Abstract
Studies of the host response to virus infection typically focus on protein-coding genes. However, non-protein-coding RNAs (ncRNAs) are transcribed in mammalian cells, and the roles of many of these ncRNAs remain enigmas. Using next-generation sequencing, we performed a whole-transcriptome analysis of the host response to severe acute respiratory syndrome coronavirus (SARS-CoV) infection across four founder mouse strains of the Collaborative Cross. We observed differential expression of approximately 500 annotated, long ncRNAs and 1,000 nonannotated genomic regions during infection. Moreover, studies of a subset of these ncRNAs and genomic regions showed the following. (i) Most were similarly regulated in response to influenza virus infection. (ii) They had distinctive kinetic expression profiles in type I interferon receptor and STAT1 knockout mice during SARS-CoV infection, including unique signatures of ncRNA expression associated with lethal infection. (iii) Over 40% were similarly regulated in vitro in response to both influenza virus infection and interferon treatment. These findings represent the first discovery of the widespread differential expression of long ncRNAs in response to virus infection and suggest that ncRNAs are involved in regulating the host response, including innate immunity. At the same time, virus infection models provide a unique platform for studying the biology and regulation of ncRNAs.
IMPORTANCE
Most studies examining the host transcriptional response to infection focus only on protein-coding genes. However, there is growing evidence that thousands of non-protein-coding RNAs (ncRNAs) are transcribed from mammalian genomes. While most attention to the involvement of ncRNAs in virus-host interactions has been on small ncRNAs such as microRNAs, it is becoming apparent that many long ncRNAs (>200 nucleotides [nt]) are also biologically important. These long ncRNAs have been found to have widespread functionality, including chromatin modification and transcriptional regulation and serving as the precursors of small RNAs. With the advent of next-generation sequencing technologies, whole-transcriptome analysis of the host response, including long ncRNAs, is now possible. Using this approach, we demonstrated that virus infection alters the expression of numerous long ncRNAs, suggesting that these RNAs may be a new class of regulatory molecules that play a role in determining the outcome of infection.
Introduction
Over the past decade, genomic projects have obtained evidence that thousands of non-protein-coding RNAs (ncRNAs) are transcribed from mammalian genomes, and it is becoming increasingly apparent that many long ncRNAs (>200 nucleotides [nt]) are biologically important (1–3). Though some small ncRNAs such as microRNAs (4) have been found to be involved in virus-host interactions, the relevance of long ncRNAs to viral infections has not been systematically studied, in part because these ncRNAs have not been easily accessible with typically available technologies. In this study, we performed whole-transcriptome analysis of severe acute respiratory syndrome coronavirus (SARS-CoV)-infected lung samples collected from four mouse strains using deep-sequencing technology. Our results show that there was a widespread differential regulation of long ncRNAs in response to viral infection, suggesting that these ncRNAs are involved in regulating the host response, including innate immunity. At the same time, virus infection models provide a unique platform for studying the biology and regulation of ncRNAs.
RESULTS
Whole-transcriptome analysis of SARS-CoV-infected mouse lung samples.
To systematically investigate the regulation of long ncRNAs during viral infection, we infected four different strains of mice with a mouse-adapted severe acute respiratory syndrome coronavirus (SARS-CoV) (5). These mice were selected due to their differential range in susceptibility phenotypes following infection with SARS-CoV or influenza virus and the capacity to pursue downstream quantitative trait locus (QTL) mapping of regulation and function in the Collaborative Cross. Weight loss in the animals was monitored over the course of the infection with SARS MA15 or influenza virus A/PR/8/34 as a measure of disease severity (Fig. 1). We then performed a whole-transcriptome analysis of collected lung tissue samples using next-generation sequencing (NGS). Directional cDNA libraries were constructed using the not-so-random (NSR) priming method (6), which enabled the profiling of polyadenylated, nonpolyadenylated, coding, and noncoding transcripts, but not small RNAs (6).
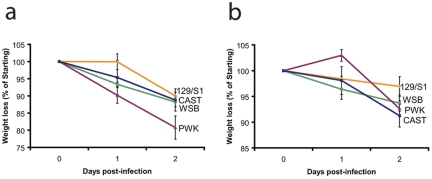
FIG 1 .
Measurement of weight loss in four strains of mice following infection with SARS MA15 or influenza virus A/PR/8/34. (a) Over the course of a 2-day SARS-CoV infection, CAST/EiJ (CAST) mice lost 12% of their starting weight, PWK/PhJ (PWK) mice lost 20% of their starting weight, WSB/EiJ (WSB) mice lost 12% of their starting weight, and 129S1/SvImJ (129/S1) mice lost 10% of their starting weight. (b) Two days after infection with 500 PFU of influenza virus strain A/PR/8/34, 129/S1 mice lost 3% of their starting weight, PWK mice lost 7% of their starting weight, WSB mice lost 6% of their starting weight, and CAST mice lost 9% of their starting weight. See supplementary material for more details.
We observed a large number of reads (1.5 to 7 million) that uniquely mapped to viral RNAs (viral genomic RNAs and transcripts) (Fig. 2) (see Table S1 in the supplemental material) in samples from virus-infected animals. From all samples, we obtained on average over 22 million reads that uniquely mapped to host genomic sites, including many that mapped to nonannotated intergenic regions (Fig. 2a; see Table S1 in the supplemental material). We reasoned that the transcriptional activities detected in nonannotated regions were largely from ncRNAs and that some could be differentially expressed in response to viral infection. To evaluate our approach for the identification of differentially expressed genes, we profiled the same samples using microarrays and compared the profiles with the profiles of the protein-coding part of the NGS data set. We observed a very good correlation (Pearson correlation coefficients of 0.73 to 0.8) between two platforms (see Fig. S2 in the supplemental material), and even better agreement between NGS and quantitative PCR (qPCR) (data not shown).
