Abstract
The facultative methylotroph Methylobacterium extorquens AM1 possesses two pterin-dependent pathways for C1 transfer between formaldehyde and formate, the tetrahydrofolate (H4F)-linked pathway and the tetrahydromethanopterin (H4MPT)-linked pathway. Both pathways are required for growth on C1 substrates; however, mutants defective for the H4MPT pathway reveal a unique phenotype of being inhibited by methanol during growth on multicarbon compounds such as succinate. It has been previously proposed that this methanol-sensitive phenotype is due to the inability to effectively detoxify formaldehyde produced from methanol. Here we present a comparative physiological characterization of four mutants defective in the H4MPT pathway and place them into three different phenotypic classes that are concordant with the biochemical roles of the respective enzymes. We demonstrate that the analogous H4F pathway present in M. extorquens AM1 cannot fulfill the formaldehyde detoxification function, while a heterologously expressed pathway linked to glutathione and NAD+ can successfully substitute for the H4MPT pathway. Additionally, null mutants were generated in genes previously thought to be essential, indicating that the H4MPT pathway is not absolutely required during growth on multicarbon compounds. These results define the role of the H4MPT pathway as the primary formaldehyde oxidation and detoxification pathway in M. extorquens AM1.
Methylotrophic bacteria growing aerobically on single-carbon (C1) substrates produce formaldehyde as a central intermediate. A key challenge for these organisms is how to maximize the flux through formaldehyde while preventing the intracellular pool of free formaldehyde from accumulating to toxic levels. It has been suggested for a typical methylotroph that the cytoplasmic formaldehyde concentration could rise to 100 mM in less than 1 min if formaldehyde consumption stopped completely (3, 33). Additionally, methylotrophs utilizing multicarbon compounds need to maintain the ability to detoxify formaldehyde that may be produced from the cometabolism of C1 substrates encountered in the environment. A number of cofactor-dependent formaldehyde oxidation pathways are present in various methylotrophs that have the potential to carry out this function, and many organisms possess more than one of these pathways (30).
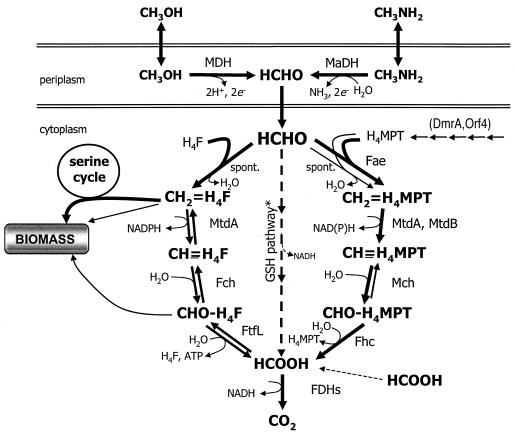
In the facultative methylotroph Methylobacterium extorquens AM1, primary oxidation of C1 substrates such as methanol or methylamine occurs in the periplasm through the action of methanol dehydrogenase (1) and methylamine dehydrogenase (8) (Fig. 1). Formaldehyde that enters the cytoplasm condenses with one of two pterin cofactors, tetrahydrofolate (H4F) or tetrahydromethanopterin (H4MPT), to form the respective methylene derivatives. The reaction of formaldehyde with H4F seems to occur spontaneously (14), and no enzyme has been found thus far that is capable of accelerating this reaction (33). Methylene-H4F can either be assimilated through the serine cycle or may be oxidized to methenyl-H4F, formyl-H4F, and ultimately formate (reviewed in reference 17). Formate can then be oxidized to CO2 through the action of formate dehydrogenases (16; L. Chistoserdova and M. E. Lidstrom, unpublished data).
FIG. 1.
Methylotrophic metabolism in M. extorquens AM1. The result of expressing the glutathione (GSH) pathway from P. denitrificans is indicated with the dashed arrows. Those reactions that can occur spontaneously or that are catalyzed by two enzymes are indicated. Arrows pointing in both directions indicate reversible enzymatic reactions. The thin arrows leading from methylene-H4F and formyl-H4F to biomass represent biosynthetic reactions directly involving these two C1-H4F derivatives. Arrows to the right of H4MPT indicate biosynthesis reactions for this cofactor; only the two indicated gene products have been implicated in this process. MDH, methanol dehydrogenase; MaDH, methylamine dehydrogenase; MtdA, NADP-dependent methylene-H4F/methylene-H4MPT dehydrogenase; Fch, methenyl-H4F cyclohydrolase; FtfL, formate-H4F ligase; FDHs, formate dehydrogenases; DmrA, putative dihydromethanopterin reductase; Orf4, putative β-RFAP synthase; Fae, formaldehyde-activating enzyme; MtdB, NAD(P)-dependent methylene-H4MPT dehydrogenase; Mch, methenyl-H4MPT cyclohydrolase; Fhc, formyltransferase/hydrolase complex.
