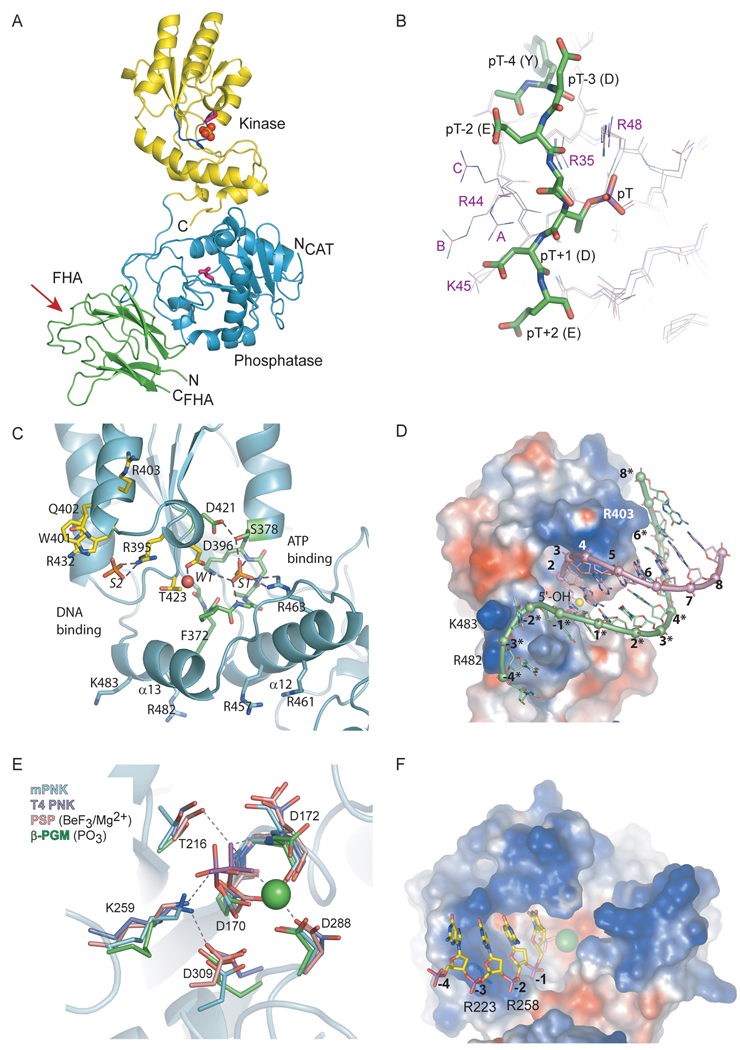
Mammalian PNK (mPNK) consists of 3 domains: the FHA (forkhead-associated) domain, the phosphatase and the kinase (Fig. 6A) [48]. The kinase and phosphatase domains together make up the catalytic fragment, which carries out the enzymatic activities of PNK. Within the catalytic fragment the kinase and phosphatase are connected by two segments of polypeptide: the intradomain linker (Leu 337 – Ala 339) and the C-terminal tail (Gln 518 – Gly 522), which interacts with the phosphatase domain. The individual catalytic domains need to be within the intact catalytic fragment to be correctly folded. The phosphatase domain expressed separately is insoluble. The kinase is soluble, but has lower specific activity than the full length PNK (Bernstein and Glover, unpublished data). The FHA domain is attached to the catalytic fragment by a flexible linker, which is completely disordered in the crystal structure. The kinase and phosphatase active sites face the same direction, although there is a small (10 degree) reorientation between the kinase and phosphatase domains in the two independent copies of PNK in the crystallographic asymmetric unit. The difference in relative domain orientation does not affect the structures of the kinase and phosphatase active sites, only their relative disposition, which may allow PNK to adjust to different sizes of DNA strand breaks. The orientation and slight flexibility of the kinase and phosphatase domains of PNK may allow this enzyme to work on either the 5’-OH or 3’-phosphate termini, one or both of which may be present at the site of a DNA strand break. The two enzymatic activities appear to be independent, as the rate of the phosphatase reaction has been shown to be faster than the kinase reaction [53]. In addition, the two active sites have mutually exclusive substrate preferences, as described below and by Bernstein et al. [48].
Figure 6.
Structural overview of mammalian PNK. (A) Full-length mouse PNK. The FHA domain is shown in green, the phosphatase is in blue and kinase is in yellow. The catalytic Asp residues in the phosphatase and kinase (Asp 170 and Asp 396, respectively) are shown in pink. The P-loop in the kinase is shown in navy, and the sulfate ion bound in the P-loop is represented by red and orange spheres. The red arrow points to the phosphopeptide binding region of the FHA domain. (B) XRCC4 phosphopeptide (green) bound to the mouse PNK FHA. The superposition of 3 independent molecules of FHA in the crystal (A, B and C) is presented to illustrate the conformational variability of R44 due to crystal packing. The phosphopeptide from complex A is shown. (C) The kinase active site. The P-loop is shown in green with the bound sulfate (S1). Also shown is W1, the water molecule bound to the catalytic Asp 396 and a sulfate (S2) bound at the DNA binding site. W1 and S2 are proposed to mimic the substrate 5’-OH and a backbone phosphate, respectively. (D) A model of the minimal preferred kinase substrate bound to the kinase active site. The substrate is an 8-bp DNA duplex with a 5-nucleotide 3’ overhang. The PNK kinase domain is shown as a surface, colored by charge (positive in blue, negative in red). (E) The active site of mouse PNK phosphatase, overlaid with active sites of T4 PNK (purple, PDB ID 1LTQ)), phosphoserine phosphatase BeF3/Mg2+ adduct (pink, PDB ID 1J97) and the phosphoaspartate form of β-phosphoglucomutase (green, PDB ID 1LVH). Residue numbering is for mPNK. (D) PNK phosphatase, in charged surface representation, with a modeled bound substrate, pC4p).