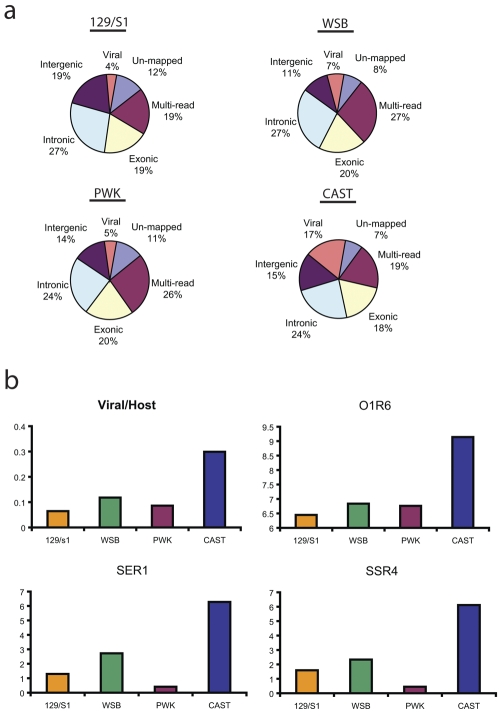
FIG 2 .
Global classification of transcriptional activity in lung samples from SARS-CoV-infected mice. (a) Global classification of transcriptional activity. Short reads were assigned to one of six nonoverlapping categories. The exonic, intronic, and intergenic categories were defined by the genomic coordinates for UCSC known genes and include only reads that map to unique genomic locations. See Fig. S1 in the supplemental material for more details on read mapping. (b) Estimation of relative abundance of viral RNAs in lung samples from SARS-CoV-infected mice. (Top left) The relative abundances of viral RNAs in lung samples from SARS-CoV-infected mice estimated as the ratio of the total number of next-generation sequencing (NGS) reads uniquely mapped to the SARS-CoV genome to the total number of reads uniquely mapped to the mouse genome. The four mouse strains are indicated below the bars. The relative abundances of viral RNAs estimated as the differences between the qPCR threshold cycle (CT) values obtained with primers designed for SARS protein-coding genes ORF1 (O1R6), SARS Envelope Reverse 1 (SER1), and SARS Spike Reverse 4 (SSR4) and those with primers designed for mouse 18S gene using the same strains as in the top left graph.
Differential expression of long ncRNAs during SARS-CoV infection.
First, we studied annotated non-protein-coding RNAs (ncRNAs); the compilation of annotated ncRNAs produced 10,986 nonoverlapping ncRNA loci (Materials and Methods). We found that 509 of these loci were differentially expressed during SARS-CoV MA15 infection (Fig. 3), 485 of which had more than 2.5-fold change in at least one of four mouse strains during infection, and 209 of which were all upregulated or all downregulated by at least 1.8-fold in three or more mouse strains (see Tables S2 and S3 in the supplemental material). Nearly all (504 of 509) were long ncRNAs (>200 nt). These results clearly show that there is widespread differential regulation of long ncRNAs in response to SARS-CoV infection.
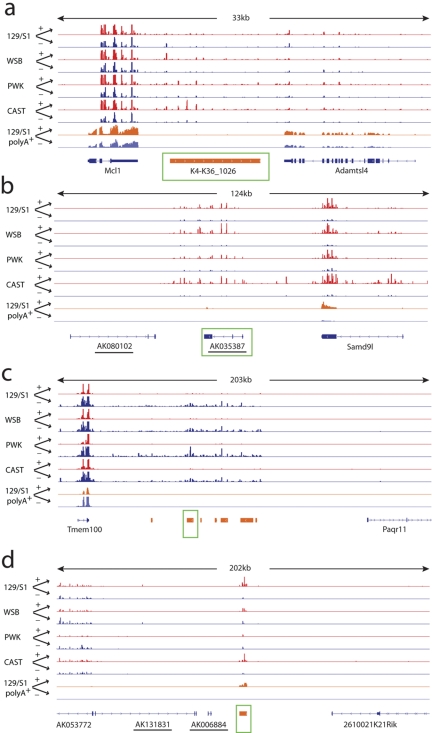
FIG 3 .
Examples of annotated ncRNA loci (a and b) and nonannotated genomic regions (c and d) differentially expressed during SARS-CoV infection. (a) An overview of short reads from whole-transcriptome analysis of mouse lung samples mapped to a 33-kb region of chromosome 3 displayed by Integrative Genome Viewer (IGV) browser (http://www.broadinstitute.org/igv). Each track represents data collected from a single mouse lung sample, with SARS-CoV-infected sample (+) and mock-infected sample (−) shown by the labeled arrows (+ or −). Infected samples are depicted in red, and mock-infected samples are depicted in blue. The strains of mice used are shown on the left. 129/S1 polyA+ represents short-read data generated from libraries separately created from the same samples with poly(A) selection. For reference, UCSC annotation of nearby protein-coding genes is shown at the bottom in blue. K4-K36_1026 is the entire K4-K36 domain of a large intervening ncRNAs (lincRNA) as identified in reference 7, which is upregulated during SARS infection, but no significant expression was observed in poly(A)-selected samples. The green box indicates that the locus was followed up using qPCR (the same for panels b, c, and d). (b) Overview similar to that in panel a for a 124-kb region of chromosome 6. The underlined UCSC annotation is noncoding. It was upregulated during SARS-CoV infection. (c) Overview similar to that in panel a for a 203-kb region of chromosome 11. The loci shown in orange indicate nonannotated genomic regions identified here as differentially expressed during SARS-CoV infection (as in panel d). These regions were downregulated. (d) Overview similar to that in panel a for a 202-kb region of chromosome 12. The locus in orange indicates an nonannotated genomic region upregulated during SARS-CoV infection.