Alternatively, formaldehyde can be oxidized through a similar C1 transfer pathway that is linked to the folate analog H4MPT. H4MPT-dependent C1 transfers were thought to be unique to methanogenic and sulfate-reducing archaea until their unexpected discovery in M. extorquens AM1 (6). Subsequently, this pathway has been found in most gram-negative methylotrophs with a few exceptions (32). In methylotrophic bacteria the initial step in this pathway is the reaction of formaldehyde with H4MPT to form methylene-H4MPT (Fig. 1). While this reaction can occur spontaneously, as is apparently the case for H4F, a specific formaldehyde-activating enzyme (Fae) has been shown to catalyze this condensation, and this enzyme is required for methylotrophic growth (33). The resulting methylene-H4MPT is oxidized to methenyl-H4MPT through the action of one of two methylene-H4MPT dehydrogenases, MtdA and MtdB. MtdA, which also catalyzes the oxidation of methylene-H4F, is strictly NADP dependent (31), whereas MtdB can use either NAD+ or NADP+ but is specific for methylene-H4MPT (10). Methenyl-H4MPT is converted to formyl-H4MPT by methenyl-H4MPT cyclohydrolase (Mch) (24). Finally, formate is released from formyl-H4MPT through the action of the formyltransferase/hydrolase complex (Fhc) (22, 23). Formate can then be oxidized to CO2 by formate dehydrogenase, as for the H4F-linked pathway. Fhc contains formyltransferase activity that is active with methanofuran purified from the methanogen Methanothermobacter marburgensis (23). The identity and function of the methanofuran analog present in M. extorquens AM1 has not yet been determined, however, and so this pathway simply will be referred to here as the H4MPT pathway.
A number of mutants defective for known or suspected H4MPT pathway functions have been generated and, based on their growth phenotype, a function in energy generation during methylotrophic growth has been proposed. Null mutants lacking mtdB (10), fae (33), and dmrA (which encodes a putative dihydromethanopterin reductase [20]) have been reported to be both incapable of growth on methanol and inhibited by either methanol or formaldehyde during growth on succinate. This methanol-sensitive mutant phenotype is thus far unique to the mutants of M. extorquens AM1 defective for the H4MPT pathway and has been proposed to be due to an inability to detoxify the formaldehyde produced from methanol. Double mutants lacking both MDH activity and MtdB activity were no longer sensitive to methanol (10), lending further support to the concept that methanol sensitivity is a proxy for formaldehyde detoxification deficiency. A null mutant lacking Orf4, a homolog of the first enzyme in the H4MPT biosynthesis pathway, β-ribofuranosylaminobenzene 5′-phosphate (β-RFAP) synthase (28), has also been generated and was incapable of growth on methanol (6). However, efforts to obtain null mutants in other genes, such as mtdA (7), mch, and fhcBADC (6), have not been successful. Mutants resulting from an incomplete allelic exchange event that separated a wild-type copy of the gene from its native promoter by the integrated vector were obtained for these genes and, where examined, this led to reduced enzymatic activity (6, 7). In all cases, this class of mutants exhibited defective growth on methanol, indicating a specific role in methylotrophy in addition to apparent essentiality. MtdA activity is likely required to produce formyl-H4F for biosynthetic needs (7). It has not been clear, however, why the other genes for which null mutants could not be obtained and have a known or predicted role in the H4MPT pathway for formaldehyde oxidation would be required for growth on a multicarbon substrate such as succinate. Therefore, the role of the H4MPT pathway has been uncertain, and in this study we have carried out experiments to define that role.
Here we present a comparative physiological analysis of H4MPT pathway mutants, analyzing the methanol-sensitive phenotype in more detail and demonstrating that it is due to formaldehyde production from methanol. In addition, through the complementation of H4MPT pathway mutants with an alternative formaldehyde oxidation system and by demonstrating that the H4MPT pathway is in fact not essential, we have defined the role of the H4MPT pathway as the primary formaldehyde oxidation and detoxification route in M. extorquens AM1.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. extorquens AM1 (21) strains were grown at 30°C on a minimal salts medium (2) containing carbon sources at the following concentrations: 35 mM formate, 125 mM methanol, or 15 mM succinate. Escherichia coli strains were grown on Luria-Bertani medium (27). Antibiotics were added to the following final concentrations: 50 μg of ampicillin/ml, 50 μg of kanamycin/ml, 50 μg of rifamycin/ml, 35 μg of streptomycin/ml, and 10 μg of tetracycline/ml. Chemicals were obtained from Sigma. Nutrient agar and Bacto-agar were obtained from Difco.
Generation of mutant strains.
