Abstract
Twitching motility is a form of surface translocation mediated by the extension, tethering, and retraction of type IV pili. Three independent Tn5-B21 mutations of Pseudomonas aeruginosa with reduced twitching motility were identified in a new locus which encodes a predicted protein of unknown function annotated PA4959 in the P. aeruginosa genome sequence. Complementation of these mutants with the wild-type PA4959 gene, which we designated fimX, restored normal twitching motility. fimX mutants were found to express normal levels of pilin and remained sensitive to pilus-specific bacteriophages, but they exhibited very low levels of surface pili, suggesting that normal pilus function was impaired. The fimX gene product has a molecular weight of 76,000 and contains four predicted domains that are commonly found in signal transduction proteins: a putative response regulator (CheY-like) domain, a PAS-PAC domain (commonly involved in environmental sensing), and DUF1 (or GGDEF) and DUF2 (or EAL) domains, which are thought to be involved in cyclic di-GMP metabolism. Red fluorescent protein fusion experiments showed that FimX is located at one pole of the cell via sequences adjacent to its CheY-like domain. Twitching motility in fimX mutants was found to respond relatively normally to a range of environmental factors but could not be stimulated by tryptone and mucin. These data suggest that fimX is involved in the regulation of twitching motility in response to environmental cues.
Pseudomonas aeruginosa is a bacterium that inhabits a wide variety of environments, including soil, and water, as well as plant and animal tissues (58). It is also a major opportunistic human pathogen that affects individuals who are immunocompromised or suffering from cystic fibrosis (6), as well as a pathogen of a wide variety of other animal and plant species, including mice, fruit flies, nematode worms, and mustard plants (15, 24, 35, 47).
Initiation and establishment of P. aeruginosa infections are dependent on a number of virulence factors (6). These factors include type IV pili (or fimbriae), which are polar filaments involved in the attachment to and translocation across epithelial cell surfaces via a process called twitching motility (8, 30). Twitching motility occurs by extension and retraction of the pili and is required for formation of biofilms (11, 32), a mode of communal organization which is observed in chronic infections (44) and which appears to provide protection against antibiotics and the host immune system (12). Twitching motility is also involved in other developmental processes, such as fruiting body formation in Myxococcus xanthus (30). In addition, retractile type IV pili act as receptors for the binding and entry of certain bacteriophages (9).
To date, around 40 genes at a number of different genomic loci have been identified as genes that are involved in the biogenesis and function of type IV pili in P. aeruginosa (30). These include the genes encoding the major structural protein (PilA) and minor proteins that may form the base and/or the tip of the pilus (PilE,PilV, PilW, PilX, PilY1, PilY2, and FimT), genes whose products are required for pilus assembly and retraction (PilB, PilC, PilD, PilF, PilM, PilN, PilO, PilP, PilQ, PilT, and PilU), and other genes whose products have unknown functions (PilF, PilZ, and FimV). In addition, there are a number of genes which encode regulatory proteins that control both the production of pili (and other virulence determinants) and the activity of twitching motility in response to environmental stimuli. The proteins that have been identified to date are (i) the classical two-component sensor-regulator pair PilS-PilR, which along with the alternative sigma factor RpoN are required for transcription of the fimbrial subunit gene pilA (26); (ii) the atypical sensor-regulator pair FimS-AlgR, which along with the alternative sigma factor AlgU regulate twitching motility and production of the exopolysaccharide alginate (30, 56, 57); (iii) the global carbon metabolism regulator CRC, which partially regulates transcription of pilA (31); (iv) PilG-PilK and ChpA-ChpE, which comprise a complex chemosensory system which appears to control the direction and rate of twitching motility and which is similar to the Che system that controls swimming motility in Escherichia coli and the Frz system that controls social gliding motility in M. xanthus (16, 17, 30); and (v) Vfr, a homolog of the E. coli catabolite repressor protein Crp, which differentially regulates twitching motility and elastase production in P. aeruginosa (5) and which has recently been shown to control expression of the majority of the genes required for type IV pilus biogenesis and twitching motility (59).
Here we describe identification of a new gene, fimX, whose product is also required for normal twitching motility. FimX has domains that are commonly present in signal transduction proteins (PAS-PAC and CheY-like domains) and are involved in cyclic di-GMP metabolism (DUF1 and DUF2), and it is located at one pole of the cell. fimX mutants have low levels of surface pili, have impaired twitching motility, and fail to respond to some (but not other) environmental signals which normally stimulate twitching motility. Therefore, FimX appears to be a new type of protein that connects environmental signals to twitching motility, involving signal sensing, phosphotransfer activity, and cyclic di-GMP metabolism.
MATERIALS AND METHODS
Media, bacterial strains, and plasmids.
E. coli and P. aeruginosa liquid cultures were grown in Luria-Bertani broth and solid medium, which was prepared by adding 1 to 1.5% Select agar (Gibco-BRL). An optically clear medium was used for light microscopy, and it contained (per liter) 4 g of tryptone, 2 g of yeast extract, 2 g of NaCl, and 8 g of GelGro (ICN). The medium used for assessment of the effects of different nutrients on twitching motility contained 0.5% (wt/vol) yeast extract, 100 mM potassium phosphate buffer (pH 7.0), and 1% agar, together with supplements at the concentrations indicated.