Next we systematically scanned the mouse genome for nonannotated regions that encoded transcripts differentially expressed during viral infection (Materials and Methods). In total, we uncovered 1,406 nonannotated genomic regions that did not overlap any annotated protein-coding genes (UCSC or Ensembl annotations) but that consistently had changes in expression of more than 1.4-fold (all upregulated or all downregulated) in at least 3 mouse strains during infection (Fig. 4; see Table S4 in the supplemental material). For 997 of these regions, we did not find overlap with any annotated loci (UCSC and Ensembl annotations), indicating that many infection-induced changes in RNA transcript abundance are not monitored by conventional microarrays. It also suggests that possibly other infection-related transcripts remain to be discovered under different experimental conditions.
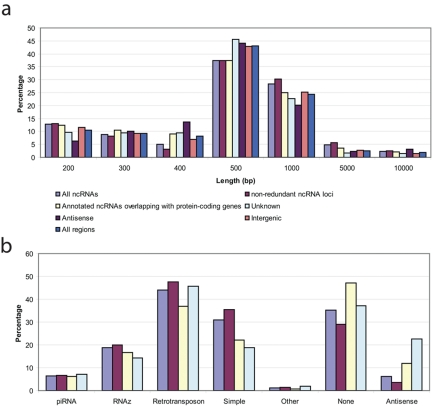
FIG 4 .
Characteristics of genomic regions differentially expressed during SARS-CoV infection. (a) The length distribution of genomic regions differentially expressed during SARS-CoV infection. The genomic regions are indicated below the graph as follows: All ncRNAs, 429 genomic regions that overlapped with one or more annotated ncRNAs; non-redundant ncRNA loci, 285 of 429 genomic regions that overlapped with the nonredundant set of 10,986 annotated ncRNAs as compiled in this study; Annotated ncRNAs overlapping with protein-coding genes, 144 of 429 genomic regions that overlapped with those annotated ncRNAs that we filtered out because of partial overlapping with a protein-coding gene (see Materials and Methods); Unknown, 977 genomic regions without any overlapping annotated genes; Antisense, 249 genomic regions that were antisense to annotated protein-coding genes; Intergenic, 1,157 genomic regions were located between annotated protein-coding genes; All regions, all 1,406 genomic regions identified. (b) Characteristics of genomic regions differentially expressed during SARS-CoV infection. The genome regions are depicted as in panel a. Annotations showing what the identified genomic regions overlap with are shown below the x axis as follows: piRNA, piwi-associated small RNAs; RNAz, conserved RNA secondary structures predicted by RNAz; Retrotransposon, retrotransposons of the SINE, LINE, LTR, and DNA superfamilies; Simple, simple repeats and low complexity; Other, remaining retrotransposons and repeats; None, no overlapping with the annotation categories above; Antisense, antisense to protein-coding genes annotated by UCSC or Ensembl. See text and Materials and Methods for details.
Differential expression of long ncRNAs in response to altered innate immunity.
We used qPCR to further evaluate the differential expression of a subset of ncRNAs in replicate samples. We selected 39 loci/regions that represented a variety of loci for the follow-up studies, including 19 nonannotated genomic regions, 13 annotated ncRNAs, 5 large intervening ncRNAs (lincRNAs [7]) partially overlapping with annotated protein-coding genes (therefore not included in our nonredundant set of annotated ncRNAs), plus two protein-coding genes (Mx1 and Ifit1) known to be regulated during viral infection. Importantly, we observed a very good agreement (Pearson correlation coefficients of 0.87 to 0.94) between SARS-CoV infection to mock infection expression log ratios obtained using NGS and the corresponding log ratios obtained using qPCR on the set of independent samples with multiple replicates (Fig. 5a; see Fig. S3 in the supplemental material).
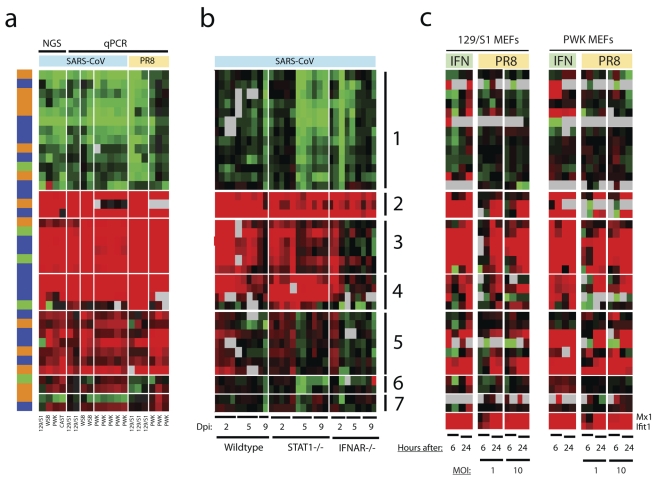
FIG 5 .