M. extorquens AM1 deletion mutants lacking mxaF, orf4, mtdB, mch, or the fhcBADC cluster were generated using the allelic exchange vector pCM184 (18). Approximately 0.5-kb regions upstream and downstream of these genes or gene clusters were amplified by PCR and cloned into pCR2.1 (Invitrogen) as follows. Cloning of the mxaF upstream and downstream flanks resulted in pCM191 and pCM192, the orf4 flanks resulted in pCM250 and pCM251, the mtdB flanks resulted in pCM255 and pCM256, the mch flanks resulted in pCM260 and pCM261, and the flank downstream of fhcC resulted in pCM264. The construct to generate ΔmxaF::kan mutants was generated by introducing the 0.5-kb ApaI-SacI fragment from pCM192 between the corresponding sites of pCM184 to produce pCM193 and, subsequently, the 0.6-kb EcoRV-Asp718I fragment from pCM191 was ligated between the PvuII and Asp718I sites of pCM193 to produce pCM194. The construct to generate Δorf4::kan mutants was generated by introducing the 0.5-kb EcoRI-Asp718I fragment from pCM250 into the same sites of pCM184 to produce pCM252 and, subsequently, the 0.7-kb ApaI-SacI fragment from pCM251 was ligated into the same sites of pCM252 to produce pCM253. The construct to generate ΔmtdB::kan mutants was generated by introducing the 0.6-kb SacII-SacI fragment from pCM256 into the same sites of pCM184 to produce pCM257 and, subsequently, the 0.5-kb AatII-Asp718I fragment from pCM255 was ligated into the same sites of pCM257 to produce pCM258. The construct to generate Δmch::kan mutants was generated by introducing the 0.6-kb AatII-Asp718I fragment from pCM260 into the same sites of pCM184 to produce pCM262 and, subsequently, the 0.7-kb ApaI-SacI fragment from pCM261 was ligated into the same sites of pCM262 to produce pCM263. Finally, the construct to generate ΔfhcBADC::kan mutants was generated by introducing the 0.5-kb EcoRI-NcoI fragment from pCM250 into the same sites of pCM184 to produce pCM265 and, subsequently, the 0.5-kb SacII-AgeI fragment from pCM264 was ligated into the same sites of pCM265 to produce pCM266.
Mutant strains of M. extorquens AM1 were generated by introducing the appropriate donor constructs by conjugation from E. coli S17-1 (29) as previously described (4). Unmarked deletion strains were generated using the cre-expressing plasmid pCM157 as described elsewhere (18), allowing the generation of double mutant strains. All mutants were confirmed by diagnostic PCR analysis. All strains and plasmids utilized in this study are described in Table 1.
TABLE 1.
M. extorquens AM1 strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| CM194.1 | ΔmxaF | This study |
| CM194K.1 | ΔmxaF::kan | This study |
| CM194-198K.1 | ΔmxaF Δfae::kan | This study |
| CM194-212K.1 | ΔmxaF ΔdmrA::kan | This study |
| CM194-253K.1 | ΔmxaF Δorf4::kan | This study |
| CM194-258K.1 | ΔmxaF ΔmtdB::kan | This study |
| CM198.1 | Δfae | 18 |
| CM198K.1 | Δfae::kan | 18 |
| CM212K.1 | ΔdmrA::kan | 20 |
| CM253.1 | Δorf4 | This study |
| CM253K.1 | Δorf4::kan | This study |
| CM253-263K.1 | Δorf4 Δmch::kan | This study |
| CM253-266K.1 | Δorf4 ΔfhcBADC::kan | This study |
| CM258.1 | ΔmtdB | This study |
| CM258K.1 | ΔmtdB::kan | This study |
| M. extorquens AM1 | Rifr derivative | 21 |
| Plasmids | ||
| pCM80 | M. extorquens AM1 expression vector (PmxaF) | 19 |
| pCM102 | pCR2.1 with flhA from P. denitrificans | This study |
| pCM103 | pCR2.1 with fghA from P. denitrificans | This study |
| pCM104 | pCM80 with fghA | This study |
| pCM106 | pCM80 with flhA-fghA | This study |
| pCM157 | Broad-host-range cre expression vector | 18 |
| pCM184 | Broad-host-range allelic exchange vector | 18 |
| pCM191 | pCR2.1 with mxaF upstream flank | This study |
| pCM192 | pCR2.1 with mxaF downstream flank | This study |
| pCM193 | pCM184 with mxaF downstream flank | This study |
| pCM194 | pCM193 with mxaF upstream flank | This study |
| pCM250 | pCR2.1 with orf4 upstream flank | This study |
| pCM251 | pCR2.1 with orf4 downstream flank | This study |
| pCM252 | pCM184 with orf4 upstream flank | This study |
| pCM253 | pCM252 with orf4 downstream flank | This study |
| pCM254 | pCR2.1 with mtdA | This study |
| pCM255 | pCR2.1 with mtdB upstream flank | This study |
| pCM256 | pCR2.1 with mtdB downstream flank | This study |
| pCM257 | pCM184 with mtdB downstream flank | This study |
| pCM258 | pCM257 with mtdB upstream flank | This study |
| pCM259 | pCM80 with mtdA | This study |
| pCM260 | pCR2.1 with mch upstream flank | This study |
| pCM261 | pCR2.1 with mch downstream flank | This study |
| pCM262 | pCM184 with mch upstream flank | This study |
| pCM263 | pCM262 with mch downstream flank | This study |
| pCM264 | pCR2.1 with fhcC downstream flank | This study |
| pCM265 | pCM184 with orf4 upstream (fhcB upstream) flank | This study |
| pCM266 | pCM265 with fhcC downstream flank | This study |
| pCR2.1 | PCR cloning vector | Invitrogen |
| pRK2073 | Helper plasmid expressing IncP tra functions | 9 |
| pWRxox451 | Plasmid with P. denitrificans flhA and fghA | 25 |
Phenotypic analyses of mutant strains.
