Abstract
Folates are essential vitamins that play a key role as one-carbon donors in a spectrum of biosynthetic pathways including RNA and DNA synthesis. The proton-coupled folate transporter (PCFT/SLC46A1) mediates obligatory intestinal folate absorption. Loss-of-function mutations in PCFT result in hereditary folate malabsorption, an autosomal recessive disorder characterized by very low folate levels in the blood and cerebrospinal fluid. Hereditary folate malabsorption manifests within the first months after birth with anemia, immune deficiency, and neurological deficits. Here we studied the role of inducible trans-activators of PCFT gene expression. Bioinformatics identified three putative nuclear respiratory factor 1 (NRF-1) binding sites in the minimal promoter. The following evidence establish that PCFT is an NRF-1-responsive gene; electrophoretic mobility shift assay showed NRF-1 binding to native but not mutant NRF-1 sites, whereas antibody-mediated supershift analysis and chromatin immunoprecipitation revealed NRF-1 binding to its consensus sites within the PCFT promoter. Moreover, mutational inactivation of individual or all NRF-1 binding sites resulted in 40–60% decrease in luciferase reporter activity. Consistently, overexpression of NRF-1 or a constitutively active NRF-1 VP-16 construct resulted in increased reporter activity and PCFT mRNA levels. Conversely, introduction of a dominant-negative NRF-1 construct markedly repressed reporter activity and PCFT mRNA levels; likewise, introduction of NRF-1 siRNA duplexes to cells resulted in decreased PCFT transcript levels. Moreover, NRF-1 silencing down-regulated genes encoding for key folate transporters and enzymes in folate metabolism. These novel findings identify NRF-1 as a major inducible transcriptional regulator of PCFT gene expression. The implications of this linkage between folate transport and metabolism with mitochondria biogenesis and respiration are discussed.
Keywords: Folate, Folate Metabolism, Gene Regulation, Mitochondria, Promoters, Folate Transport, Hereditary Folate Malabsorption, NRF-1
Introduction
Folates are essential B9 vitamins that play a critical role as one-carbon donors in a multitude of biosynthetic pathways including RNA synthesis and DNA replication, mitochondrial protein synthesis, amino acid metabolism, and methyl group biogenesis (1, 2). Mammalians lack the enzymatic capacity for folate biosynthesis and must therefore obtain folates from their diet. Folates are negatively charged at physiological pH and thus cannot traverse the plasma membrane by passive diffusion. Accordingly, three transport systems are currently known to accommodate folate uptake: (a) the reduced folate carrier (RFC)2 (RFC/SLC19A1) 1 (3–6): RFC displays high affinity for reduced folates but poor affinity for folic acid, an oxidized folate (Km = 200–400 μm) (5, 7). (b) Folate receptors (FRs): FRα and FRβ are high affinity (Kd = 0.1–10 nm) folic acid-binding proteins (8–14). (c) The proton-coupled folate transporter (PCFT/SLC46A1) (PCFT) was initially described as a low-affinity heme carrier protein-1 (HCP1) (influx Km = 125 μm) (15). However, although the role that PCFT might play in heme transport is less dominant, it is well established that PCFT is a high-affinity folate influx transporter with optimal transport activity at acidic pH (16). PCFT functions as a folate-proton co-transporter in the acidic microclimate of the upper small intestine. The dominant role that PCFT plays in intestinal folate absorption was recently established with the demonstration that patients suffering from hereditary folate malabsorption (HFM; OMIM 229050) harbor loss-of-function mutations in the PCFT gene (16–19). HFM is a rare autosomal recessive disorder caused by impaired intestinal folate absorption (16, 19–22). HFM patients present with very low folate levels in the blood and cerebrospinal fluid. HFM manifests within the first months after birth with anemia, an immune deficiency with hypogammaglobulinemia, thereby resulting in severe infections, recurrent or chronic diarrhea, and consequent failure to thrive. We recently identified the promoter of the human PCFT gene and demonstrated that it harbors a 1085-bp CpG island (nucleotides −600 through +485), which plays a key role in modulation of PCFT gene expression (23); we showed that this CpG island was densely methylated in human leukemia cell lines and the PCFT gene was consequently silenced. Using a sequential deletion analysis of the PCFT promoter we found that a 271-bp fragment upstream to the first ATG drives the same promoter activity obtained with the entire 3.1-kb fragment (24). We further showed that the minimal PCFT promoter localizes to only 157-bp (24); we also found that the basal promoter is rich in functional GC-box sites that play a key role in regulation of PCFT gene expression (24).