Comparison of infection to mock infection expression ratios for 37 differentially expressed ncRNAs and genomic regions. (a) Comparison of the log2 infection/mock infection expression ratios for 37 differentially expressed ncRNAs and genomic regions originally obtained by NGS and then by qPCR on independently collected lung samples from mice infected with SARS-CoV or influenza virus. The influenza virus used was A/PR/8/34 (PR8). Each column represents an independent sample collected from one infected mouse. For qPCR, individual replicate samples were compared to the average of 2 or 3 matched mock-infected samples to form infection/mock infection expression ratios. All samples were collected 2 days after infection. The color red on the heat map indicates upregulation during infection, while the color green indicates downregulation during infection. In the colored bar to the left of the heat map, 13 annotated ncRNAs are shown in dark orange, 19 nonannotated genomic regions are shown in blue, and 5 lincRNAs partially overlapping with annotated protein-coding genes are shown in green. The genomic locations of ncRNAs and genomic regions are shown in Fig. S4 in the supplemental material. The mouse strains are shown at the bottom of the panel. (b) Temporal expression profiles of the same selected ncRNAs and genomic regions (in the same order as in panel a) measured by qPCR in wild-type (129/S6), STAT1 knockout (STAT1−/−), and type 1 interferon receptor knockout (IFNAR−/−) mice infected by SARS-CoV. The values shown are the log2 ratios of the expression levels in infected replicate samples to the average expression levels in strain-matched mock-infected samples. The color red on the heat map indicates up regulation during infection, while the color green indicates downregulation during infection. The samples were collected 2, 5, and 9 days after infection (dpi 2, 5, and 9, respectively). The numbered lines to the right of the heat map indicate groups of ncRNAs and genomic regions based on their temporal expression changes during infection to guide visualization: 1, mostly downregulated in STAT1−/− mice; 2, consistently upregulated in all three types of mice; 3, mostly upregulated in wild-type and STAT1−/− mice, but not in IFNAR−/− mice, especially for the later time points; 4, highly upregulated in STAT1−/− mice, but not in wild-type and IFNAR−/− mice; 5, transiently upregulated at early time points in all three types of mice; 6, downregulated in STAT1−/− mice only and, as shown in panel a, upregulated in SARS-CoV-infected mice but downregulated in influenza virus-infected mice; and 7, no significant changes observed in all three types of mice. (c) Temporal expression profiles of the same selected ncRNAs and genomic regions (in the same order as in panel a) measured by qPCR in mouse embryonic fibroblasts (MEFs) from mice of strain 129S1/SvlmJ (129/S1 MEFs) and PWK/PhJ (PWK MEFs) treated with 500 U of mouse beta interferon (IFN) or influenza A virus A/Pr/8/34 (PR8) (MOI of 1 or 10). The values shown are the log2 ratios of the expression levels in samples from virus-infected or IFN-treated MEFs to the average expression levels in time- and strain-matched samples from mock-infected MEFs. The color red on the heat map indicates upregulation during infection, while the color green indicates downregulation during infection. The samples were collected 6 and 24 hours after IFN treatment or virus infection (shown below the heat maps). Mx1 and Ifit1, two protein-coding genes that are known to be upregulated during viral infection, are positive controls for in vitro treatment.
To investigate whether the observed differential expression of long ncRNAs was specific to SARS-CoV infection or represented a more general host response to viral infection, we infected the same strains of mice with influenza virus A/PR/8/34 and used qPCR to quantify expression changes of the 37 selected ncRNAs and genomic regions in lung samples from infected animals. Interestingly, we found that most (35 of 37) of the selected ncRNAs and genomic regions were similarly differentially expressed during influenza virus infection (Fig. 5a). Thus, many long ncRNAs are differentially regulated during both SARS-CoV and influenza virus infections, suggesting that the differential regulation of long ncRNAs may be a common host response to respiratory viral infection.
To determine the relationship between differential expression of long ncRNAs and innate immune signaling, we performed qPCR on lung samples obtained from a previous study in which mice lacking the type I interferon receptor (IFNAR−/−) or STAT1 (signal transducer and activator of transcription factor 1) (STAT−/−) were infected with SARS-CoV. In that study, we found that SARS-CoV infection resulted in the death of STAT−/− mice, but not IFNAR−/− mice (8). As shown in Fig. 5b, even for the set of 37 ncRNAs examined here, we observed unique patterns of expression changes over time. As expected, most (35 of 37 [95%]) of the selected ncRNAs and genomic regions were differentially expressed (P < 0.05) during SARS-CoV infection under one or more conditions studied. Interestingly, the response to viral infection also displayed temporal changes, as 35 (95%) of the selected ncRNAs and genomic regions showed significant changes in expression (P < 0.05) between at least two consecutive time points. Twenty-six (70%) of the ncRNAs and genomic regions were differentially expressed (P < 0.05) among knockout and wild-type mice under one or more conditions during infection. These findings strongly indicate that the differential expression of long ncRNAs during viral infection is affected by perturbations to innate immune signaling and, importantly, is associated with pathogenic outcome.
Because lung samples contain heterogeneous cell types, the observed differential regulation of long ncRNAs could, in part, be expressed by infiltrating immune cells during infection. We therefore infected cultured mouse embryonic fibroblasts (MEFs) from the same strains of mice with the mouse-adapted influenza virus A/PR/8/34, as SARS-CoV does not infect MEFs. Importantly, we found that about 43% (16 of 37) of the selected ncRNAs and genomic regions were differentially expressed (P < 0.05) in infected MEFs similarly to ncRNAs and genomic regions in lung tissue from infected animals (Fig. 5c). To investigate whether these ncRNAs were also regulated by the interferon response, we treated MEFs separately with beta interferon and found patterns of expression changes that were similar to those observed in influenza virus-infected MEFs. The consistent changes in expression in MEFs in response to both influenza virus infection and interferon treatment convincingly argue that differential regulation of long ncRNAs was neither artifactual nor a result of immune infiltration but instead represents a bona fide host response regulated by innate immunity.
Putative functions of long ncRNAs.