In order to compare the growth of wild-type M. extorquens AM1 with that of mutants in liquid medium, cultures were grown to mid-exponential phase, centrifuged, and then resuspended into fresh medium containing the carbon source described. To test for sensitivity to methanol, methanol was added to one set of succinate flasks to the reported final concentration after 2 h. Mutant phenotypes were also assessed on solid medium by comparing the relative rate of colony formation. Sensitivity to methanol or formaldehyde was assayed using succinate medium to which methanol or formaldehyde was added immediately before pouring plates at the following tested concentrations: 125, 10, and 1 mM and 100, 10, 1, and 0.1 μM for methanol; 1, 0.5, 0.1, 0.05, 0.01, and 0.005 mM for formaldehyde. Because an undetermined fraction of the methanol will volatilize, the reported MIC for methanol is a maximum value. All phenotypic analyses were performed at least twice.
Generation of a plasmid overexpressing mtdA.
The coding region of mtdA was amplified by PCR and cloned into pCR2.1 (Invitrogen) to produce pCM254. The 1.0-kb HindIII-XbaI fragment of pCM254 was cloned between the same sites of the expression plasmid pCM80 (19) to generate pCM259. Plasmids were introduced into appropriate strains using the helper strain pRK2073 (9).
Construct for the heterologous expression of the GSH-dependent formaldehyde oxidation pathway from Paracoccus denitrificans.
The two primary genes comprising the glutathione (GSH)-dependent formaldehyde oxidation pathway of P. denitrificans, flhA (26) and fghA (11), were amplified by PCR using pWRxox451 (25) as a template and cloned into pCR2.1 (Invitrogen) to produce pCM102 and pCM103. The 1.0-kb EcoRI fragment of pCM103 was introduced into the EcoRI site of pCM80 to generate pCM104, into which the 1.4-kb XbaI fragment from pCM102 was inserted into the corresponding site to produce pCM106.
Enzyme assays.
The activities of MtdA (31), FlhA (26), and FghA (11) were assayed in two to three replicates as described using cell extracts prepared using a French press from cell material harvested from exponential-phase cultures. Variation in enzyme activities between cultures was less than 20%. Total protein concentration in the extracts was assayed spectrophotometrically (13, 34) using a Beckmann DU 640B spectrophotometer.
RESULTS
Mutants defective for the H4MPT pathway have varying degrees of sensitivity to methanol and formaldehyde.
M. extorquens AM1 mutants defective for mtdB (10), fae (33), and dmrA (20) of the H4MPT pathway are all unable to grow on C1 compounds as their sole source of carbon and energy and exhibit sensitivity to methanol or formaldehyde during growth on multicarbon compounds such as succinate. It has been hypothesized that this unique phenotype is due to an inability to detoxify formaldehyde (10, 20, 33). In order to test this hypothesis and to better understand the role of the H4MPT pathway in M. extorquens AM1, the phenotypes of these mutants were examined in more detail. For this work, mutants were employed in which the genes in question were deleted from the chromosome. Some of these deletions were subsequently unmarked via cre-lox-based allelic exchange (18), and the resulting strains were used to construct new strains bearing more than one mutation. In addition to the Δfae::kan strain CM198K.1 (18) and the ΔdmrA::kan strain CM212K.1 (20) available from previous studies, new mutants were generated defective for orf4 (CM253K.1; Δorf4::kan) and mtdB (CM258K.1; ΔmtdB::kan) so that all comparisons would involve similar genetic constructions.
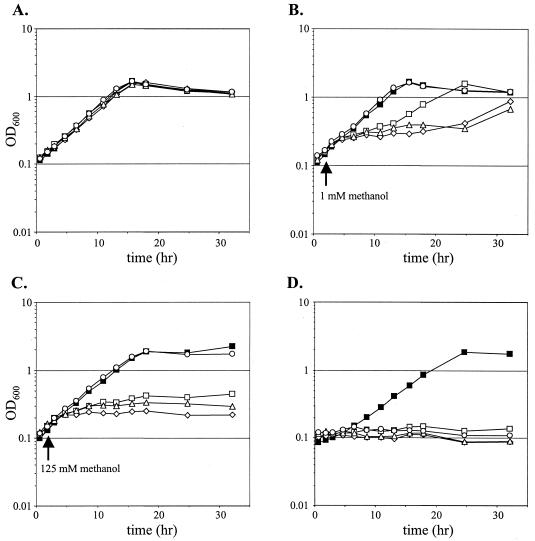
All four mutants in the H4MPT pathway employed in this study grew with wild-type characteristics in liquid medium containing succinate, indicating that the respective functions are not required for general heterotrophic growth (Fig. 2A), but they were unable to grow in medium containing methanol (Fig. 2D). Analogous results were observed for growth on solid medium. Additionally, the H4MPT pathway mutant strains grew like wild type on formate, indicating that they are not required for the metabolism of the more-oxidized C1 compound formate. In order to compare the inhibitory effect of methanol on the growth of mutants defective for the H4MPT pathway on succinate, methanol was added to a set of succinate flasks after 2 h to either a 1 or 125 mM final concentration (Fig. 2B and C). Under these conditions, the mtdB mutant CM258K.1 grew like wild type. However, the three other mutants were inhibited at both methanol concentrations. Addition of methanol at 1 mM caused a more severe inhibition of the dmrA and orf4 mutant strains relative to the fae mutant strain, whereas 125 mM methanol caused cessation of growth in all three strains.
FIG. 2.