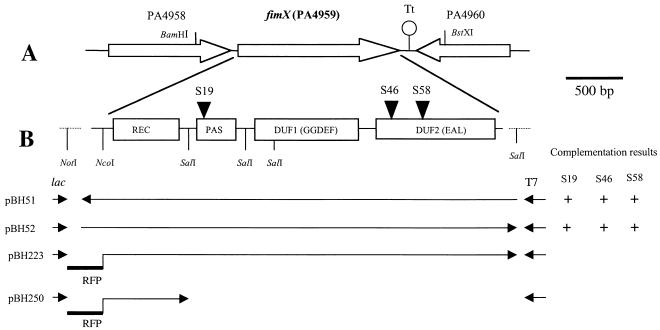
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strain DH5α (recA endA1 gyrA96 hsdR17 thi-1 supE44 relA1 φ80 dlacZΔM15) was used for all genetic manipulations. The P. aeruginosa strains used were PAK (D. Bradley, Memorial University of Newfoundland, St. John's, Canada); Tn5-B21 mutants of this strain (26), including the pilV mutant R306; and PAKpilA::Tcr (54). The wild-type fimX gene was isolated from the P. aeruginosa PAO1 minimal tiling path cosmid library (27). A 2.7-kb BamHI-BstXI fragment from cosmid pMO012502, covering positions −462 to 186 upstream and downstream of the start and stop codons, respectively, of the fimX coding sequence, was subcloned (after the BstXI end was blunted with T4 DNA polymerase) into the BamHI and EcoRV sites of the vectors pUCPSK and pUCPKS (53), producing the constructs pBH51 (wild-type fimX in the opposite orientation with respect to the lac promoter) and pBH52 (wild-type fimX in the forward orientation with respect to the lac promoter) (Fig. 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid(s) | Description | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | E. colia | Lab collection |
| PAK | Wild-type P. aeruginosa strain K | Lab collection |
| PAKΔpilA | PAK pilA::Tcr; previously referred to as AWK | 54 |
| PAKΔpilV | PAK Tn5-B21 mutant of pilV; previously referred to as R306 | 3 |
| S19 | PAK Tn5-B21 fimX mutant | This study |
| S46 | PAK Tn5-B21 fimX mutant | This study |
| S58 | PAK Tn5-B21 fimX mutant | This study |
| Plasmids | ||
| pUCPKS, pUCPSK | P. aeruginosa T7 expression vectors | 53 |
| pBluescript II KS | Ampr cloning vector | Stratagene |
| pMO012502 | pLA2917 cosmid from PAO1 library found to contain fimX | 27 |
| pBH51 | 2.7 kb of BamHI-BstXI fragment cloned in pUCPKS vector (fimX in direction of T7 promoter) | This study |
| pBH52 | 2.7 kb of BamHI-BstXI fragment cloned in pUCPSK vector (fimX in direction of lac promoter) | This study |
| pBH203 | 0.8-kb amplified RFP fragment cloned in pGEM-T Easy vector (Promega) | This study |
| pBH210 | 0.8-kb RFP as NotI fragment from pBH203 subcloned in pUCPSK | This study |
| pBH223 | 0.8-kb NotI-NcoI fragment of RFP cloned in frame in pBH52 before fimX | This study |
| PBH250 | Truncated fimX containing RFP and REC domain, derived from digestion of pBH223 with SalI and self-ligation | This study |
See text.
FIG. 1.
Schematic representation of the fimX locus. The relevant restriction sites are indicated. (A) Overall topography of the fimX region. The open arrows indicate the relative transcriptional orientations of fimX and its neighboring genes, and Tt indicates the predicted transcription terminator following fimX. (B) Expanded view of fimX. The rectangles indicate the predicted domains in FimX. The transposon insertion sites in fimX mutants S19, S46, and S58 are indicated by solid triangles. The orientations of fimX in derived plasmid constructs are indicated by arrows; the arrows on the left indicate that the fimX coding region is in the same orientation as the adjacent lac promoter, and the arrows on the right indicate that the fimX coding region is in the same orientation as the adjacent T7 promoter. Note that the NotI and last SalI restriction sites were derived from the multiple cloning site of the pUCPKS and pUCPSK vectors, which were used to construct translational fusions with RFP for subcellular localization studies.
To examine the subcellular localization of fimX, a red fluorescent protein (RFP), which has been successfully employed previously for Pseudomonas labeling (49), was used in fusion experiments. The RFP was obtained from plasmid pUT-RFP (GenBank accession no. AF102233), kindly provided by S. Molin, Department of Microbiology, Technical University of Denmark. Primers RFP-1 and RFP-2 (sequences available on request) were used to amplify the RFP coding sequence and to introduce adjacent NotI and NcoI restriction sites. The RFP was cloned in frame by using the NotI site on the plasmid vector and the NcoI site before the start codon of fimX (Fig. 1) in pBH52, which produced the construct pBH223, in which the RFP coding region was under control of the inducible lac promoter. pBH223 was also digested with SalI to remove most of the domains of FimX, leaving RFP fused only to the N-terminal CheY-like domain, which produced construct pBH250. As a positive control, the amplified RFP was cloned into the vector pGEM-T Easy (Promega), from which the NotI fragment containing RFP was subcloned into pUCPSK, which produced the construct pBH210. All constructs were transformed into PAK and mutants derived from this strain to examine the localization of fimX by fluorescence microscopy.
Preparation and transformation of competent P. aeruginosa cells were carried out by using MgCl2 treatment as described previously (3). The following antibiotic concentrations were used for selection in E. coli: 100 μg of ampicillin ml−1, 10 μg of tetracycline ml−1, and 40 μg of tetracycline ml−1 for cosmid selection. Selection in P. aeruginosa was carried out with 250 μg of carbenicillin ml−1.
Recombinant DNA techniques and sequence analysis.
Preparation of cosmid and plasmid DNAs, restriction endonuclease digestion, DNA extraction from agarose gels, and ligation reactions were carried out by using standard protocols (39) and the manufacturers' instructions. The enzymes used for DNA manipulation were purchased from Roche and New England Biolabs.