As the functions of long ncRNAs are largely unknown, we performed computational analyses to gain insight into the potential biological roles of these identified ncRNAs. Interestingly, we observed that ~37% (189 of 509) of differentially expressed ncRNA loci overlapped with previously discovered mouse lincRNAs (7). Khalil et al. reported that many human lincRNAs can affect gene expression through their associations with chromatin-modifying complexes (9). We found that 20 mouse loci orthologous to human lincRNAs bound by chromatin-modifying complexes exhibited differential expression in this study (see the supplementary material), suggesting that some of our identified ncRNAs may also interact with chromatin-modifying complexes during viral infection.
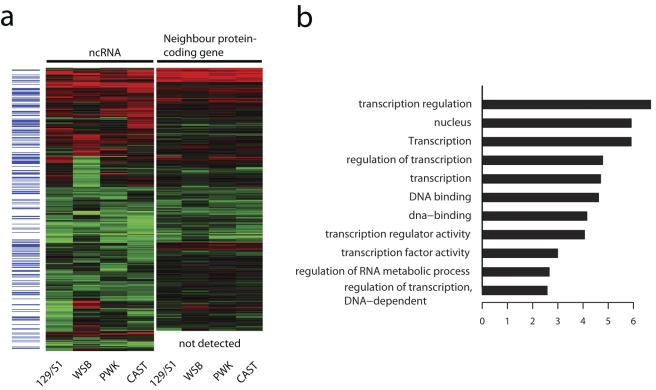
Another approach for inferring putative functions of long ncRNAs is to examine protein-coding genes located near ncRNAs of interest (7, 10). For each mouse strain, we examined the infection-induced patterns of expression of ncRNAs and their paired neighbor protein-coding genes (see the supplementary material). Interestingly, we found that the changes in expression of neighbor protein-coding genes (fold changes) were significantly associated with the fold changes in expression of the corresponding ncRNAs during infection (P values =1.8e−22 to 2.4e−32, analysis of variance [ANOVA] F test, Fig. 6a, and the supplementary material). We utilized the DAVID Functional Annotation Tool (11) for functional enrichment analysis on those neighbor protein-coding genes. The most significant functional group identified using DAVID consisted of 11 similar annotation terms related to gene expression (Fig. 6b). Interestingly, previous studies also reported that the genes in neighboring long ncRNAs exhibit a bias toward transcription-related factors (7, 10). We therefore hypothesize that long ncRNAs might also be able to modulate host responses through neighboring protein-coding genes.
FIG 6 .
Differential expression of 509 annotated ncRNAs and corresponding neighbor protein-coding genes. (a) Heat maps of the infected/mock-infected expression ratios (log2 scale) of 509 annotated ncRNAs and their corresponding neighbor protein-coding genes in four mouse strains. The blue bands on the left indicate those ncRNAs overlapping with long intergenic noncoding RNAs as reported in reference 7. On the heat map, red indicates upregulation during infection, while green indicates downregulation during infection. Below the heat map for neighbor protein-coding genes, “not detected” indicates protein-coding genes with less than 20 uniquely mapped reads in all samples. (b) Functional annotation of protein-coding genes neighboring differentially expressed ncRNAs as shown in panel a. The annotation terms in the most significant functional annotation cluster identified by using the DAVID functional annotation tool are shown, and plotted as the –log10 P value for the enrichment of each annotation term.
DISCUSSION
Previous studies on virus-host interactions and viral pathogenesis have largely focused on protein-coding genes. However, a number of recent studies have begun to suggest that non-protein-coding RNAs (ncRNAs) also function in pathogen-host interactions. For example, Pang et al., using a custom 70-mer microarray, showed that long ncRNA probes had altered expression during CD8+ T cell differentiation upon antigen recognition (12). In additional studies using cDNA microarrays, Ahanda et al. identified eight mRNA-like ncRNAs that were differentially expressed in virus-infected birds (13), and Ravasi et al. showed that 70 ncRNAs were dynamically regulated in mouse macrophages activated by lipopolysaccharide (14). Likewise, Guttman et al., using a custom large intervening ncRNA (lincRNA) array, found that lincRNAs were associated with diverse biological processes across different tissues, including immune surveillance (7). To our knowledge, our study is the first to use comprehensive deep-sequencing technology to clearly demonstrate that long ncRNAs are involved in the host response to viral infection and innate immunity.
As noted, the functions of ncRNAs remain largely unexplored, indicating the need for future studies in this area. For example, the differential regulation of some ncRNAs could simply be by-products of global transcriptional profile changes imparted by interferon and/or viral infection, and they may not play a significant role in the context of infection. Alternatively, ncRNAs may represent a whole new class of innate immunity signaling molecules and interferon-dependent regulators, or even a new layer of gene expression regulation responsible for modulating host responses during viral infection. Similarly, ncRNAs may also represent a new potential class of biomarkers for infectious diseases. The similar differential regulation of ncRNAs in response to SARS-CoV and influenza virus infection indicates that a ncRNA-based signature of respiratory virus infection may exist, suggesting additional diagnostic potential. Finally, using viruses to perturb host systems, such as described here, also presents a valuable platform for future studies of ncRNA biology in general. In the future, it is likely that a detailed knowledge of ncRNA regulation and function will be necessary for a full understanding of viral pathogenesis.
MATERIALS AND METHODS
Mouse lines and virus infection.