Growth of wild-type M. extorquens AM1 and mutant strains pregrown on succinate, harvested, and resuspended in medium containing succinate (A) or succinate with methanol added to 1 mM (B) or 125 mM (C) at 2 h, or 125 mM methanol (D). The strains represented are wild type (filled squares), the fae mutant CM198K.1 (open squares), the dmrA mutant CM212K.1 (open diamonds), the orf4 mutant CM253K.1 (open triangles), and the mtdB mutant CM258K.1 (open circles).
The MICs of methanol or formaldehyde during growth on succinate plates were also examined for the four H4MPT pathway mutants. On solid medium a distinct inhibitory effect of methanol was observed for the mtdB mutant CM258K.1, with an MIC of 10 mM and an MIC of formaldehyde of 0.5 mM. In comparison, the wild type had an MIC of formaldehyde of 1 mM and was not inhibited by 125 mM methanol. The other mutants were observed to be significantly more sensitive, with MICs for methanol and formaldehyde, respectively, of 10 and 100 μM for the fae mutant CM198K.1 and 1 and 10 μM for the dmrA and orf4 mutants CM212K.1 and CM253K.1.
Overexpression of mtdA provides partial complementation of the mtdB mutant phenotype.
We hypothesized that the relatively moderate sensitivity of the mtdB mutant strain CM258K.1 to methanol may be due to the presence of another enzyme, MtdA, whose substrate specificity overlaps with that of MtdB. Even though the presence of MtdA is insufficient for wild-type resistance to methanol, it may contribute to the removal of formaldehyde by converting methylene-H4MPT to methenyl-H4MPT. To test this hypothesis, the region encoding mtdA was cloned and introduced into the expression vector pCM80 (19) to allow for overexpressed levels of MtdA. The plasmid containing mtdA expressed from the strong promoter PmxaF resulted in an over-sevenfold increase in MtdA activity from 270 to 1,970 mU during growth on methanol. Neither CM258K.1 bearing the empty vector pCM80 nor pCM259 was capable of growth on methanol plates. However, the MIC for methanol in the presence of succinate was 125 mM for CM258K.1 containing pCM259, compared to 10 mM for CM258K.1 with pCM80. Therefore, a substantial increase in MtdA activity provides partial complementation of the mtdB mutant phenotype.
Methanol sensitivity of H4MPT pathway mutants requires formaldehyde production.
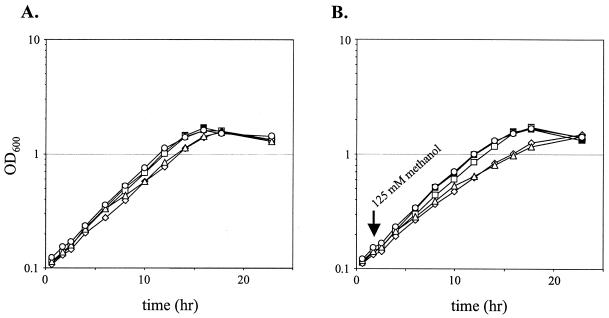
Previous work (10) on mtdB mutants indicated that the sensitivity to methanol could be alleviated if a mtdB::kan mutant was generated in a strain that contained a mutation in the gene (mxaF) encoding the large subunit of MDH (21). This demonstrated that the sensitivity of this strain required the production of formaldehyde and was not simply a consequence of methanol itself. We extended this analysis to characterize the other three mutants with greater sensitivity to methanol. A series of double mutants were constructed in the ΔmxaF strain CM194.1. The resulting strains were resistant to the addition of 125 mM methanol to succinate cultures, with only the ΔmxaF ΔdmrA::kan and ΔmxaF Δorf4::kan mutants showing even a slight growth inhibition (Fig. 3). Similar results were obtained using solid media. The ΔmxaF ΔmtdB::kan strain CM194-258K.1 was not inhibited by 125 mM methanol, compared to an MIC of 10 mM for the ΔmtdB::kan strain CM258K.1. The ΔmxaF Δfae::kan strain CM194-198K.1 had an MIC of methanol of 10 mM, compared to 10 μM for the Δfae::kan strain CM198K.1. Finally, the ΔmxaF ΔdmrA::kan strain CM194-212K.1 and the ΔmxaF Δorf4::kan strain CM194-253K.1 exhibited an MIC for methanol of 100 μM, compared to 1 μM for the corresponding MDH+ strains. This residual sensitivity to methanol suggests either a low-level alternate methanol oxidation activity or a direct effect of methanol at higher levels. However, these data suggest that the extreme sensitivity to methanol observed in all tested H4MPT pathway mutants is not due to methanol itself but, rather, requires the production of formaldehyde.
FIG. 3.
Growth of mutant strains pregrown on succinate, harvested, and resuspended in medium containing succinate (A) or succinate plus methanol added to 125 mM at 2 h (B). The strains represented are the ΔmxaF::kan strain CM194K.1 (filled squares), the ΔmxaF Δfae::kan mutant CM194-198K.1 (open squares), the ΔmxaF ΔdmrA::kan mutant CM194-212K.1 (open diamonds), the ΔmxaF Δorf4::kan mutant CM194-253K.1 (open triangles), and the ΔmxaF ΔmtdB::kan mutant CM194-258K.1 (open circles).
Methanol sensitivity of H4MPT pathway mutants is alleviated and growth on C1 compounds is achieved by expressing a heterologous GSH-dependent formaldehyde oxidation pathway.