We can infer much about the substrate binding and catalytic mechanism of mammalian PNK from a structural comparison to its homolog, phage T4 PNK, for which significantly more structural and functional information is available. The bacteriophage T4 uses its PNK, also a bifunctional kinase and phosphatase, to repair its tRNA after damage by bacterial defense systems [54]. While superficially the structures of mammalian and T4 PNKs differ, their functions and active sites share remarkable similarities. In T4 PNK, the positions of the kinase and phosphatase domains are reversed in sequence relative to mammalian PNK. However, in the three-dimensional structure the two domains have a comparable relative orientation in the two enzymes. Secondly, the biological unit of T4 PNK is a tetramer, while for mPNK it is a monomer [55, 56]. Finally, the kinase portions of the two enzymes have different substrate specificities, as described below.
Mammalian PNK contains three distinct domains with different functions that potentially could be targeted for drug design. The structural and functional details of these domains and the prospects for the development of specific inhibitors for each are outlined in the following sections.
The FHA domain
The FHA (forkhead-associated) domain is a phosphothreonine-binding signaling module that is found in a variety of proteins, including protein kinases ChK2 and Rad53. Of all the known FHA domains, the FHA domain of PNK shares the highest sequence similarity with aprataxin, a product of the APTX gene mutated in the neurological disorder ataxia-oculomotor apraxia, that is involved in SSB repair [57–61]. In PNK the FHA domain serves a targeting role, directing PNK to the site of DNA damage by binding to phosphorylated XRCC1 and XRCC4, key components of the BER and NHEJ pathways, respectively.
The structure of the isolated FHA domain of mouse PNK in complex with a phosphopeptide derived from XRCC4 (Ac-YDES(pT)DEESEKK-CONH2) has been determined (Fig. 6A and B) [48]. The FHA domain is folded into a β-sandwich consisting of a 3-stranded and a 4-stranded anti-parallel β-sheets. The phosphopeptide is bound by 2 loops on one edge of the β-sandwich.
The structures of a number of FHA domains have been elucidated, some in complex with ligands [62–65]. All FHA domains that have been structurally characterized to date share a common mode of binding to the phosphothreonine. They also exhibit additional selectivity for another portion of the phosphopeptide, generally determined by the residue at pT+3 position. PNK FHA, however, recognizes negatively charged residues N-terminal to the pT. Whereas in Rad53 and Chk2 the pT+3 side chain fits into a pocket that is complementary to it in size and shape, in PNK there appears to be a more diffuse electrostatic interaction between the negatively charged peptide and a positively charged surface of the FHA (Fig. 6B). Arg 44 and Arg 48 constitute the positively charged patch, in which the negatively-charged peptide is cradled. Mutation of either of these two residues was found to eliminate binding of the phosphopeptide to the FHA domain of PNK. Arg 48 is also involved in ionic interaction with the phosphothreonine, and its position is fixed. Since Arg 44 is on the surface, it has more conformational freedom. In the crystal structure its side chain samples different orientations due to crystal packing effects. One out of the three FHA molecules in the asymmetric unit does not make any symmetry contacts in the peptide-binding region, and its Arg 44 points toward Glu (pT-2) in the phosphopeptide (molecule C in Fig. 6B). In this complex the FHA does not make any contacts with peptide residues C-terminal to the phosphothreonine. In fact, this part of the peptide is disordered and undefined in the structure [48].
Since FHA binding to the phosphorylated XRCC4 or XRCC1 directs PNK to the site of DNA damage, disruption of this interaction will interfere with multiple DNA repair pathways, and could thereby sensitize cells to DNA-targeting therapies. The fact that the mPNK FHA is structurally distinct from other FHA domains offers hope that it may be possible to design inhibitors that are highly selective for mPNK, but not for other FHA domains. An important exception however is the aprataxin FHA, which is very similar at the amino acid sequence level. While the structure of APTX has not yet been determined, experimental results suggest that the pT+3 position may be important for XRCC1 binding to APTX [66]. Analysis of the APTX sequence in light of the PNK FHA structure reveals that Lys 75 of APTX may be at the position corresponding to Pro 81 of PNK, placing a positive charge within electrostatic interaction distance of a modeled pT+3 residue of the phosphopeptide. Thus it may prove possible to design inhibitors that differentiate between the FHA interactions of PNK and APTX.