Genomic sequences flanking the site of transposon insertion in the remaining transposon (Tn5-B21) mutants of PAK were obtained by marker rescue cloning (26). Chromosomal DNA from each of the mutants was digested with EcoRI (which cut on one side of the tetracycline resistance gene located in Tn5-B21), followed by ligation into the EcoRI site of pBluescript II KS and selection on medium containing both ampicillin and tetracycline. The resulting plasmids contained an insert that spanned the junction between Tn5-B21 and PAK chromosomal DNA in the mutants. These plasmids were then sequenced outward from the transposon by using primer Ollie2 (26), which was complementary to a region near the terminus of the transposon Tn5-B21 cassette, to determine the sequence adjacent to the point of transposon insertion. Automated DNA sequencing was performed by the Australian Genome Research Facility (University of Queensland, Brisbane, Australia) with a Big Dye sequencing kit from Applied Biosystems. BLAST searches of the P. aeruginosa PAO1 genome sequence at the National Center for Biotechnology Information (Bethesda, Md.) were carried out to identify the position of transposon insertion. Further information on the interrupted gene and the adjacent genomic landscape was obtained from the P. aeruginosa interactive databases at http://www.bit.uq.edu.au/pseudomonas (13) and http://www.pseudomonas.com. Information on the domain structure of predicted proteins was obtained by searching the FimX protein sequence against the SMART database at http://smart.embl-heidelberg.de (29).
Western blotting and ELISA.
One hundred microliters of an overnight broth culture was spread onto a freshly prepared Luria-Bertani agar plate and incubated at 37°C for 24 h. Surface pili were isolated by harvesting the resulting cells in 2 ml of phosphate-buffered saline and vortexing for 2 min. The suspension was centrifuged at low speed (2,300 × g for 5 min) to remove the whole cells, after which the supernatant was collected and subjected to high-speed centrifugation (15,000 × g for 20 min) to remove the cell debris. The resulting supernatant was incubated overnight at 4°C in the presence of 100 mM MgCl2 to precipitate pili, as described previously (2). The precipitate was collected by centrifugation (15,000 × g for 20 min) and suspended in gel loading buffer as described above. The whole-cell fraction was prepared as previously described (3). Western blotting to detect pili in the surface and whole-cell fractions and quantification of the surface pili in P. aeruginosa cultures by an enzyme-linked immunosorbent assay (ELISA) were carried out as described previously (3, 41).
Twitching motility assays and microscopy.
The twitching motility activities of P. aeruginosa strains and mutants were assayed by the subsurface agar stab method, as described previously (3). After 24 h of incubation at 37°C, the size of the twitching zone around the inoculation site at the interface between the agar and the petri dish surface was measured by eye and/or after staining with 0.05% Coomassie brilliant blue R250 to increase the contrast (3).
To investigate the influence of nutrients on mutant twitching motility under standard conditions, 15-ml portions of medium containing 1% agar were poured into 9-cm-diameter petri dishes and dried at 43°C for 15 min prior to stab inoculation of wild-type and mutant strains. The plates were then incubated in a humidified incubator at 37°C, and the diameters of the twitching zones at the agar-petri dish interfaces were measured after 24 and 48 h of incubation. The diameter of each zone was measured by using two cross sections (at right angles), and five replicate plates were used in each assay. The average area of the twitching zone for each plate was calculated, and when a significant difference was observed in the ratio of the diameter of the twitching zone of PAK to the diameter of the twitching zone of a mutant under particular conditions, the experiment was repeated two more times.
Light microscopy of twitching zones was performed as described previously (41). Briefly, sterile microscope slides were submerged in molten GelGro medium at approximately 60°C to coat them with a thin layer of medium. The slides were set in a horizontal position and air dried for 2 min before use. Each of the slides was then inoculated with a small loopful of bacteria taken from an overnight plate culture. A sterile glass coverslip was placed over the point of inoculation, and the slide transferred to a large petri dish containing a moist tissue and sealed with Parafilm to maintain humid conditions. After incubation at 37°C for 2 h, slide cultures were examined with an Olympus AX70 microscope at a magnification of ×200.
The transformants of PAK and mutants with RFP-fimX translational fusions were incubated at 4°C for 2 days for maturation of RFP before they were examined with the Olympus AX70 fluorescence microscope with the standard rhodamine filter. Dural light sources were used in the examination to outline the bacterial cells; this involved bright-field light from underneath the slide and fluorescent light from above the slide.
RESULTS
fimX is required for normal twitching motility.
Marker rescue cloning was used to obtain the DNA sequence adjacent to the transposon insertion site in each of the 93 remaining twitching motility mutants from the Tn5-B21 transposon library that was generated previously (26). Database searches with these sequences revealed that three of the mutants (S19, S46, and S58) had independent transposon insertions in the same open reading frame (ORF) (Fig. 1), which was designated ORF PA4959 (encoding a hypothetical protein) in the sequenced PAO1 genome (http://www.pseudomonas.com) (45).
Twitching motility in the S19, S46 and S58 mutants was reduced but not absent (Fig. 2). The diameters of the twitching zones of the mutants were slightly less than one-half the diameter of the twitching zone of the wild-type parent strain PAK, whereas no twitching zone was observed for the PAKΔpilA mutant, which lacks pili and twitching motility altogether (54) (Fig. 2B). The growth rate of the mutants was the same as the growth rate of the PAK parental strain (data not shown), suggesting that the impaired twitching motility was not simply due to a growth defect. On this basis, the PA4959 gene was designated fimX.
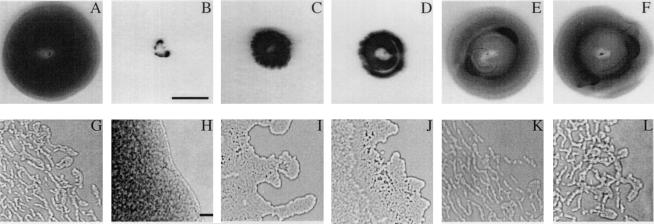
FIG. 2.