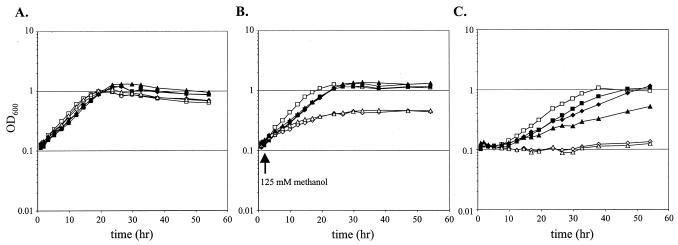
The data presented thus far suggest that the methanol-sensitive phenotype observed for H4MPT pathway mutants is due to an inability to detoxify the formaldehyde produced from methanol, either directly or as one of its derivatives. As a final test of this hypothesis, a heterologous formaldehyde oxidation system was cloned and expressed in H4MPT pathway mutants. The two primary genes of the glutathione (GSH)-dependent formaldehyde oxidation pathway of P. denitrificans, flhA (encodes GSH- and NAD-dependent formaldehyde dehydrogenase [26]) and fghA (encodes S-formyl-GSH hydrolase [11]) were cloned by PCR amplification and introduced together into the expression vector pCM80 (19) to generate the plasmid pCM106. Introduction of pCM106 resulted in activities of 2,500 and 2,300 mU for FlhA and FghA, respectively, whereas these activities were undetectable in wild-type M. extorquens AM1 carrying pCM80 without an insert. Mutants defective for mtdB, fae, dmrA, and orf4 bearing pCM106 were insensitive to 125 mM methanol present in succinate plates. Additionally, the presence of pCM106 allowed the mtdB mutant to grow like wild type in the presence of 1 mM formaldehyde and raised the MIC of formaldehyde to 0.5 mM for fae, dmrA, and orf4 mutant strains. The protective effect of expressing the GSH-dependent formaldehyde oxidation pathway was also tested in liquid medium for two representative mutants that lacked either fae or dmrA. Addition of 125 mM methanol to succinate growth medium did not inhibit growth in strains bearing pCM106 (Fig. 4A and B). Finally, beyond alleviating methanol sensitivity, the expression of the GSH pathway in the H4MPT pathway mutants allowed growth in methanol liquid medium (Fig. 4C) and on methanol plates, albeit the complementation of the dmrA and orf4 mutants was less robust than for the other two mutants. The ability of the heterologous GSH-dependent formaldehyde oxidation system to alleviate the methanol sensitivity of H4MPT pathway mutants provides strong evidence that the cause of the methanol-sensitive phenotype is the inability to detoxify intracellular formaldehyde produced from methanol.
FIG. 4.
Growth of wild-type M. extorquens AM1 and mutant strains with plasmids pregrown on succinate, harvested, and resuspended in medium containing succinate (A), succinate with methanol added to 125 mM at 2 h (B), or 125 mM methanol (C). All media also contained tetracycline for plasmid maintenance. The strains represented are wild type (squares), the fae mutant CM198K.1 (diamonds), and the orf4 mutant CM253K.1 (triangles), with the empty vector pCM80 (open symbols) or the pCM106 plasmid expressing flhA-fghA (filled symbols).
Null mutants lacking mch or fhcBADC can only be obtained in an H4MPT biosynthesis-negative background.
One important question regarding the role of the H4MPT pathway in M. extorquens AM1 is why it has not been possible to obtain null mutations in genes that encode the two enzymes catalyzing the final reactions of the pathway, Mch and Fhc (6). A few scenarios may be suggested to explain this phenomenon: (i) a C1-H4MPT intermediate is required for growth on multicarbon compounds; (ii) these mutants are even more sensitive to methanol, such that ambient concentrations are lethal; or (iii) accumulation of an intermediate of the H4MPT pathway causes a toxic effect or a regulatory problem. To test these hypotheses, we attempted constructing deletion versions of these mutations in various backgrounds. No mutants were obtained in the wild type using the deletion constructs for Mch (Δmch::kan) or FhcBADC (ΔfhcBADC::kan), in agreement with our laboratory's previous results for insertion mutants (6). Likewise, no deletion mutants were obtained in the backgrounds lacking MDH (ΔmxaF strain CM194.1), Fae (Δfae strain CM198.1), or MtdB (ΔmtdB strain CM258.1). Both deletions were readily generated, however, in the Δorf4 strain CM253.1. The Δorf4 Δmch::kan strain CM253-263K.1 and Δorf4 ΔfhcBADC::kan strain CM253-266K.1 grew normally on succinate or formate but exhibited defective growth on methanol or methylamine, as had been observed for the orf4 mutant CM253K.1. Furthermore, the MICs of methanol and formaldehyde during growth on succinate were the same as those observed for CM253K.1. These data indicate that none of the H4MPT pathway enzymes are essential for metabolism of multicarbon compounds. The inability to generate Δmch or ΔfhcBADC mutants in a wild-type background is discussed below.