The kinase domain
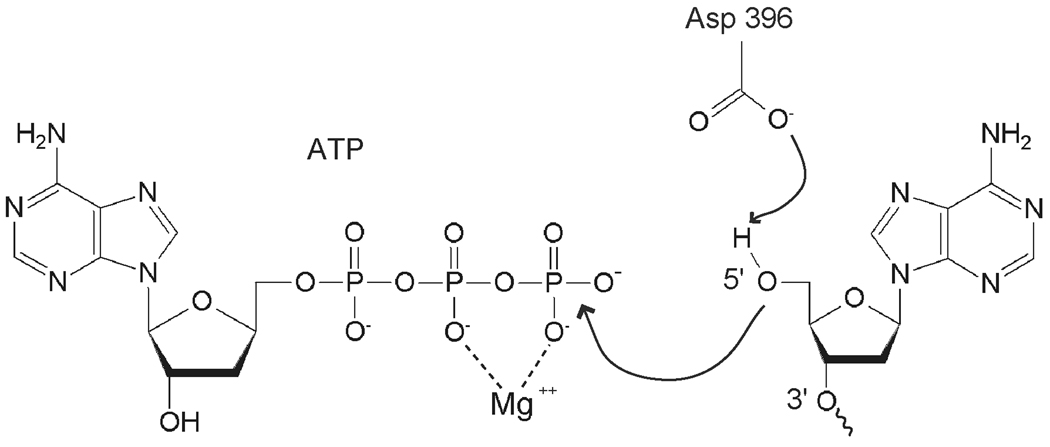
The kinase in PNK catalyzes transfer of the γ-phosphate of an ATP molecule to the 5’-OH of the DNA substrate. It does so by a sequential mechanism in which the enzyme binds both the substrate and ATP in a ternary complex before releasing the phosphorylated product and ADP (Fig. 7) [67]. By analogy with phage T4 PNK, Asp 396 is believed to provide general base assistance by activating the 5’-OH for nucleophilic attack on the ATP γ-phosphate. As with most processes involving ATP, Mg2+ is required for this reaction. The kinase domain of PNK has an adenylate kinase fold (Fig. 6A and C). The active site is located in a long cleft with the ATP binding site at one end and the DNA binding site at the other. The ATP binding site comprises the P-loop or Walker A motif (371GFPGAGKS378), the Walker B motif (Asp421 at the C-terminus of a hydrophobic β-strand) and the α-helical lid subdomain. The lid, consisting of α-helices 12 and 13, folds over the P-loop, sequestering the ATP in the active site. In the crystal structure of T4 PNK, the active site contains ADP formed by hydrolysis of ATP in the crystallization solution. Mammalian PNK was crystallized without nucleotide at the active site, but a sulfate ion from the crystallization medium is bound by the P-loop, in the position that corresponds to the ATP β-phosphate. The sulfate in PNK forms hydrogen bonds with the P-loop and with Arg 463 from helix 12. In order for ATP to bind, however, some structural rearrangement may be required. When the T4 and mammalian PNK structures are superimposed, the Arg 463 side chain of mPNK has a steric clash with the ribose ring of ADP.
Figure 7.
Mechanism of 5’ DNA phosphorylation catalyzed by the PNK kinase domain. The role of the catalytic Asp 396 (Asp 397 in human PNK) is highlighted.
The DNA binding site, deduced by comparison of mPNK and T4 PNK structures, is located at the other end of the active site cleft (Fig. 6C). Whereas mPNK and T4 PNK most likely utilize a very similar mode of ATP binding, their methods of DNA binding likely differ, reflecting their contrasting substrate preferences. mPNK is selective for DNA, and prefers recessed 5’-OH ends with a 3’ overhang of at least 4 nucleotides [27, 48]. T4 PNK, on the other hand, is more promiscuous, acting on DNA, RNA and nucleotide substrates. Unlike mammalian PNK, which prefers the more occluded recessed 5’ ends, T4 PNK prefers single-stranded polynucleotide substrates [68].
We proposed a model for the substrate binding to the kinase active site (Fig. 6D). This model takes into account various experimental results and the structures of T4 PNK bound to several single-stranded DNA substrates [27, 32, 48, 68]. Our kinase activity assays helped to define the minimal preferred kinase substrate as an 8-base pair DNA duplex with a 5’-OH group recessed by more than 3 nucleotides. In the model, the 5’-OH forms a hydrogen bond with the catalytic Asp 396. The first backbone phosphate group (3’ to the reactive 5’-OH) is located in a position similar to that observed in T4 PNK. This phosphate group is within hydrogen bonding distance of the conserved Thr 423. The corresponding residue in T4 PNK, Thr 86, together with Arg 38, form a binding pocket for the first backbone phosphate. Mutation of Thr 86 in T4 PNK does not cause any loss of kinase activity [68, 69], but Arg 38 is acutely sensitive to substitution, even by lysine [70]. Thus it would appear that in T4 PNK Arg 38 makes a stronger contribution to substrate binding than Thr 86. While mammalian PNK lacks a direct counterpart of Arg 38 the guanidinium group of Arg 395 is positioned such that it might make ionic interactions with a phosphate group in contact with Thr 423. In the crystal structure, a sulfate ion from the crystallization medium is bound between Arg 395 and the partially positively charged edge of the indole ring of Trp 401. Trp 401, in turn, is held in position by cation-π stacking with the guanidinium moiety of Arg 432. In modeling substrate binding, we hypothesized that the bound sulfate mimics the second phosphate in the DNA chain. Beyond this point, the modeled DNA substrate does not contact the protein until backbone phosphates at positions 6–8 on the complementary strand to the one bearing the reactive 5’-OH interact with Arg 403 on the protein surface. Intriguingly, this proposed protein-DNA contact is consistent with the minimal length of DNA substrate required by mammalian PNK [27, 48]. On the other side of the 5’-OH, the single-stranded DNA side, 4 backbone phosphate groups of the 3’ overhang can be placed in the vicinity of Arg 482 and Lys 483, which together form a positive surface patch. This may explain the minimal length requirement for the 3’ overhang.
Kinases constitute a large and highly diverse enzyme family and inhibitors could, in theory, also interact with other kinases. We have used protein structure similarity searches to identify those kinases which are most similar to PNK, which might be most likely to be targeted by PNK kinase inhibitors. The most similar kinases identified in a DALI [71] search include 6-phosphofructo-2-kinase (PFK-2), an enzyme involved in the regulation of glycolysis, and uridine/cytidine kinase (UCK), which functions in the pyrimidine salvage pathway. Although these enzymes have a common fold and share significant structural resemblance to PNK at the ATP binding site, their substrate binding sites differ. The substrate binding site in PNK provides a more polar milieu than those of PFK-2 or uridine/cytidine kinase. The 431DNTN motif in PNK, where the threonine OH points into the putative substrate binding pocket is replaced by EGIL in UCK. The residue that aligns with threonine is the β-branched, but bulkier and more hydrophobic isoleucine. Further, PFK-2 lacks the general base (Asp 396 in PNK), and its reaction is believed to proceed through a dissociative transition state, where the developing negative charge during ATP hydrolysis is stabilized by electrostatic interaction with a lysine (174) [72]. In short, the kinase active site of mammalian PNK appears to be structurally unique in the DNA binding region, which suggests that it should be possible to design inhibitors that are highly selective for this enzyme.