Nuclear respiratory factor-1 (NRF-1) is a central transcription factor that promotes the expression of nuclear-encoded respiratory genes (25); NRF-1 was found to regulate the expression of a spectrum of genes required for mitochondrial respiratory function, including most of the nuclear genes that encode subunits of the five enzymatic respiratory complexes, components of mitochondrial DNA (mtDNA) replication and transcription, mitochondrial and cytosolic enzymes of the heme biosynthetic pathway, components of the protein import, and the assembly apparatus (for reviews, see Refs. 26–28). Although NRF-1 displays a major role in regulation and coordination of the well timed expression of both nuclear as well as mitochondrial genes in response to a growing need in mitochondrial biosynthesis (26–28), it is currently clear that it also plays a key role in cell growth.
Here we show for the first time that NRF-1 is a key transactivator of PCFT gene expression. We demonstrate that NRF-1 binds to the PCFT promoter using electrophoretic mobility shift assays (EMSA), antibody-mediated supershift analysis, and chromatin immunoprecipitation. We further show that the promoter activity of PCFT is markedly transactivated by constitutively active NRF-1 constructs and repressed by dominant-negative NRF-1 constructs, thereby leading to decreased PCFT mRNA levels. We finally show that silencing of NRF-1 via siRNA technology results in a marked decrease in PCFT mRNA levels, along with a decrease in key folate transporters and enzymes in the folate metabolic pathway. The physiological implications involving PCFT as an NRF-1-responsive gene are discussed.
EXPERIMENTAL PROCEDURES
Cell Culture
Human cervical cancer HeLa cells were maintained in RPMI 1640 medium (Invitrogen) containing 10% fetal calf serum, 2 mm glutamine, 100 μg/ml of penicillin, and 100 μg/ml of streptomycin (Biological Industries, Israel) in a humidified atmosphere of 5% CO2.
Plasmids
Construction and cloning of the following reporter plasmids harboring the PCFT promoter pGL3-3.1kb, pGL3-271bp, and pGL3-157bp was previously described (24). pCDNA3 was purchased from Invitrogen, whereas pCDNA3-Flag-NRF-1 WT, which contains the full-length human NRF-1 and a dominant-negative pCDNA3-Flag-NRF-1 DN construct, which expresses amino acids 1–342 of NRF-1 but lacks the transactivation domain, were a generous gift from Prof. Kimitoshi Kohno (University of Occupational and Environment Health, Fukuoka, Japan). pCDNA 3.1 hygro NRF-1 VP16 encodes for a constitutively active fusion protein, consisting of the full-length human NRF-1 and the herpes simplex virus VP16 transactivation domain, was kindly provided by Dr. T. Gulick (Sanford-Burnham Medical Research Institute, CA). pCDNA3-HA-hPGC-1α was a gift from Dr. A. Kralli (The Scripps Research Institute, CA).
Site-directed Mutagenesis
To inactivate the potential consensus binding sites of NRF-1 via site-directed mutagenesis we utilized the QuikChange XL kit, according to the instructions of the manufacturer (Stratagene Inc.). All putative NRF-1 binding sites were point mutated on both sides of the palindromic binding site using specially designed oligonucleotides detailed under supplemental Table S1. All mutations were verified by DNA sequencing (Hy-labs services, Rehovot, Israel).
RNA Extraction and RT-PCR
Total RNA was extracted either using the TriReagent protocol (Sigma) or the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Any residual genomic DNA was digested with RQ1 RNase-Free DNase (Promega Corporation, Madison, WI). cDNA synthesis was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Quantification of Gene Expression by Real Time PCR Analysis
mRNA levels of various genes were determined by SYBR Green quantitative real time PCR using an Applied Biosystems 7300 Real-time PCR system (Applied Biosystems, Foster City, CA). Quantitative PCR (20 μl) contained: 10 ng of cDNA, 70 nm of the primer mixture, and 1× Power SYBR Green (Applied Biosystems). Primers for genes examined using real time PCR are depicted under supplemental Table S1. All primers were validated to be suitable for relative quantification analysis. The fold-change in expression of each target mRNA relative to β2-microglobulin mRNA was calculated as 2−Δ(ΔCt), where ΔCt = Cttarget − Ctβ2M and Δ(ΔCt) = ΔCtsiRNA − ΔCtMock. Results represent mean ± S.D. of three independent experiments, where all experiments were performed in duplicates.