Macroscopic and microscopic examination of twitching motility in fimX mutants. (A to F) Twitching zones observed in the subsurface stab assay on agar plates after 24 h of growth. Bar = 1 cm. (G to L) Light microscopy of the edges of the twitching zones at the interstitial surfaces between the glass coverslips and GelGro medium. Bar = 10 μm. (A and G) Wild-type strain PAK; (B and H) PAKΔpilA mutant; (C and I) fimX mutant S19; (D and J) S19(pUCPSK); (E and K) S19(pBH51); (F and L) S19(pBH52). The medium used for the subsurface twitching assay in complementation studies with the control vector or vectors containing fimX sequences contained 250 μg of carbenicillin ml−1. Similar results were obtained for complementation of mutants S46 and S58 (data not shown).
The fimX gene is located at the 3′ end of a cluster of five ORFs (PA4955 to PA4959), which are in the same transcriptional orientation and are separated by no more than 75 nucleotides (except PA4957 and PA4958, which are separated by 678 nucleotides). fimX encodes a 691-amino-acid protein with a predicted molecular weight of approximately 76,000, which was confirmed by protein expression studies performed with an inducible T7 promoter in the presence of radiolabeled methionine (data not shown). The fimX coding sequence is followed closely by a likely rho-independent transcriptional terminator sequence, 5′-ATGAAGAACGGGCGCCCTGGGCGCCCGTTCTTTT-3′ (http://www.tigr.org/software/TransTermResults/ntpa03.html). Wild-type fimX cloned in the forward and reverse directions with respect to the lac promoter in the vectors pUCPSK and pUCPKS (Fig. 1) restored normal twitching motility to all three fimX mutants (Fig. 2E and F), suggesting that the level of fimX expression is not critical to functioning of the gene and that the cloned sequence may include an endogenous promoter. In addition, expression of cloned fimX did not impair twitching motility in wild-type cells, confirming that the activity of FimX was not dose sensitive (data not shown), in contrast to the activities of some other proteins which affect twitching motility, such as FimV (41).
Analysis of the phenotype of fimX mutants.
Twitching motility in the S19 mutant and its complemented transformants was analyzed in more detail by light microscopy (Fig. 2G to L). Normal twitching motility in wild-type cells involves outward movement of broad rafts of cells, followed by a breaking up of the rafts into a thinner lattice-like network, within which cells traverse up and down with frequent reversals of movement (Fig. 2G), whereas pilA mutants (which lack type IV pili and twitching motility) have smooth and relatively static colony edges (Fig. 2H) and exhibit no network formation (41). In contrast, S19 and transformant S19(pUCPSK) exhibited significantly reduced outward movement of the rafts and a lack of the lattice-like network (Fig. 2I and J). Complementation of S19 with cloned fimX restored the normal micromorphology of the twitching zone in S19(pBH51) and S19(pBH52) (Fig. 2K and L). This suggests that the absence of FimX interferes with the signal transduction systems which control the frequent reversals in cell movement that are involved in lattice formation and which are typical of the leading edge of twitching motility-mediated colony expedition(30).
fimX mutants were also analyzed for expression of pilin and the assembly of surface pili by Western blotting and ELISA. fimX mutants exhibited relatively normal levels of intracellular pilin, similar to the levels in both the wild type and pilV mutants (Fig. 3C), but the amounts of surface-assembled pili were significantly reduced. Complementation of fimX mutants with cloned fimX restored the surface pilus levels to levels that appeared to be quantitatively higher than the wild-type levels (Fig. 3D), although twitching motility appeared to be normal in these complemented cells.
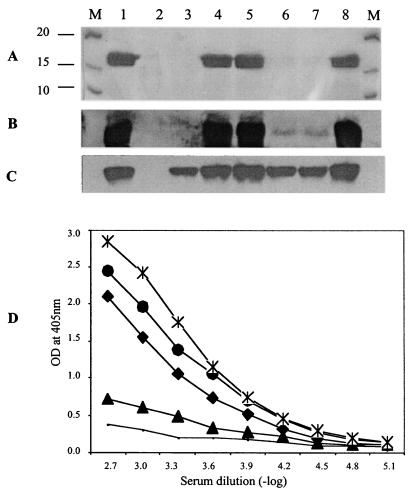
FIG. 3.
Western and ELISA analyses of pilus production in fimX mutants. (A) Surface pili extracted from PAK (lane 1), PAKΔpilA (lane 2), PAKΔpilV (lane 3), PAK(pUCPSK) (lane 4), PAK(pBH52) (lane 5), S19 (lane 6), S19(pUCPSK) (lane 7), and S19(pBH52) (lane 8). The gel was stained with Coomassie brilliant blue R250. (B) Western blotting of the surface pili from the same strains that were used in panel A. (C) Western blotting of the whole-cell proteins from the same strains that were used in panel A. (D) Quantitative analysis of the level of surface pili by ELISA for PAK (♦), PAKΔpilA (solid line), S19 (▴), S19(pBH51) (•), and S19(pBH52) (*). OD, optical density.
Analysis of the fimX sequence.
Database searches showed that FimX exhibits significant homology over its entire sequence (691 amino acids) to hypothetical proteins encoded in a number of type IV piliated members of the γ subclass of the Proteobacteria, including Microbulbifer degradans (Mdeg2095, 39% identity and 60% similarity) (23), Xanthomonas axonopodis (XAC2398, 30% identity and 48% similarity) (18), Xanthomonas campestris (XCC2291, 30% identity and 49% similarity) (18), and Xylella fastidiosa (XF2624, 29% identity and 48% similarity) (43), but not in some other sequenced type IV piliated bacteria, such as Neisseria gonorrhoeae, suggesting that regulation of pilus function by environmental signals (see below) is different in the latter organisms, which would not be surprising given the different ecology of these species as free-living organisms versus obligate pathogens. FimX contains four recognizable domains (identified by using the SMART database [http://smart.embl-heidelberg.de]) which are found in various combinations in a variety of signal transduction proteins in a wide range of bacteria (see below), which provides further evidence that protein domain shuffling has occurred during prokaryotic evolution, as well as during eukaryotic evolution. The N-terminal region of FimX (residues 8 to 119) contains a predicted but unusual REC (CheY-like) domain, which normally receives a phosphoryl group from histidine phosphotransfer domains in other proteins, as part of a signal transduction cascade (42), although in the case of FimX the critical aspartate residue and several other normally conserved features of this domain are missing (Fig. 4A).