The phosphatase domain
The phosphatase domain of PNK catalyzes the dephosphorylation of the 3’ terminus of DNA. This domain belongs to the haloacid dehalogenase (HAD) superfamily, members of which catalyze a range of reactions, including phosphate transfer (phosphoserine phosphatase (PSP), β-phosphoglucomutase (β-PGM), Ca++ ATPase, RNA polymerase II carboxy terminal domain (CTD) phosphatase), phosphonate transfer (phosphonatases) and dehalogenation (haloacid dehalogenase) [73]. The HAD architecture consists of an α/β domain with a Rossmann fold (Fig. 6A). A comparison of available HAD phosphotransferase structures reveals a strong conservation of active site residues involved in phosphate binding and catalysis. The active site is located in a deep pocket in the HAD α/β domain (Fig. 6E). Despite a generally low sequence similarity, the HAD phosphotransferase members can be identified by the conserved motif Dx(D/T)x(T/V), where the first Asp (D170 in mPNK) participates in covalent catalysis. Additional conserved residues include a serine or threonine (T216), a lysine (K259) and aspartates 288 and 309. Structural differences among the HAD phosphotransferases are generally confined to the capping structure, which binds the rest of the substrate, determining substrate specificity. In phosphoserine phosphatase, for example, the capping structure is a flexible helical domain, which encloses the small bound substrate, phospho-l-serine, in the active site. In CTD phosphatase, which dephosphorylates phosphoserine in the context of a protein chain, the cap is much smaller, and the active site is more accessible. In both T4 and mPNK, which work on quite large substrates in vivo, the capping structures consist of loops, leaving the active site deep, but open.
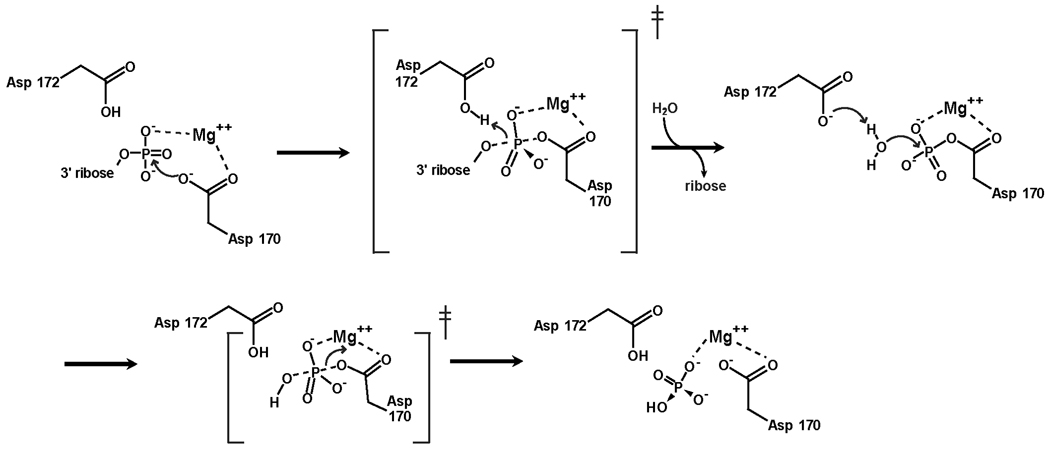
From structural and functional studies of various HAD-related phosphotransferases, the mechanism for phosphate transfer has been elucidated, and is presented here for PNK (Fig. 8). The reaction, which requires Mg2+, proceeds through a phosphoaspartate intermediate. The phosphate group of the substrate is stabilized at the active site by contacts with Lys 259, Thr 216 and Mg2+ (Figs. 6E and 8). The phosphate is transferred to Oδ1 of Asp 170 in an in-line nucleophilic attack, producing the phosphoaspartate intermediate and releasing the leaving group. During this reaction, Asp 172 may provide general acid assistance by protonating the alcohol leaving group. The phosphoaspartate intermediate has been observed in β-PGM and in PSP (as a BeF3 adduct) [74, 75], and can therefore be envisioned for mPNK (Fig. 6E). In this intermediate, the phosphate interacts with the side chains Lys 269 and Thr 216, with backbone NH of Leu 171 and Asp 172 and with the Mg2+. The metal ion is coordinated by a phosphate oxygen atom, Asp 170 Oδ2, Asp 172 O, Asp 288 Oδ2 and two water molecules. Asp 309 helps to maintain the configuration of the active site: its Oδ1 forms hydrogen bonds with backbone NH of Leu 171 and Asp 172, while its Oδ2 forms a salt bridge with Nζ of the conserved Lys 259. The precise location of this carboxylate is crucial, as mutation of the corresponding Asp (218) in the S. cerevisiae homolog, Tpp1, to Glu severely impairs phosphatase activity [76]. In T4 PNK and in β-PGM the carboxylate group at this position is contributed by an Asp from a different part of the peptide chain (Asp 277 in T4 PNK). In the last step, the phosphoaspartate is hydrolyzed, releasing phosphate and regenerating the free enzyme. Again, Asp 172 provides general base assistance, activating a water molecule to serve as a nucleophile in an SN2 attack on the phosphorus atom. The role of Asp172 is interesting. This aspartate is conserved in all the phosphatases and phosphomutases, where the leaving group is an alcohol. In ATPases, where the ADP leaving group does not require protonation, the corresponding residue is threonine [74]. In phosphonoacetaldehyde hydrolase, which catalyzes P–C bond cleavage, the corresponding residue is an alanine, and the leaving group is stabilized by a lysine via Schiff-base formation [77]. In mPNK, T4 PNK and PSP, this aspartate is found in a conformation appropriate for activation of the nucleophilic water, where it interacts with Gln 218 Nε2, Arg 213 Nε and Nη1, and Thr 21 N, respectively. In β-PGM, which has an unusually stable phosphoaspartate intermediate, the side chain of this aspartate is rotated by ∼60° in χ1 away from the phosphate due to a steric clash with the cap domain. This Asp is thus unable to assist in phosphoaspartate hydrolysis (Fig. 6E) [74]. Substrate binding induces a conformational change in the cap domain, allowing the Asp to assume the reactive position.