Transient Transfections with pGL3-PCFT Expression Vectors and Luciferase Reporter Gene Assay
For the luciferase reporter assays, cells were first grown in 24-well plates (2.5 × 104 cells/well). The following day, monolayer cells were transiently co-transfected with 1 μg of the various pGL3-PCFT constructs (including the empty vector pGL3-Basic) along with 20 ng of the well established pROL control plasmid (Renilla luciferase) using the jetPEI transfection reagent (Polyplus-transfection Inc., New York) according to the manufacturer's instructions. Twenty-four hours after transfection, cells were harvested and luciferase and Renilla activities were determined using the Dual Luciferase Reporter Assay System (Promega Corporation, Madison, WI) as described in the manufacturer's protocol. Results presented were obtained from three independent experiments performed in duplicate cultures.
Stable Transfections
Exponentially growing cells (2 × 107) were transfected by electroporation (1000 μF, 234 V) with 10 μg of the following expression vectors: pCDNA 3, pCDNA-NRF-1 WT, pCDNA-NRF-1 DN, and pCDNA 3.1 hygro-NRF-1 VP16. Twenty-four hours after transfection, cells were selected using 0.6 mg/ml of active G-418 (Calbiochem, San Diego, CA) or 0.2 mg/ml of hygromycin (Sigma).
Electrophoretic Mobility Shift Assay and Antibody-mediated Supershift Analysis
Exponentially growing cells (2 × 107 cells) were harvested and nuclear protein extract was isolated as previously described (29). Following determination of the protein concentration using the colorimetric assay of Bradford (30), DNA-protein complexes were allowed to form as detailed elsewhere (31) with some minor modifications: rather than using dI-dC, which can compete out NRF-1 binding, we used 1 μg of dA-dT per reaction. The oligonucleotides used for both binding and competition assays are detailed under supplemental Table S1. Competition EMSA experiments were performed with a 250-fold molar excess of non-radiolabeled oligonucleotides. For supershift analysis, an aliquot of nuclear proteins (6 μg) was incubated for 20 min on ice either with the unpurified polyclonal anti-NRF-1 antibody (a generous gift from Prof. Kimitoshi Kohno, University of Occupational and Environment Health, Fukuoka, Japan) or with 2 μg of anti-ABCG2 BXP-21 monoclonal antibody (kindly provided by Dr. G. L. Scheffer, VU University Medical Center, Amsterdam, The Netherlands) before allowing for protein-DNA complexes to form. Complexes consisting of DNA, nuclear protein(s), and a specific antibody were resolved by electrophoresis on 6% non-denaturing polyacrylamide gels in Tris borate-EDTA, pH 8.4, at 4 °C. Gels were then dried, and DNA-protein complexes were visualized by phosphorimaging (FLA-5000, FUJIFILM, Tokyo, Japan).