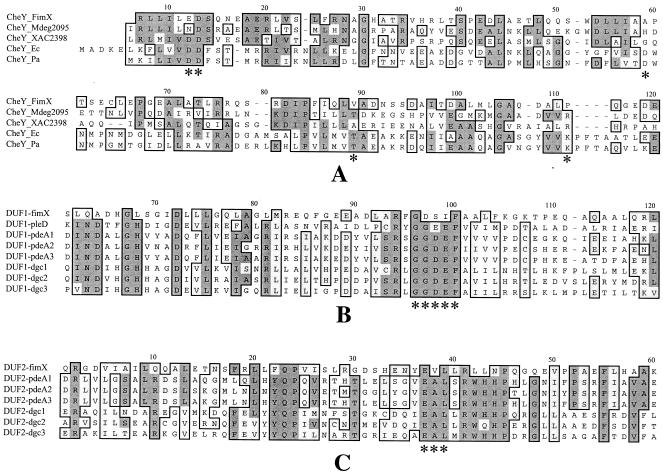
FIG. 4.
Comparison of the CheY-like, DUF1, and DUF2 domains of FimX with the domains of other proteins. (A) Alignment of the CheY-like domain of FimX with domains of other typical CheY-containing proteins. The five conserved functional sites identified by Volz (51) are indicated by asterisks. The sources of the CheY sequences (with their accession numbers) are as follows: CheY_Mdeg2095, putative CheY-like domain of Mdeg2095 in M. degradans; CheY_XAC2398, putative CheY-like domain of XAC2398 in X. axonopodis; CheY_Pa, CheY of P. aeruginosa (AAG04845); CheY_Ec, CheY of E. coli (NP_416396). (B and C) Alignment of the DUF1 and DUF2 domains of FimX with the corresponding domains of PleD, PdeA1 to PdeA3 and Dgc1 to Dgc3. The conserved functional sites in DUF1 and DUF2 are indicated by asterisks. The sequences are from references 25 and 46.
FimX residues 144 to 210 encompass a predicted PAS-PAC domain, which is present in a wide range of proteins involved in light, oxygen, and redox sensing, as well as in some ion channel proteins (34, 60). Residues 256 to 428 encompass a predicted DUF1 (or GGDEF) domain whose exact function is unknown but which was first recognized in the Caulobacter crescentus signaling protein PleD, which is involved in flagellum rotation and cell differentiation (25). Residues 439 to 683 encompass a predicted DUF2 (or EAL) domain, whose function also is not known. Both DUF1 and DUF2 domains were first recognized in the diguanylate cyclase gene clusters, including pdeA1 to pdeA3 and dgc1 to dgc3, which control the cellular synthesis and turnover of cyclic di-GMP in Acetobacter xylinum (46). Like the CheY-like domain, FimX shows significant deviation from the normal consensus of DUF1 and DUF2 domains (Fig. 4B and C).
Environmental assays.
The fact that FimX contains four domains which have known or inferred functions in signal transduction and the fact that a number of other proteins that affect twitching motility are also predicted to be part of sensory signaling pathways (30) suggest that environmental and cellular signals are important in determining the twitching motility activity, which would not be unexpected. A range of nutrients and other compounds which either stimulate or inhibit twitching motility in P. aeruginosa have been identified (C. B. Whitchurch, A. B. T. Semmler, and J. S. Mattick, unpublished data). Twitching motility in wild-type cells is stimulated by mucin (0.05%), bovine serum albumin (BSA) (0.1%), and tryptone (5%), whereas it is inhibited by high-osmolarity conditions, including 300 mM NaCl, 300 mM KCl, 50 mM KNO3, 5% sucrose, 10% glucose, and 2% polyvinylpyrrolidone.
Twitching motility in fimX mutants responded like twitching motility in the wild type under most conditions tested, except in the case of added tryptone and mucin; under these conditions twitching was greatly stimulated in the wild type but not in the mutant (Table 2). Other compounds which stimulate (0.1% BSA) or inhibit (300 mM NaCl) twitching motility had similar effects on the wild type and the fimX mutants.
TABLE 2.
Twitching zone diameters and sizes of PAK and S19 in environmental assays
| Medium | Strain | Diam (cm) | Area (cm2) | Ratio of PAK area to S19 area | Relative change in zone area (added nutrient/ base medium) |
|---|---|---|---|---|---|
| Base mediuma | PAK | 2.45 ± 0.26 (15)b | 4.77 ± 1.04 | 4.1 | 1 |
| S19 | 1.21 ± 0.09 (15) | 1.16 ± 0.17 | 1 | ||
| Mucin (0.05%) | PAK | 5.28 ± 0.14 (18) | 21.86 ± 1.17 | 10.0 | 4.58 |
| S19 | 1.66 ± 0.18 (18) | 2.18 ± 0.48 | 1.88 | ||
| Tryptone (5%) | PAK | 5.37 ± 0.18 (15) | 22.68 ± 1.55 | 12.5 | 4.75 |
| S19 | 1.51 ± 0.18 (15) | 1.82 ± 0.45 | 1.57 | ||
| BSA (0.1%) | PAK | 5.24 ± 0.13 (18) | 21.58 ± 1.06 | 4.0 | 4.52 |
| S19 | 2.63 ± 0.09 (18) | 5.42 ± 0.38 | 4.67 | ||
| NaCl (300 mM) | PAK | 1.13 ± 0.17 (16) | 1.03 ± 0.32 | 3.2 | 0.22 |
| S19 | 0.63 ± 0.09 (16) | 0.32 ± 0.09 | 0.28 |
The base medium contained 0.5% yeast extract and 100 mM potassium phosphate buffer (pH 7.0).