Figure 8.
Mechanism of 3’ DNA dephosphorylation carried out by the PNK phosphatase domain. The mechanism is predicted to proceed via a covalent phospho-aspartate intermediate involving Asp 170. Putative transition states are indicated by ‡.
The PNK phosphatase is much less strict in its substrate preference than the kinase, as it can accept single and double stranded DNA substrates, including nicks and gaps, with equal efficiency [48]. The phosphatase appears to work most effectively on substrates that possess at least 4 backbone phosphates (including the 3’-phosphate). We modeled a single-stranded DNA substrate in the phosphatase active site, and found that up to three backbone phosphates of the minimal substrate can bind at a positive patch near the rim of the active site, formed by Arg 258 and Arg 223 (Fig. 6F). For longer substrates, the DNA chain may not make any additional protein contacts, and thus no improvement in activity is observed. At least one phosphate from the substrate must be bound at the positive patch, since 3’-monophosphodeoxynucleotides are not substrates for the phosphatase, while nucleotides which place a phosphate at the positive patch, dinucleotides and 3’,5’-diphosphodeoxynucleotides are weak substrates.
The active site appears to be too narrow to fit a double-stranded substrate, but 3’-dephosphorylation proceeds at equal rates in single-stranded oligonucleotides as in nicked or gapped substrates. These observations suggest that either several base pairs of double-stranded DNA are unwound, or that the active site pocket, which is delineated by loops, may be flexible enough to accommodate a bulkier double-stranded substrate.
The rather unique characteristics of the substrate binding pocket of the PNK phosphatase domain, compared to other members of the HAD family, again offer promise that small molecule inhibitors that are highly selective for PNK could be developed.
Cellular inhibition of PNK
From the above discussion it is clear that a number of approaches and targets are available to reduce hPNK activity in cells. These include targeting (a) the complete protein by reducing its expression, (b) the kinase activity, (c) the phosphatase activity and (d) the interaction of hPNK with either XRCC1 or XRCC4.
Reduction of hPNK expression by siRNA
hPNK expression has been successfully downregulated in cultured human lung adenocarcinoma and glioblastoma cells by an siRNA expression vector [45, 52]. This approach could potentially be modified by using a system suitable for therapeutic delivery of the siRNA, either as the oligonucleotide directly or in an expressible form. The former approach is still very much at the development stage. A major problem that needs to be overcome is one of siRNA stability, i.e. protection against nucleases. A number of delivery vehicles are being explored including the use of nanoparticles [78]. The latter approach is also still at an early stage although adenoviral and other viral vectors have been developed for gene therapy and are being modified for use with siRNA. For example, adenoviral vectors expressing siRNAs against matrix metalloproteinase-2 and vascular endothelial growth factor effectively suppressed tumour growth in mice [79, 80].
Small molecule inhibition of hPNK activity
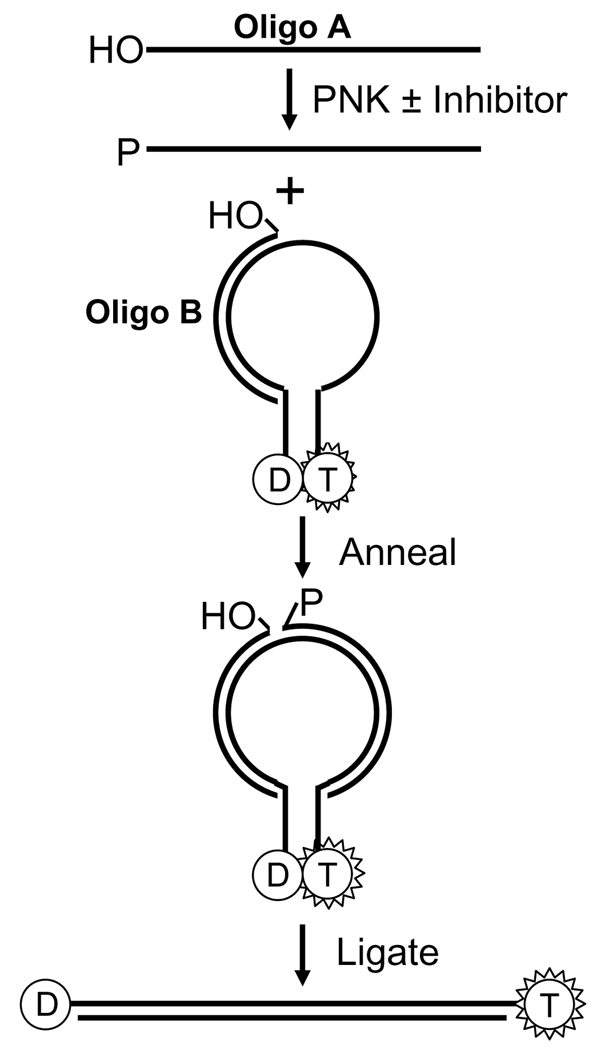
Small molecule inhibitors have now been generated for several DNA repair proteins, most notably O6-methylguanine DNA methyltransferase, poly(ADP-ribose) polymerase, DNA-PK and APE1 [81–84]. At present, no small molecule inhibitors have been synthesized for hPNK. Both enzymatic activities are potential targets, although there is an indication that the phosphatase activity may take precedence over the kinase activity [53]. The first step is the development of an assay suitable for high-throughput screening of chemical libraries. One such assay has recently been described for the kinase activity of PNKs based on molecular beacon technology [85]. The assay (Fig. 9) involves the phosphorylation of an oligonucleotide, which is then hybridized to part of a stem loop DNA structure. The stem termini of the stem loop structure carry a tetramethyl rhodamine (TMR) and a dabcyl group, which are held in close juxtaposition by the stem structure and as a result the fluorescence of the TMR group is quenched by the dabcyl group. A second oligonucleotide already annealed to the remaining section of the stem loop is enzymatically ligated to the phosphorylated oligonucleotide, which forces the stem loop to linearize, thus separating the dabcyl group from the TMR resulting in a substantial increase in the fluorescence signal. Of course, one of the drawbacks of this approach is the dependence on a DNA ligase and the potential for small molecule inhibition of this enzyme rather than PNK. Presumably this assay could be modified to screen for inhibitors of the phosphatase activity of PNK. However, several other phosphatase assays are available. Most have been developed for protein phosphatases and fall into two types; those that follow the release of inorganic phosphate using dyes such as malachite green in a colorimetric assay [86] or those that make use of fluorogenic substrates such as 6,8-difluoro-4-methylumbiliferyl phosphate [87]. To this point no fluorogenic substrate has been identified or developed for the phosphatase activity of PNK.