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as previously described (32) with the following minor modifications: HeLa cells were grown to ∼80% confluence in 60-mm Petri dishes. Nuclear proteins were then cross-linked to DNA with 1% formaldehyde following which cross-linking was arrested by adding glycine at a final concentration of 125 mm. Cells were scraped off using a rubber policeman, sedimented by centrifugation, resuspended in cell lysis buffer consisting of: 25 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 1 mm DTT, and 0.1% Nonidet P-40, supplemented with the protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 0.5 mm) and incubated on ice for 10 min. Following centrifugation, the crude nuclear pellet was resuspended in nuclei lysis buffer containing: 50 mm HEPES, pH 7.9, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and 0.1% SDS supplemented with 0.5 mm PMSF. Chromatin was sheared at 4 °C, using sonication for 15 s (at 25% power; ULTRASONIC-Vibra Cell sonicator, Sonics & Materials Inc., CT) to obtain DNA fragments of ∼200 to 1000 bp, a process that was repeated a total of 10 times. Cells were then sedimented by centrifugation at 16,000 × g in 4 °C for 10 min. Polyclonal anti-NRF-1 antibody (2 μl) or 1 μg of the following antibodies (anti-mouse IgG (Upstate Biotechnology Inc., Lake Placid, NY; 12–371B), anti-rabbit IgG (Sigma; I5006), or anti-RNA polymerase II antibody (Upstate Biotechnology Inc.; 05-623B)) were incubated at room temperature for 90 min with gentle rotation. Dynabeads-protein G (10 μl; Invitrogen) was then added to the lysate and subsequently incubated at room temperature for 90 min with gentle rotation. The antibody-containing nucleoprotein complexes were then washed extensively with the following buffers: twice with nuclear lysis buffer, once with nuclear lysis buffer supplemented with 350 mm NaCl, once with washing buffer containing: 10 mm Tris, pH 8.0, 250 mm LiCl2, 1 mm EDTA, 0.5% Nonidet P-40, and 0.5% sodium deoxycholate, and finally with TE buffer containing 10 mm Tris, pH 7.5, and 1 mm EDTA. Dissociation of the beads from the nucleoprotein complexes was carried out in elution buffer containing: 50 mm Tris, pH 8.0, 10 mm EDTA, and 1% SDS for 1 h at 65 °C. Nucleoprotein complexes were then de-cross-linked overnight at 65 °C and DNA was purified using the QIAquick PCR purification kit (Qiagen). Immunoprecipitated DNA as well as input DNA served as templates for promoter-specific PCR. Primers used to amplify the proximal promoter region of the PCFT gene and the intergenic region of chromosome 12 are described under supplemental Table S1. PCR was performed in a total volume of 25 μl with 0.4 μm primers and 1× ReddyMix PCR Master Mix (Thermo Fisher Scientific Inc., Waltham, MA). PCR was conducted as follows: initial denaturation at 95 °C for 2 min, 35 cycles each of 30 s of denaturation at 95 °C, 30 s of annealing at 60 °C, and 1 min elongation at 72 °C, followed by a final extension period of 5 min at 72 °C. PCR products were resolved by electrophoresis on 2% agarose gels in Tris borate EDTA, pH 8.4.
RNA Interference
siRNA transfections were performed using HiPerFect (Qiagen) according to the supplier's instructions. HeLa cells were transfected 48 h prior to the isolation of RNA. 75 ng of dsRNA/well (12-well plates) were used in transfections. All double-stranded siRNAs contained 19 bp corresponding to the targeted mRNA as well as two dT extensions on each strand. Human sense siRNA sequences were: GAPDH, 5′-GUCAACGGAUUUGGUCGUAdTdT-3′; NRF1–1, 5′-GAAACGGCCUCAUGUAUUUdTdT-3′; NRF1–2, 5′-UAGUAUAGCUCAUCUUGUAdTdT-3′; NRF1–3, 5′-CACAUUGGCUGAUGCUUCAdTdT-3′. A fluorescent siRNA (5′-AAACAUGCAGAAAAUGCUGdTdT-3′) that fails to recognize any human target was used as a negative control and also to optimize transfection conditions.
Western Blot Analysis
Nuclear extracts were prepared from exponentially growing cells (2 × 107 cells) as previously described (29). Nuclear proteins (≤30 μg) were resolved by electrophoresis on 10% polyacrylamide gels containing SDS, electroblotted onto Protran BA83 cellulose nitrate membranes (Schleicher & Schuell), and reacted with rabbit anti-NRF-1 (1:2,500) (a generous gift from Prof. K. Kohno, University of Occupational and Environment Health, Fukuoka, Japan) and rabbit anti-hnRNP A1 (1:500) (Aviva Systems Biology, CA) antibodies for 1 h at room temperature. Following three 10-min washes in Tris-buffered saline supplemented with 0.5% Tween 20 (TBST) at room temperature, blots were reacted with a goat anti-rabbit secondary antibody (Jackson ImmunoResearch), rewashed, and enhanced chemiluminescence (ECL) detection was performed according to the manufacturer's instructions (Biological Industries). ECL was recorded using the LAS-3000 imaging system (Fujifilm Global, Tokyo, Japan). Protein band intensity was quantified using the EZQuant-Gel software (EZQuant LTD, Tel Aviv, Israel).