Because human severe acute respiratory syndrome coronavirus (SARS-CoV) isolates replicate but do not cause severe clinical disease in mice, we used the mouse-adapted strain MA15 that is lethal in BALB/c mice and that causes 10 to 15% weight loss in young C56BL/6 mice (5). In this study, we infected four of the founder mouse strains used in generating the Collaborative Cross (CC), a newly emerging recombinant inbred mouse resource for mapping complex traits (15). These strains included 129S1/SvImJ (129/S1), CAST/EiJ (CAST), PWK/PhJ (PWK), and WSB/EiJ (WSB) mice, and the animals were provided by Fernando Pardo-Manuel de Villena or obtained from the Jackson Laboratory (Bar Harbor, ME). A benefit of using these strains is that it allows for downstream quantitative trait locus (QTL) and expression QTL (eQTL) mapping of the regulation and function of non-protein-coding RNAs (ncRNAs) in pathogenesis and innate immunity in the final panel of 400 CC recombinant inbred mouse lines. Mice were bred at the University of North Carolina (UNC) mouse facility (Chapel Hill, NC). Animal housing, care, and experimental protocols were in accordance with all UNC-Chapel Hill Institutional Animal Care and Use Committee guidelines. All animal studies were conducted in animal biosafety level 3 laboratories using Sealsafe HEPA-filtered caging, and personnel wore personal protective equipment, including Tyvek suits and hoods as well as positive-pressure HEPA-filtered air respirators. Ten-week-old mice were anesthetized with isoflurane. Mice were intranasally infected with phosphate-buffered saline (PBS) alone or with 1 × 105 PFU of SARS recombinant MA15 (rMA15) in 50 µl of PBS (Invitrogen, Carlsbad, CA) or 500 PFU of influenza A virus strain A/Pr/8/34 (H1N1) in 50 µl of PBS. The mice were weighed once per day and observed twice per day over the course of the infection. For each virus, three to five virus-infected and three mock-infected mice from each strain were euthanized at 2 days postinfection (dpi) with tissues taken for determination of the viral titer and for expression analysis. In this study, one SARS rMA15-infected and one mock-infected mouse from each of the four strains was euthanized at 2 dpi for both the whole-transcriptome analysis using high-throughput sequencing and microarray-based expression profiling. The remaining replicate samples from matched infections were evaluated by qPCR.
Lung samples from rMA15-infected or mock-infected 129S6/SvEv wild-type mice, STAT1 knockout (STAT1−/−) mice, and type I interferon receptor knockout (IFNAR1−/−) mice were obtained from a previously published study (8). The infected samples were collected 2, 5, and 9 days after infection.
Interferon treatment and influenza virus infection of MEFs in vitro.
PWK/PhJ and 129S1/SvlmJ mouse embryonic fibroblasts (MEFs) were obtained from D. Threadgill and F. Manuel-Pardo de Villena at UNC, Chapel Hill, NC. The cells were maintained in complete medium (Dulbecco modified Eagle medium [DMEM] supplemented with 1% glutamine, 10% fetal bovine serum [FBS], and penicillin-streptomycin). As SARS-CoV does not infect MEFs, 1 × 105 cells were plated in each well of a 12-well plate and treated the following day with 300 µl of infection medium alone (DMEM supplemented with 1% glutamine, 2% heat-inactivated calf serum, 50 mM HEPES, and penicillin-streptomycin) or 300 µl of infection medium supplemented with either negative allantoic fluid (Charles River Laboratories, Wilmington, MA), influenza A virus strain A/Pr/8/34 (H1N1) (multiplicity of infection [MOI] of 1 or 10), or 500 U of mouse beta interferon (PBL InterferonSource). The cells were incubated for 1 h at 4°C while being rocked. Mock-infected and virus-infected cells were washed twice and maintained in complete medium. The cells were harvested at 0, 6, 12, 24, and 48 hours after treatment and lysed in 1 ml of Trizol reagent. RNA was further purified using the RNeasy minikit (Qiagen), and the RNA quality was assessed using an Agilent 2100 bioanalyzer. RNA (200 ng) was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen).
RNA preparation.
Both lobes of the right lung were removed and homogenized in Trizol using the MagNA Lyser system (Roche) according to the manufacturer’s instructions. RNA was further purified using the miRNeasy minikit (Qiagen) according to the manufacturer’s instructions. The purity of the RNA samples was verified spectroscopically, and the quality of the intact RNA was assessed using an Agilent 2100 Bioanalyzer. This assay also confirmed that the RNA samples were free of genomic DNA contamination.
Sequencing and read mapping.
We generated cDNA libraries for sequencing analysis using the “not-so-random” (NSR) priming method (6). Briefly, the NSR method uses a set of computationally selected random hexamers to deplete rRNA from total RNA, while still allowing the acquisition of full-length, strand-specific, polyadenylated and nonpolyadenylated transcripts. We purified PCR products without additional manipulation to generate clusters for sequencing by synthesis using the Illumina GA2 platform. Single-end sequencing produced 36-nucleotide (nt) antisense reads containing a dinucleotide bar-coded sequence (CT) at the 5′ terminus. We truncated raw reads as 25 nt before mapping against the mouse genome (mm9, July 2007, NCBI Build 37) combined with SARS viral genomic sequence (MA15 [GenBank accession no. DQ497008]) using Bowtie (16). For global classification, reads mapping to single genomic sites were classified into exonic, intronic, and intergenic categories using the coordinates defined by the UCSC Genes (knownGene) Track (http://genome.ucsc.edu/). Read sequences that mapped to multiple genomic sequences were excluded from subsequent analyses. For the visualization, WIG files were generated using TopHat (17) with UCSC known gene annotations and displayed using the Integrative Genomics Viewer (http://www.broadinstitute.org/igv) or the UCSC genome browser (http://genome.ucsc.edu).