Figure 9.
Scheme showing the assay for the kinase activity of PNK using a molecular beacon approach. Oligo A is 5’-phosphorylated by PNK and then annealed to the stem loop construct in which the fluorescence of the tetramethyl rhodamine dye (T) is quenched by the proximity of the dabcyl group (D). Ligation of oligo A to oligo B, which can only occur if oligo A is phosphorylated, forces the opening of the stem loop structure and enhancement of the fluorescence signal. (Adapted from reference [82]).
In addition to the enzymatic activities of hPNK, another potential target is the binding site of hPNK in the FHA domain for phosphorylated XRCC1 or XRCC4 [37,43,48]. Although phosphopeptides based on the respective binding sites of these proteins may potentially act as small molecule inhibitors themselves, their relatively large size (∼7 kDa) may reduce their cellular uptake. Nonpeptidic small molecule inhibitors may however be developed to disrupt the protein-protein interactions. For example, fluorescein-labeled phosphopeptides have been used in fluorescence polarization-based binding assays to identify members of small molecule chemical libraries that inhibit phosphopeptide interactions with SH2 [88, 89] and BRCT domains [90]. A similar approach could be used to identify small molecule inhibitors of hPNK.
Conclusion
The concept of targeting DNA repair enzymes in order to enhance the potency of radiation and antineoplastic chemotherapeutic agents is now well established. The involvement of hPNK in several key DNA repair pathways, especially those required by cells to resist the genotoxic effects of ionizing radiation and topoisomerase I inhibitors, suggests that this enzyme may be a useful target for inhibition. This objective will be facilitated by our knowledge of the structure and function of the enzyme.
Acknowlegements
We would like to acknowledge Drs. Sankar Mitra and Lee Wiederhold (UTMB, Galveston) for useful discussions and Mesfin Fanta for technical assistance.
Financial support for this work was provided by grants from the Canadian Institutes of Health Research, the Alberta Cancer Board and the National Cancer Institute of Canada.
References
- 1.Gatti L, Zunino F. Methods Mol. Med. 2005;111:127. doi: 10.1385/1-59259-889-7:127. [DOI] [PubMed] [Google Scholar]
- 2.Madhusudan S, Middleton MR. Cancer Treat. Rev. 2005;31:603. doi: 10.1016/j.ctrv.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Murray D, Begg AC. In: In DNA Repair in Cancer Therapy. Panasci LC, Alaoui-Jamali MA, editors. Totowa: Humana Press; 2004. pp. 211–256. [Google Scholar]
- 4.Ding J, Miao ZH, Meng LH, Geng MY. Trends Pharmacol. Sci. 2006;27:338. doi: 10.1016/j.tips.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez-Perez I. Clin. Transl. Oncol. 2006;8:642. doi: 10.1007/s12094-006-0034-8. [DOI] [PubMed] [Google Scholar]
- 6.Von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair. Berlin: Springer-Verlag; 2006. [Google Scholar]
- 7.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. J. Biol. Chem. 1983;258:711. [PubMed] [Google Scholar]
- 8.Buchko GW, Weinfeld M. Biochemistry. 1993;32:2186. doi: 10.1021/bi00060a009. [DOI] [PubMed] [Google Scholar]
- 9.Jones GD, Weinfeld M. Cancer Res. 1996;56:1584. [PubMed] [Google Scholar]
- 10.Coquerelle T, Bopp A, Kessler B, Hagen U. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1973;24:397. doi: 10.1080/09553007314551251. [DOI] [PubMed] [Google Scholar]
- 11.Lennartz M, Coquerelle T, Bopp A, Hagen U. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1975;27:577. doi: 10.1080/09553007514550611. [DOI] [PubMed] [Google Scholar]
- 12.Ward JF. Int. J. Radiat. Biol. 1994;66:427. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Proc. Natl. Acad. Sci. U S A. 2000;97:103. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dianov GL, O’Neill P, Goodhead DT. Bioessays. 2001;23:745. doi: 10.1002/bies.1104. [DOI] [PubMed] [Google Scholar]
- 15.Meer L, Janzer RC, Kleihues P, Kolar GF. Biochem. Pharmacol. 1986;35:3243. doi: 10.1016/0006-2952(86)90419-3. [DOI] [PubMed] [Google Scholar]
- 16.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Cancer Treat. Rev. 1997;23:35. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 17.Dolan ME, Pegg AE. Clin. Cancer Res. 1997;3:837. [PubMed] [Google Scholar]
- 18.Lown JW, McLaughlin LW. Biochem. Pharmacol. 1979;28:1631. doi: 10.1016/0006-2952(79)90176-x. [DOI] [PubMed] [Google Scholar]