DISCUSSION
In order to determine the role of the H4MPT-linked formaldehyde oxidation pathway in M. extorquens AM1, we have carried out a physiological analysis of four mutants of M. extorquens AM1 defective for the H4MPT-linked formaldehyde oxidation pathway. These mutants fall into three phenotypic classes that correlate with the biochemical roles of the respective enzymes in the pathway. The most severe defect is found for mutants defective for dmrA and orf4. Both mutants are predicted to lack H4MPT (20, 28) and would thus lack both the H4MPT cofactor and any C1 intermediates linked to this cofactor. Therefore, the flux of formaldehyde through the H4MPT pathway should be zero, and the full burden of formaldehyde production would fall on these mutants upon exposure to methanol.
Mutants defective for fae exhibit an intermediate level of sensitivity to methanol or formaldehyde. Fae catalyzes the condensation of formaldehyde with H4MPT, but this reaction also proceeds nonenzymatically at a lower rate (33). The fact that the fae mutant has a less severe phenotype than the H4MPT biosynthesis mutants is consistent with the nonenzymatic condensation of formaldehyde with H4MPT occurring at sufficient levels to allow a low level of formaldehyde oxidation through this pathway in the absence of Fae activity, but not enough to handle the full formaldehyde flux of methylotrophic growth.
Mutants lacking MtdB activity have the least severe phenotype of the H4MPT pathway mutants investigated in this work. Two methylene-H4MPT dehydrogenases are present in M. extorquens AM1, MtdA (NADP dependent, but also utilizes methylene-H4F) and MtdB (H4MPT specific, but utilizes either NAD+ or NADP+). The sensitivity of mtdB mutants to methanol or formaldehyde and the inability to grow on methanol indicated that this enzyme plays a critical role in formaldehyde oxidation (10). The relatively moderate sensitivity of the mtdB mutant compared to that of either the fae or H4MPT biosynthesis mutants indicates that, despite being insufficient for growth on C1 compounds or complete resistance to methanol or formaldehyde, MtdA activity can support a moderate formaldehyde flux in the absence of MtdB. To further address this question, mtdA was cloned and overexpressed to levels 7.4-fold higher than in the wild type. This level of MtdA activity was insufficient to allow growth on methanol; however, it largely alleviated the sensitivity to methanol. These data suggest that despite the normal high level of MtdA activity in the wild type, the enzyme level is limiting in the absence of MtdB activity. It has been suggested that the requirement for MtdA to use NADP+, rather than NAD+, in methylene-H4MPT reduction limits its in vivo activity (31).
The mutant phenotypes discussed above are correlated with the magnitude of the decreased formaldehyde flux through the H4MPT pathway. For the mutants with the greatest defect, the H4MPT biosynthesis mutants (dmrA and orf4), the impact is remarkable considering that the MIC drops at least 5 orders of magnitude compared to the wild type. Our demonstration that this phenotype can be at least partially compensated with an alternate NAD- and GSH-linked formaldehyde oxidation system demonstrates that this H4MPT pathway not only serves as the main energy-generating pathway during methylotrophic growth, it also must be the major formaldehyde detoxification pathway. It is notable that an analogous methanol-sensitive phenotype has been observed for P. denitrificans mutants lacking flhA (26) or fghA (11), which demonstrates the widespread importance for methylotrophic bacteria to maintain the capacity for formaldehyde detoxification. The growth inhibition observed for M. extorquens AM1 H4MPT pathway mutants may be due directly to formaldehyde accumulation. Alternatively, growth inhibition may be caused by a reactive conjugate of formaldehyde with another compound, analogous to what has been described previously for GSH-dependent oxidation of dichloromethane (15), or perhaps even a regulatory circuit poised to sense an imbalance of formaldehyde production and utilization.
Our results suggest that in these mutants, formaldehyde may accumulate in the cytoplasm. The relative resistance of the wild type to formaldehyde added to the medium, as well as the ability of a cytoplasmic formaldehyde oxidation system to alleviate the phenotype, support the idea of cytoplasmic formaldehyde rather than periplasmic formaldehyde being responsible for toxicity. However, it is not possible at this time to measure cytoplasmic formaldehyde distinct from periplasmic formaldehyde. In addition, proteins and nucleic acids inside the cell will serve as a large sink for formaldehyde, and it is likely that formaldehyde will damage the cell substantially before it accumulates internally.
Our demonstration that it is possible to obtain null mutants in the H4MPT pathway shows that this pathway is not required for growth on multicarbon compounds. We suggest that the explanation for the inability to completely block the H4MPT-dependent formaldehyde oxidation pathway in wild-type cells is due to the accumulation of a C1-H4MPT intermediate(s), since the identical mutations are tolerated in the absence of H4MPT biosynthesis. This scenario implies that accumulation of a C1-H4MPT intermediate(s) is either toxic and/or interferes with normal regulatory circuits. Further work will be required to test this hypothesis and distinguish between these possibilities.
The work presented here demonstrates that M. extorquens AM1 relies on the H4MPT pathway to oxidize formaldehyde both during growth on C1 substrates and to detoxify formaldehyde during growth on multicarbon compounds. Remarkably, the heterologous GSH-dependent pathway from P. denitrificans is able to largely replace this function. This result indicates that these pathways comprise analogous metabolic modules (5, 12). Although they use entirely different enzymes and cofactors, they can fulfill the same cellular function, namely, the NAD(P)-dependent oxidation of formaldehyde to formate.
Acknowledgments
We thank M. Kalyuzhnaya, N. Korotkova, S. Stolyar, R. K. Thauer, and J. Vorholt for their thoughtful discussion of our work, Y. Okubo for assistance with growth curves, and N. Harms for providing pWRxox451.