The numbers in parentheses are the numbers of zones examined.
Localization of FimX.
The RFP-fimX translational fusion construct (pBH223) was transformed into PAK and S19 to examine the subcellular location of FimX. We previously determined that the activity of FimX is not significantly affected by fusion to RFP because the RFP-fimX construct is able to complement the fimX mutant S19 (data not shown). In both the wild type and S19 mutants the RFP-labeled FimX was localized at one pole of the cell (Fig. 5A and B). We attempted to orient this pole with respect to the position of the flagella, but the methods that we utilized for flagellum staining either did not give reliable results (21) or eliminated the fluorescence signal (55).
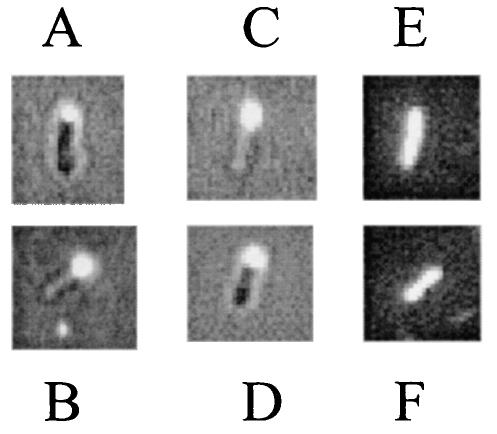
FIG. 5.
Localization of RFP fused with full-length FimX and with the CheY-like domain of FimX in PAK and S19. (A) PAK(pBH223) (RFP fused to FimX); (B) S19(pBH223); (C) PAK(pBH250) (RFP fused to the N-terminal region of FimX containing the CheY-like domain and adjacent sequences); (D) S19(pBH250); (E) PAK(pBH210) (RFP control); (F) S19(pBH210). Panels A to D were photographed with a dual light source to reveal the location of fluorescence in relation to the cell as a whole. The background is reduced in panels E and F, as only red fluorescent light and no background bright-field light were used in these cases.
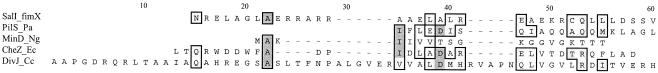
In order to examine which part of FimX might be responsible for the polar localization, we performed deletion experiments in which fragments of FimX were fused to RFP. We found that the polar location signal in FimX is located in the N-terminal region of the protein (encoded by a SalI fragment [Fig. 1 and 5C and D]), which includes the CheY-like domain and a subsequent stretch of 35 amino acids that exhibits homology to protein sequences implicated in polar localization of other proteins (Fig. 6), including PilS in P. aeruginosa (20), DivJ in C. crescentus (40), MinD in N. gonorrhoeae (36), and CheZ in E. coli (10). Although the truncated fusion protein (pBH250) contained the localization sequence, it failed to complement the mutants (data not shown), suggesting that some or all of the other domains of FimX (PAS-PAC, DUF1, and DUF2) are required for its proper function, as might be expected. In order to identify proteins that might interact with FimX, 42 other known mutants (5, 30) with mutations which affect type IV pilus biogenesis and/or twitching motility and its regulation were transformed with pBH223 to examine whether the absence of any of the proteins might eliminate the polar location of FimX. These mutants were pilA-X, pilY1, pilY2, pilZ, fimH, fimL, fimS, fimT, fimU, fimV, chpA-E, rpoN, algR, vfr, and tonB3 mutants, and three of them (fimH, fimL, and tonB3) have not been described. In all of these mutants except the vfr mutant, FimX remained localized at the pole. In the vfr mutant no fluorescence signal was observed despite repeated attempts, indicating that the RFP-FimX fluorescence signal was dispersed and/or that the fusion product was unstable in the vfr mutant, since RFP itself was stably detected in P. aeruginosa (Fig. 5E and F). This suggests that Vfr or, more likely, a gene product regulated by Vfr may be required for the polar localization of FimX, presumably in a complex with other proteins. Alternatively, since Vfr has been recently shown to control expression of fimX (PA4959) (59), it is also possible that RFP-FimX was not expressed at detectable levels in the vfr mutant despite the fact that the RFP-FimX fusion was cloned in the direction of the lac promoter.
FIG. 6.
Alignment of amino acid sequence adjacent to the CheY-like domain of FimX with amino acid sequences implicated in polar localization in other bacteria. The amino acid sequence between the SalI site and the end of the CheY-like domain of FimX (SalI_FimX) was compared to the equivalent regions of other proteins implicated in polar localization in P. aeruginosa and other species, including PilS in P. aeruginosa (PilS_Pa) (20), MinD in N. gonorrhoeae (MinD_Ng) (36), CheZ in E. coli (CheZ_Ec) (10), and DivJ in C. crescentus (DivJ_Cc) (40).
DISCUSSION
In this study we identified a new gene, fimX, which is required for twitching motility, bringing to 40 the number of reported genes known to be required for the biogenesis and functioning of type IV pili and twitching motility in P. aeruginosa (30). FimX has a polar location, like type IV pili and like PilS, which is the sensor in a two-component regulatory system that controls transcription of the pilin subunit PilA (7, 26), and it appears to be a signal transduction protein that connects environmental signals to control of twitching motility through an unknown mechanism.