- 19.Povirk LF. Mutat. Res. 1996;355:71. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 20.Claussen CA, Long EC. Chem. Rev. 1999;99:2797. doi: 10.1021/cr980449z. [DOI] [PubMed] [Google Scholar]
- 21.Porter SE, Champoux JJ. Nucleic Acids Res. 1989;17:8521. doi: 10.1093/nar/17.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markovits J, Pommier Y, Mattern MR, Esnault C, Roques BP, Le Pecq JB, Kohn KW. Cancer Res. 1986;46:5821. [PubMed] [Google Scholar]
- 23.Pommier Y. Nat. Rev. Cancer. 2006;6:789. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 24.Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti MA, Patchan MW, Thompson RB, Osheroff N. J. Biol. Chem. 1996;271:29238. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 25.Pheiffer BH, Zimmerman SB. Biochem. Biophys. Res. Commun. 1982;109:1297. doi: 10.1016/0006-291x(82)91918-0. [DOI] [PubMed] [Google Scholar]
- 26.Habraken Y, Verly WG. Eur. J. Biochem. 1988;171:59. doi: 10.1111/j.1432-1033.1988.tb13758.x. [DOI] [PubMed] [Google Scholar]
- 27.Karimi-Busheri F, Weinfeld M. J. Cell. Biochem. 1997;64:258. [PubMed] [Google Scholar]
- 28.Friedman ECWGC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 29.Chen DS, Herman T, Demple B. Nucleic Acids Res. 1991;19:5907. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. Mol. Cell. 2004;15:209. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. DNA Repair (Amst) 2006;5:1439. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimi-Busheri F, Lee J, Tomkinson AE, Weinfeld M. Nucleic Acids Res. 1998;26:4395. doi: 10.1093/nar/26.19.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. J. Biol. Chem. 1999;274:24176. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 34.Karimi-Busheri F, Daly G, Robins P, Canas B, Pappin DJ, Sgouros J, Miller GG, Fakhrai H, Davis EM, Le Beau MM, Weinfeld M. J. Biol. Chem. 1999;274:24187. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 35.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. Cell. 2001;104:107. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 36.Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC. EMBO J. 2002;21:2827. doi: 10.1093/emboj/21.11.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. EMBO J. 2004;23:3874. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demple B, DeMott MS. Oncogene. 2002;21:8926. doi: 10.1038/sj.onc.1206178. [DOI] [PubMed] [Google Scholar]
- 39.Suh D, Wilson DM, Povirk LF., 3rd Nucleic Acids Res. 1997;25:2495. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaudhry MA, Dedon PC, Wilson DM, Demple B, Weinfeld M., 3rd Biochem. Pharmacol. 1999;57:531. doi: 10.1016/s0006-2952(98)00327-x. [DOI] [PubMed] [Google Scholar]
- 41.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. J. Biol. Chem. 2002;277:27162. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 42.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. DNA Repair (Amst) 2003;2:1087. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 43.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW. Cell. 2004;117:17. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 44.Parsons JL, Dianova, Boswell E, Weinfeld M, Dianov GL. FEBS J. 2005;272:5753. doi: 10.1111/j.1742-4658.2005.04962.x. [DOI] [PubMed] [Google Scholar]
- 45.Karimi-Busheri F, Rasouli.l-Nia A, Allalunis-Turner J, Weinfeld M. Cancer Res. 2007;67:6619. doi: 10.1158/0008-5472.CAN-07-0480. [DOI] [PubMed] [Google Scholar]
- 46.Sekiguchi JM, Ferguson DO. Cell. 2006;124:260. doi: 10.1016/j.cell.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Lees-Miller SP, Meek K. Biochimie. 2003;85:1161. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 48.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, Durocher D, Weinfeld M, Glover JN. Mol. Cell. 2005;17:657. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Audebert M, Salles B, Calsou P. J. Biol. Chem. 2004;279:55117. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 50.Audebert M, Salles B, Weinfeld M, Calsou P. J. Mol. Biol. 2006;356:257. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 51.Meijer M, Karimi-Busheri F, Huang TY, Weinfeld M, Young D. J. Biol. Chem. 2002;277:4050. doi: 10.1074/jbc.M109383200. [DOI] [PubMed] [Google Scholar]
- 52.Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Proc. Natl. Acad. Sci. U S A. 2004;101:6905. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobson CJ, Allinson SL. Nucleic Acids Res. 2006;34:2230. doi: 10.1093/nar/gkl275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amitsur M, Levitz R, Kaufmann G. EMBO J. 1987;6:2499. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lillehaug JR. Eur. J. Biochem. 1977;73:499. doi: 10.1111/j.1432-1033.1977.tb11343.x. [DOI] [PubMed] [Google Scholar]
- 56.Mani RS, Karimi-Busheri F, Cass CE, Weinfeld M. Biochemistry. 2001;40:12967. doi: 10.1021/bi011383w. [DOI] [PubMed] [Google Scholar]