Since the available mouse reference genomic sequences are from the mouse strain C57BL/6 and the sequence differences between the C57BL/6 strain and the four mouse strains used in this study are unknown, we wanted to allow a certain number of mismatches between read sequences and the reference genomic sequences for efficiently mapping reads onto genomic sites. We investigated various numbers of mismatches ranging from zero (i.e., a perfect match between read and genomic sequence) to four. We then looked for a value under which the increase of the percentages of uniquely mapped reads tended to reach a plateau for all samples and allowing a larger number of mismatches did not change the overall read mapping significantly. We selected the same number of maximum mismatches for all subsequent analyses (two mismatches was selected at the end for this study).
ncRNA annotations and the estimation of expression levels.
Annotations of long noncoding RNAs (>200 nt) were compiled from UCSC known genes and three published studies (7, 10, 18). As it is not trivial to differentiate short reads mapped to the regions shared by overlapping transcripts, we clustered the overlapping annotated transcripts into single loci. We then filtered out those loci that overlap with any protein-coding transcripts as annotated or predicted by UCSC or Ensembl to minimize the possibility of the inclusion of protein-coding genes. Obviously, many genuine ncRNAs were excluded from our consideration because of this conservative approach. We obtained 10,986 nonoverlapping ncRNA loci (8,008 of which were larger than 200 nt), in addition to 21,565 protein-coding loci. We estimated the transcript abundance of a locus by counting all reads mapped to the locus, instead of only exonic regions as is typically done when using RNA-Seq for protein-coding loci (19), as the gene structures of many ncRNAs were unknown. We then normalized the raw read counts by the length of the locus and the total uniquely mapped reads for each sample and represented the normalized expression levels similarly as typical RPKMs (reads per kilobase per million reads). An offset of 0.05 was added to all RPKMs before calculating log ratios to avoid taking the log of 0 and to decrease the variability of the log ratios for loci with low read counts.
To balance individual strain differences, we used two complementary criteria that differed in stringency to select differentially expressed ncRNAs: the first being a relatively large fold change (>2.5-fold) in normalized expression during infection in at least one mouse strain and the second being consistent up- or downregulation during infection across multiple strains (3 or more strains here) but with a slightly smaller difference in expression (>1.8-fold) within each strain. In both cases, we also required that the locus must have at least 20 uniquely mapped reads in at least one sample when calculating the ratios between samples from virus-infected and mock-infected animals.
Expression profiling using oligonucleotide microarray.
cRNA probes were generated from each sample using the Agilent one-color Quick-Amp labeling kit. Individual cRNA samples were hybridized to Agilent 4 × 44 mouse whole-genome oligonucleotide microarrays according to the manufacturer’s instructions. Slides were scanned with an Agilent DNA microarray scanner, and the resulting images were analyzed using Agilent Feature Extractor software. Data were warehoused in the Katze LabKey system (LabKey, Inc., Seattle, WA) and preprocessed using Agi4x44PreProcess (version 1.4.0), a Bioconductor package in R (20).
Comparison of differential expressions from next-generation sequencing and microarray.
We first mapped all probes represented on the array to the mouse reference genomic sequences and selected only those that mapped uniquely and perfectly. We then mapped those selected probes to the assembled loci as described above for both protein-coding and noncoding loci. When more than one probe mapped to the same locus, we selected the probe with the highest normalized intensity averaged over all eight samples. For each mouse strain, for samples from virus-infected and mock-infected animals, we then compared the log2 ratios of mapped probe intensities to the log2 ratios of the RPKMs for the same loci.
Identification of novel transcripts by a genome-wide scan.
Briefly, we first assigned reads that were mapped uniquely in the genome to their site of origin. To identify regions differentially expressed during viral infection, we employed a sliding window approach to compare the expression levels between a pair of samples (infected versus mock-infected samples in this study): we slid windows, scored each window based on the number of uniquely mapped reads, and selected intervals with fold changes between two samples above a threshold level. Specifically, we did the following. (i) We fixed a window size (w) and slid it across the genome with a moving step (s). For each window, we computed a score, Sw, as the number of reads aligned within the window, normalized by the total number of uniquely mapped reads for each sample. (ii) To identify differentially expressed windows, we created ratios of scores between the pair of samples and selected those windows passing a threshold (fs) for fold change. (iii) We merged overlapping windows into large intervals if they were differentially expressed in the same direction. (iv) To obtain larger intervals, we joined identified neighboring intervals if there were a low number of reads in between and the larger intervals formed by neighboring ones were also differentially expressed, judged by a threshold fj. (v) To increase the confidence, we then selected only those intervals that were differentially expressed consistently in at least k pairs of samples (here all upregulated or all downregulated in at least 3 out of 4 mouse strains). (vi) We then removed those intervals overlapping protein-coding genes annotated by UCSC or Ensembl, merged remaining overlapping intervals identified from all scans into nonoverlapping genomic regions, and recalculated expression ratios.
We searched the identified genomic regions against different annotations, including noncoding RNA annotations from ncRNA.org (http://www.ncrna.org/). Annotation of piwi-associated small RNAs (piRNAs) were obtained from the functional RNA database (21). Conserved RNA secondary structures (P > 0.5) were predicted based on the 30-way multiple alignments downloaded from the UCSC genome browser (http://genome.ucsc.edu) using RNAz (22). The repeat information was downloaded from RepeatMasker Track of the UCSC genome browser. For simplicity, the different classes of repeats were grouped similarly as previously described (23), and denoted as “retrotransposon” for short interspersed nuclear elements (SINE), long interspersed nuclear elements (LINE), long terminal repeat elements (LTR), and DNA repeat elements (DNA) superfamilies; “Simple” for simple repeats and low complexity; and “Others” for the rest.