This work was supported by a grant from the National Institutes of Health (GM 36296).
REFERENCES
- 1.Anthony, C., and L. J. Zatman. 1964. The microbial oxidation of methanol. 2. The methanol-oxidizing enzyme of Pseudomonas sp. M27. Biochem. J. 92:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attwood, M. M., and W. Harder. 1972. A rapid and specific enrichment procedure for Hyphomicrobium spp. Antonie Leeuwenhoek 38:369-377. [DOI] [PubMed] [Google Scholar]
- 3.Attwood, M. M., and J. R. Quayle. 1984. Formaldehyde as a central intermediary metabolite of methylotrophic metabolism, p. 315-323. In R. L. Crawford and R. S. Hanson (ed.), Microbial growth on C1 compounds. American Society for Microbiology, Washington, D.C.
- 4.Chistoserdov, A. Y., L. V. Chistoserdova, W. S. McIntire, and M. E. Lidstrom. 1994. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J. Bacteriol. 176:4052-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chistoserdova, L., S. W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 7.Chistoserdova, L. V., and M. E. Lidstrom. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J. Bacteriol. 176:1957-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eady, R. R., and P. J. Large. 1968. Purification of an amine dehydrogenase form Pseudomonas AM1 and its role in growth on methylamine. Biochem. J. 106:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagemeier, C. H., L. Chistoserdova, M. E. Lidstrom, R. K. Thauer, and J. A. Vorholt. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 267:3762-3769. [DOI] [PubMed] [Google Scholar]
- 11.Harms, N., J. Ras, W. N. Reijnders, R. J. van Spanning, and A. H. Stouthamer. 1996. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178:6296-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartwell, L. H., J. J. Hopfield, S. Leibler, and A. W. Murray. 1999. From molecular to modular cell biology. Nature 402:C47-C52. [DOI] [PubMed] [Google Scholar]
- 13.Kalb, V. F., and R. W. Bernlohr. 1977. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82:362-371. [DOI] [PubMed] [Google Scholar]
- 14.Kallen, R. G., and W. P. Jencks. 1966. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J. Biol. Chem. 241:5851-5863. [PubMed] [Google Scholar]
- 15.Kayser, M. F., and S. Vuilleumier. 2001. Dehalogenation of dichloromethane by dichloromethane dehalogenase/glutathione S-transferase leads to formation of DNA adducts. J. Bacteriol. 183:5209-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laukel, M., L. Chistoserdova, M. E. Lidstrom, and J. A. Vorholt. 2003. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 270:325-333. [DOI] [PubMed] [Google Scholar]
- 17.Lidstrom, M. E. 2 November 2001, posting date. Aerobic metylotrophic prokaryotes. In M. Dworkin (ed.), Prokaryotes. [Online.] http://link.springer.de/link/service/books/10125/.
- 18.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 19.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 20.Marx, C. J., B. N. O'Brien, J. Breezee, and M. E. Lidstrom. 2003. Novel methylotrophy genes of Methylobacterium extorquens AM1 identified by using transposon mutagenesis including a putative dihydromethanopterin reductase. J. Bacteriol. 185:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunn, D. N., and M. E. Lidstrom. 1986. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J. Bacteriol. 166:581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomper, B. K., O. Saurel, A. Milon, and J. A. Vorholt. 2002. Generation of formate by the formyltransferase/hydrolase complex (Fhc) from Methylobacterium extorquens AM1. FEBS Lett. 523:133-137. [DOI] [PubMed] [Google Scholar]
- 23.Pomper, B. K., and J. A. Vorholt. 2001. Characterization of the formyltransferase from Methylobacterium extorquens AM1. Eur. J. Biochem. 268:4769-4775. [DOI] [PubMed] [Google Scholar]
- 24.Pomper, B. K., J. A. Vorholt, L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1999. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur. J. Biochem. 261:475-480. [DOI] [PubMed] [Google Scholar]
- 25.Ras, J., W. N. Reijnders, R. J. Van Spanning, N. Harms, L. F. Oltmann, and A. H. Stouthamer. 1991. Isolation, sequencing, and mutagenesis of the gene encoding cytochrome c553i of Paracoccus denitrificans and characterization of the mutant strain. J. Bacteriol. 173:6971-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ras, J., P. W. Van Ophem, W. N. Reijnders, R. J. Van Spanning, J. A. Duine, A. H. Stouthamer, and N. Harms. 1995. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J. Bacteriol. 177:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Scott, J. W., and M. E. Rasche. 2002. Purification, overproduction, and partial characterization of β-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J. Bacteriol. 184:4442-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 30.Vorholt, J. A. 2002. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178:239-249. [DOI] [PubMed] [Google Scholar]
- 31.Vorholt, J. A., L. Chistoserdova, M. E. Lidstrom, and R. K. Thauer. 1998. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 180:5351-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorholt, J. A., L. Chistoserdova, S. M. Stolyar, R. K. Thauer, and M. E. Lidstrom. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitaker, J. R., and P. E. Granum. 1980. An absolute method for protein determination based on the difference at 235 and 280 nm. Anal. Biochem. 109:156-159. [DOI] [PubMed] [Google Scholar]