FimX has an unusual domain composition; it has a PAS-PAC sensor domain (PAS) and a CheY-like domain fused with DUF1 and DUF2 domains, whose functions are not fully understood but which are implicated in cyclic di-GMP metabolism. These four domains are widely present in signal transduction proteins (22), which suggests that FimX may integrate multiple regulatory signals. However, while the CheY-like domain exhibits sufficient homology to other well-characterized CheY domains (approximately 15% identity and 26% similarity) to be predicted to be a REC domain by SMART, it lacks certain critical residues (Fig. 4A). There are five conserved residues at the active site in the CheY domain superfamily, including D12, D13, D57, T87, and K109 (51). Except for D13, these conserved active site residues are not conserved in FimX (which has E12, A57, V87, and L109); this includes the phospho-accepting aspartate residue (D57A), suggesting that this domain in FimX is not active in phosphotransfer cascades but rather inhibits or competes with an analogous CheY domain in another protein, which receives and/or donates phosphoryl groups in another pathway. This pathway may be the Chp chemosensory system, which also controls twitching motility and which includes two conventional CheY proteins (PilG and PilH) and another protein, ChpA, which contains a conventional CheY domain at its carboxy-terminal end (30). This is also consistent with the polar location of FimX.
In the P. aeruginosa genome, there are 38 ORFs containing DUF1 and/or DUF2 domains (13); two of these ORFs have only the DUF1 domain (PA0169 and PA3177), and one has only the DUF2 domain (PA2133). All of the rest also contain other known or suspected signaling domains, including REC, PAS-PAC, GAF, HAMP, PBPb, and CBS domains (13, 22). In addition, 23 proteins containing DUF1 and/or DUF2 domains (but not FimX) have one to several predicted transmembrane domains, suggesting that the majority of the DUF1 and/or DUF2 domain-containing proteins may be membrane bound for environmental signal transduction. It is apparent that DUF1- and DUF2-containing proteins are all members of a signal transduction system whose precise function(s) remains unknown but which appear to be connected to cyclic di-GMP metabolism (see below), which also implies that cyclic di-GMP, along with other guanine nucleotides, may be part of a global regulatory network in P. aeruginosa that intersects with twitching motility.
A range of studies have suggested that proteins containing DUF1 and DUF2 domains are involved in the biosynthesis and degradation of cyclic diguanylate, an intracellular signal regulating production of extracellular cellulose (22, 46). This system may be a widespread means of physiological regulation in bacteria (22, 33). The DUF1 domain has a fold similar to that of the eukaryotic cyclase catalytic domain, which is involved in the formation of cAMP, an important signal transduction messenger in both prokaryotic and eukaryotic cells (28, 33). DUF1 domain-containing proteins have diguanylate cyclase activity and are interchangeable in bacterial species (4, 46). The DUF2 domain has been suggested to have a phosphodiesterase activity and a possible role in degrading cyclic diguanylate (22, 46). Recently, the functions of the DUF1 domain-containing protein WspR and the DUF2 domain-containing protein PvrR in P. aeruginosa have been reported (14, 19). WspR is a suppressor that controls an autoaggregation phenotype and is linked to regulation of cup genes that encode a putative fimbrial adhesin required for biofilm formation (14). PvrR regulates the conversion between antibiotic-resistant rough small-colony variants and antibiotic-susceptible wild-type forms (19). PvrR is also involved in autoaggregation, adhesiveness of the bacterial cell surface, and biofilm formation (14, 19). The function of PleD, which is also a DUF1 domain-containing protein, has been studied in depth in C. crescentus (1, 25). PleD is required both for differential development of the swarmer- to-stalked-cell transition and for turning off flagellum rotation. FimX may have a similar function, as twitching motility is implicated in developmental phenomena, such as fruiting body formation in M. xanthus and biofilm formation in P. aeruginosa (30, 32, 52), possibly by affecting the rate of pilus assembly or retraction, which would be consistent with our observation that FimX mutants have strongly reduced levels of extracellular pili (Fig. 3).
It has been reported that certain environmental conditions, such as the concentrations of NaCl, glycerol, carbon, nitrogen, and phosphate, can influence mucoidy in P. aeruginosa (48). In our laboratory, we have observed that certain polypeptides, notably tryptone, mucin, and BSA, can stimulate twitching motility in vitro. Our results show that compared to the wild type, fimX mutants are unable to respond to stimulation by mucin or tryptone but are able to respond relatively normally to BSA. The mucin signal is not due to a low-molecular-weight contaminant, as extensive dialysis of the mucin solution failed to eliminate its stimulatory effect on twitching in wild-type cells. Mucin is a major component of respiratory and stomach secretions (50) and is a glycoprotein that consists of a polypeptide core with branched oligosaccharide side chains, each of which contains 8 to 10 sugars. P. aeruginosa has been reported to exhibit preferential binding to mucin, which is regarded as an important molecule in the initial colonization by this organism of the airways of cystic fibrosis patients (38). Pili are not essential for mucin binding as pilin-deficient mutants have binding ability similar to that of the wild type (37, 38); rather, it appears that mucin is bound through the flagellar cap protein FliD (37). However, as shown here, addition of very low concentrations of mucin to the medium dramatically increases twitching motility in P. aeruginosa, which may accelerate surface colonization of the cells in infected tissue.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., A. J. Bodero, P. D. Free, and J. S. Mattick. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:46-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. [DOI] [PubMed] [Google Scholar]
- 4.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 5.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, J. M. 2000. Localization of the histidine kinase PilS to the poles of Pseudomonas aeruginosa and identification of a localization domain. Mol. Microbiol. 36:153-162. [DOI] [PubMed] [Google Scholar]