Quantitative real-time PCR.
Quantitative real-time PCR was used to validate expression of noncoding RNA. For each sample, total RNA input of approximately 100 ng was used, and cDNA was synthesized by reverse transcription using the QuantiTect reverse transcription kit (Qiagen). Primer sets for SYBR green quantitative reverse transcription-PCR (qRT-PCR) were designed using Primer3 (24). For each locus of interest, we designed two or more pairs of primers, and we selected the one with the best amplification efficiency in samples across all mouse strains for the subsequent quantification. Primer sequences are available in Table S5 in the supplemental material. qPCR was performed using an ABI 7900HT real-time PCR system, and each assay was run in triplicate using Power SYBR green PCR master mix (Applied Biosystems). We chose the 18S rRNA gene for normalization using geNORM (25) and assaying multiple endogenous controls across all samples. These endogenous controls were the 18S rRNA gene, actin beta (Actb), beta-2 microglobulin (B2m), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), hypoxanthine guanine phosphoribosyl transferase (Hprt1), phosphoglycerate kinase 1 (Pgk1), and transferrin receptor (Tfrc). We selected 39 candidates representing a variety of loci for follow-up by qPCR. We required that candidate loci have genomic locations containing unique sequences for designing PCR primers and a reasonable read coverage suggesting efficient amplification.
ACKNOWLEDGMENTS
Influenza A virus strain A/Pr/8/34 (H1N1) was kindly provided by Peter Palese (Mt. Sinai University, New York), and viral stocks were prepared by Shinobu Yamamoto (University of Washington, Seattle) in pathogen-free eggs (Charles River). We thank Shujun Luo, Gary P. Schroth, and Irina Khrebtukova from Illumina, Inc., for generating the short-read data from selected samples with poly(A) selection. We thank Victoria Carter (University of Washington, Seattle) for generating microarray data.
This work was supported by the National Institutes of Health grant U54 AI081680 (PNWRCE) and by funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, contract no. HHSN272200800060C.
Footnotes
Citation Peng, X., L. Gralinski, C. D. Armour, M. T. Ferris, M. J. Thomas, et al. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1(5):e00206-10. doi:10.1128/mBio.00206-10.
SUPPLEMENTAL MATERIAL
REFERENCES
- 1. Wilusz J. E., Sunwoo H., Spector D. L. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23:1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponting C. P., Oliver P. L., Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136:629–641 [DOI] [PubMed] [Google Scholar]
- 3. Mercer T. R., Dinger M. E., Mattick J. S. 2009. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10:155–159 [DOI] [PubMed] [Google Scholar]
- 4. Gottwein E., Cullen B. R. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts A., Deming D., Paddock C. D., Cheng A., Yount B., Vogel L., Herman B. D., Sheahan T., Heise M., Genrich G. L., Zaki S. R., Baric R., Subbarao K. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armour C. D., Castle J. C., Chen R., Babak T., Loerch P., Jackson S., Shah J. K., Dey J., Rohl C. A., Johnson J. M., Raymond C. K. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods 6:647–649 [DOI] [PubMed] [Google Scholar]
- 7. Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., Cabili M. N., Jaenisch R., Mikkelsen T. S., Jacks T., Hacohen N., Bernstein B. E., Kellis M., Regev A., Rinn J. L., Lander E. S. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frieman M. B., Chen J., Morrison T. E., Whitmore A., Funkhouser W., Ward J. M., Lamirande E. W., Roberts A., Heise M., Subbarao K., Baric R. S. 2010. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 6: e1000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B. E., van Oudenaarden A., Regev A., Lander E. S., Rinn J. L. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ponjavic J., Oliver P. L., Lunter G., Ponting C. P. 2009. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5:e1000617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang D. W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 12. Pang K. C., Dinger M. E., Mercer T. R., Malquori L., Grimmond S. M., Chen W., Mattick J. S. 2009. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 182:7738–7748 [DOI] [PubMed] [Google Scholar]
- 13. Ahanda M. L., Ruby T., Wittzell H., Bed’Hom B., Chausse A. M., Morin V., Oudin A., Chevalier C., Young J. R., Zoorob R. 2009. Non-coding RNAs revealed during identification of genes involved in chicken immune responses. Immunogenetics 61:55–70 [DOI] [PubMed] [Google Scholar]
- 14. Ravasi T., Suzuki H., Pang K. C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M. C., Gongora M. M., Grimmond S. M., Hume D. A., Hayashizaki Y., Mattick J. S. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 16:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Churchill G. A. 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36:1133–1137 [DOI] [PubMed] [Google Scholar]
- 16. Langmead B., Trapnell C., Pop M., Salzberg S. L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trapnell C., Pachter L., Salzberg S. L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dinger M. E., Pang K. C., Mercer T. R., Crowe M. L., Grimmond S. M., Mattick J. S. 2009. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 37:D122–D126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 20. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mituyama T., Yamada K., Hattori E., Okida H., Ono Y., Terai G., Yoshizawa A., Komori T., Asai K. 2009. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 37:D89–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Washietl S., Hofacker I. L., Stadler P. F. 2005. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 102:2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Faulkner G. J., Kimura Y., Daub C. O., Wani S., Plessy C., Irvine K. M., Schroder K., Cloonan N., Steptoe A. L., Lassmann T., Waki K., Hornig N., Arakawa T., Takahashi H., Kawai J., Forrest A. R., Suzuki H., Hayashizaki Y., Hume D. A., Orlando V., Grimmond S. M., Carninci P. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41:563–571 [DOI] [PubMed] [Google Scholar]
- 24. Rozen S., Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 25. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.