- 57.Rass U, Ahel I, West SC. J. Biol. Chem. 2007 doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 58.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. Nature. 2006;443:713. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 59.Hirano M, Yamamoto A, Mori T, Lan L, Iwamoto TA, Aoki M, Shimada K, Furiya Y, Kariya S, Asai H, Yasui A, Nishiwaki T, Imoto K, Kobayashi N, Kiriyama T, Nagata T, Konishi N, Itoyama Y, Ueno S. Ann. Neurol. 2007;61:162. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- 60.Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, Lavin MF. Hum. Mol. Genet. 2004;13:1081. doi: 10.1093/hmg/ddh122. [DOI] [PubMed] [Google Scholar]
- 61.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. Nat. Genet. 2001;29:189. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 62.Mahajan A, Yuan C, Pike BL, Heierhorst J, Chang CF, Tsai MD. J. Am. Chem. Soc. 2005;127:14572. doi: 10.1021/ja054538m. [DOI] [PubMed] [Google Scholar]
- 63.Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Mol. Cell. 2002;9:1045. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 64.Stavridi ES, Huyen Y, Loreto IR, Scolnick DM, Halazonetis TD, Pavletich NP, Jeffrey PD. Structure. 2002;10:891. doi: 10.1016/s0969-2126(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 65.Liao H, Yuan C, Su MI, Yongkiettrakul S, Qin D, Li H, Byeon IJ, Pei D, Tsai MD. J. Mol. Biol. 2000;304:941. doi: 10.1006/jmbi.2000.4291. [DOI] [PubMed] [Google Scholar]
- 66.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, Mu JJ, Chen D, Songyang Z, Wang Y, Qin J. Mol. Cell. Biol. 2004;24:8356. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mani RS, Karimi-Busheri F, Fanta M, Cass CE, Weinfeld M. Biochemistry. 2003;42:12077. doi: 10.1021/bi030127b. [DOI] [PubMed] [Google Scholar]
- 68.Eastberg JH, Pelletier J, Stoddard BL. Nucleic Acids Res. 2004;32:653. doi: 10.1093/nar/gkh212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang LK, Shuman S. J. Biol. Chem. 2001;276:26868. doi: 10.1074/jbc.M103663200. [DOI] [PubMed] [Google Scholar]
- 70.Wang LK, Shuman S. Nucleic Acids Res. 2002;30:1073. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holm L, Sander C. Trends Biochem. Sci. 1995;20:478. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 72.Rider MH, Bertrand L, Vertommen D, Michels PA, Rousseau GG, Hue L. Biochem. J. 2004;381:561. doi: 10.1042/BJ20040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lahiri SD, Zhang G, Dai J, Dunaway-Mariano D, Allen KN. Biochemistry. 2004;43:2812. doi: 10.1021/bi0356810. [DOI] [PubMed] [Google Scholar]
- 74.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. Biochemistry. 2002;41:8351. doi: 10.1021/bi0202373. [DOI] [PubMed] [Google Scholar]
- 75.Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, Wemmer D, Kustu S, Yan D. Proc. Natl. Acad. Sci. U S A. 2001;98:8525. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deshpande RA, Wilson TE. Biochemistry. 2004;43:8579. doi: 10.1021/bi049434n. [DOI] [PubMed] [Google Scholar]
- 77.Morais MC, Zhang W, Baker AS, Zhang G, Dunaway-Mariano D, Allen KN. Biochemistry. 2000;39:10385. doi: 10.1021/bi001171j. [DOI] [PubMed] [Google Scholar]
- 78.Toub N, Malvy C, Fattal E, Couvreur P. Biomed. Pharmacother. 2006;60:607. doi: 10.1016/j.biopha.2006.07.093. [DOI] [PubMed] [Google Scholar]
- 79.Chetty C, Bhoopathi P, Joseph P, Chittivelu S, Rao JS, Lakka S. Mol. Cancer Ther. 2006;5:2289. doi: 10.1158/1535-7163.MCT-06-0169. [DOI] [PubMed] [Google Scholar]
- 80.Yoo JY, Kim JH, Kwon YG, Kim EC, Kim NK, Choi HJ, Yun CO. Mol. Ther. 2007;15:295. doi: 10.1038/sj.mt.6300023. [DOI] [PubMed] [Google Scholar]
- 81.Dolan ME, Moschel RC, Pegg AE. Proc. Natl. Acad. Sci. U S A. 1990;87:5368. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ratnam K, Low JA. Clin. Cancer Res. 2007;13:1383. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 83.Hardcastle IR, Cockcroft X, Curtin NJ, El-Murr MD, Leahy JJ, Stockley M, Golding BT, Rigoreau L, Richardson C, Smith GC, Griffin RJ. J. Med. Chem. 2005;48:7829. doi: 10.1021/jm050444b. [DOI] [PubMed] [Google Scholar]
- 84.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, Dianov GL, Hickson ID. Nucleic Acids Res. 2005;33:4711. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang Z, Wang K, Tan W, Ma C, Li J, Liu L, Guo Q, Meng X. Nucleic Acids Res. 2005;33:e97. doi: 10.1093/nar/gni096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cogan EB, Birrell GB, Griffith OH. Anal. Biochem. 1999;271:29. doi: 10.1006/abio.1999.4100. [DOI] [PubMed] [Google Scholar]
- 87.Welte S, Baringhaus KH, Schmider W, Muller G, Petry S, Tennagels N. Anal. Biochem. 2005;338:32. doi: 10.1016/j.ab.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 88.Schust J, Berg T. Anal. Biochem. 2004;330:114. doi: 10.1016/j.ab.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 89.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Chem. Biol. 2006;13:1235. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 90.Lokesh GL, Rachamallu A, Kumar GD, Natarajan A. Anal. Biochem. 2006;352:135. doi: 10.1016/j.ab.2006.01.025. [DOI] [PubMed] [Google Scholar]