- 8.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, D. E., and T. L. Pitt. 1974. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J. Gen. Virol. 24:1-15. [DOI] [PubMed] [Google Scholar]
- 10.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, P., and L. L. Burrows. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 9:50-52. [DOI] [PubMed] [Google Scholar]
- 13.Croft, L., S. A. Beatson, C. B. Whitchurch, B. Huang, R. L. Blakeley, and J. S. Mattick. 2000. An interactive web-based Pseudomonas aeruginosa database: discovery of new genes, pathways and structures. Microbiology 146:2351-2364. [DOI] [PubMed] [Google Scholar]
- 14.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darzins, A. 1993. The pilG gene product, required for Pseudomonas aeruginosa pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J. Bacteriol. 175:5934-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darzins, A., and M. A. Russell. 1997. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system—a review. Gene 192:109-115. [DOI] [PubMed] [Google Scholar]
- 18.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 19.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 20.Ethier, J., and J. M. Boyd. 2000. Topological analysis and role of the transmembrane domain in polar targeting of PilS, a Pseudomonas aeruginosa sensor kinase. Mol. Microbiol. 38:891-903. [DOI] [PubMed] [Google Scholar]
- 21.Forbes, L. 1981. Rapid flagella stain. J. Clin. Microbiol. 13:807-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, J. M., and R. M. Weiner. 2000. Phylogenetic characterization of marine bacterium strain 2-40, a degrader of complex polysaccharides. Int. J. Syst. E vol. Microbiol. 50:831-834. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, H. P. 1997. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene 192:99-108. [DOI] [PubMed] [Google Scholar]
- 25.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer- to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobbs, M., E. S. Collie, P. D. Free, S. P. Livingston, and J. S. Mattick. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7:669-682. [DOI] [PubMed] [Google Scholar]
- 27.Huang, B., C. B. Whitchurch, L. Croft, S. A. Beatson, and J. S. Mattick. 2000. A minimal tiling path cosmid library for functional analysis of the Pseudomonas aeruginosa PAO1 genome. Microb. Comp. Genomics 5:189-203. [DOI] [PubMed] [Google Scholar]
- 28.Hunter, T. 2000. Signaling—2000 and beyond. Cell 100:113-127. [DOI] [PubMed] [Google Scholar]
- 29.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 33.Pei, J., and N. V. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210-216. [DOI] [PubMed] [Google Scholar]
- 34.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 35.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez-Arcos, S., J. Szeto, J. A. Dillon, and W. Margolin. 2002. Conservation of dynamic localization among MinD and MinE orthologues: oscillation of Neisseria gonorrhoeae proteins in Escherichia coli. Mol. Microbiol. 46:493-504. [DOI] [PubMed] [Google Scholar]
- 37.Ramphal, R. 1999. Molecular basis of mucin-Pseudomonas interactions. Biochem. Soc. Trans. 27:474-477. [DOI] [PubMed] [Google Scholar]
- 38.Ramphal, R., L. Koo, K. S. Ishimoto, P. A. Totten, J. C. Lara, and S. Lory. 1991. Adhesion of Pseudomonas aeruginosa pilin-deficient mutants to mucin. Infect. Immun. 59:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 40.Sciochetti, S. A., T. Lane, N. Ohta, and N. A. 2002. Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus. J. Bacteriol. 184:6037-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semmler, A. B., C. B. Whitchurch, A. J. Leech, and J. S. Mattick. 2000. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146:1321-1332. [DOI] [PubMed] [Google Scholar]
- 42.Silversmith, R. E., and R. B. Bourret. 1999. Throwing the switch in bacterial chemotaxis. Trends Microbiol. 7:16-22. [DOI] [PubMed] [Google Scholar]
- 43.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, M. V. Marques, E. A. Martins, E. M. Martins, A. Y. Matsukuma, C. F. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteriro-Vitorello, D. H. Moon, M. A. Nagai, A. L. Nascimento, L. E. Netto, A. J. Nhani, F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. Pereira, H. A. J. Pereira, J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, A. J. de M Rosa, V. E. J. de Rosa, R. G. de Sa, R. V. Santelli, H. E. Sawasaki, A. C. da Silva, A. M. da Silva, F. R. da Silva, W. A. da Silva, Jr., J. F. da Silveira, M. L. Silvestri, W. J. Siqueira, A. A. de Souza, A. P. de Souza, M. F. Terenzi, D. Truffi, S. M. Tsai, M. H. Tsuhako, H. Vallada, M. A. Van Sluys, S. Verjovski-Almeida, A. L. Vettore, M. A. Zago, M. Zatz, J. Meidanis, and J. C. Setubal. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 44.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terry, J. M., S. E. Pina, and S. J. Mattingly. 1991. Environmental conditions which influence mucoid conversion in Pseudomonas aeruginosa PAO1. Infect. Immun. 59:471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and M. S. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vishwanath, S., and R. Ramphal. 1984. Adherence of Pseudomonas aeruginosa to human tracheobronchial mucin. Infect. Immun. 45:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 52.Ward, M. J., H. Lew, A. Treuner-Lange, and D. R. Zusman. 1998. Regulation of motility behavior in Myxococcus xanthus may require an extracytoplasmic-function sigma factor. J. Bacteriol. 180:5668-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson, A. A., R. A. Alm, and J. S. Mattick. 1996. Construction of improved vectors for protein production in Pseudomonas aeruginosa. Gene 172:163-164. [DOI] [PubMed] [Google Scholar]
- 54.Watson, A. A., J. S. Mattick, and R. A. Alm. 1996. Functional expression of heterologous type 4 fimbriae in Pseudomonas aeruginosa. Gene 175:143-150. [DOI] [PubMed]
- 55.West, M., N. M. Burdash, and F. Freimuth. 1977. Simplified silver-plating stain for flagella. J. Clin. Microbiol. 6:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitchurch, C. B., R. A. Alm, and J. S. Mattick. 1996. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 93:9839-9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitchurch, C. B., T. E. Erova, J. A. Emery, J. L. Sargent, J. M. Harris, A. B. Semmler, M. D. Young, J. S. Mattick, and D. J. Wozniak. 2002. Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J. Bacteriol. 184:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, R., and R. B. Dowling. 1998. Lung infections. 3. Pseudomonas aeruginosa and other related species. Thorax 53:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell. 4:253-263. [DOI] [PubMed] [Google Scholar]
- 60.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]