Statistical Analysis
Statistical analysis was performed using Student's t test. Two-tailed p values ≤0.05 were considered to be statistically significant.
RESULTS
Identification and Functional Characterization of NRF-1 Binding Sites in the Minimal Promoter of the Human PCFT Gene
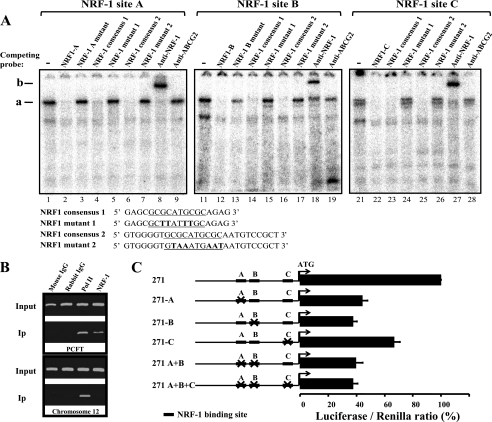
Recently we identified the minimal promoter of the human PCFT gene and showed that constitutive elements including GC-box sites play a key role in PCFT gene expression (24). Specifically, a 1085-bp (nucleotides −600 through +485) CpG island was identified that harbors the minimal promoter; dense methylation of this CpG island resulted in complete silencing of the PCFT gene in tumor cells (23). PCFT displays a restricted tissue and cell line expression pattern (16, 23). We therefore hypothesized here that PCFT gene expression may also be regulated by inducible transacting factors, apart from the regulation by constitutive transactivators like Sp1 that bind to GC-box sites in the minimal PCFT promoter (24). Toward this end, we first undertook a bioinformatic analysis on the 3.1-kb PCFT promoter region for the presence of putative NRF-1 binding sites. This analysis identified three putative NRF-1 binding sites termed NRF-1A (−108/−97), NRF-1B (−93/−82), and NRF-1C (−10/+1), all of which reside within the 157-bp minimal promoter region of PCFT, with NRF-1A displaying the highest bioinformatic score (Fig. 1). Hence, we first explored the degree of evolutionary conservation of these putative NRF-1 binding sites in mammalians using the putative promoter regions of the human, monkey, cow, mouse, and rat PCFT genes (Fig. 1); this analysis revealed that the putative NRF-1A binding site, which displayed the highest bioinformatic score, was extremely conserved in the PCFT promoter region of these mammals. To assess the functional role of these putative NRF-1 binding sites, we first performed EMSA using nuclear extracts isolated from HeLa cells (Fig. 2A). EMSA was carried out using oligonucleotides containing the three authentic NRF-1 binding sites from the PCFT promoter (detailed under “Experimental Procedures”). With all three oligonucleotides, these EMSA experiments exhibited a distinct nuclear protein-oligonucleotide complex, termed complex a, which was absent upon competition with 250-fold molar excess of the corresponding nonradioactive oligonucleotide (Fig. 2A, compare lanes 1, 11, and 21 to 2, 12, and 22, respectively). Furthermore, competition was also performed with two established NRF-1 consensus oligonucleotides (33, 34), both of which efficiently eliminated the specific nuclear protein-oligonucleotide complexes (Fig. 2A, compare lanes 1, 11, and 21 to 4, 6, and 14 as well as to 16, 23, and 25, respectively). In contrast, an attempt to achieve competition with either one of the two NRF-1 mutant consensus oligonucleotides (33, 34) (Fig. 2A, compare lanes 1, 11, and 21 to 5, 7, and 15 as well as to 17, 24, and 26, respectively) or with the authentic oligonucleotides from the PCFT promoter in which the core NRF-1 binding site was mutated, failed to eliminate complex a (Fig. 2A, compare lanes 1 and 11 to 3 and 13, respectively). To provide direct evidence that complex a contained NRF-1, antibody-mediated supershift analysis was performed; an NRF-1-specific antibody recognized complex a, hence resulting in a major supershift to a higher molecular weight termed complex b (Fig. 2A, compare lanes 1, 11, and 21 to 8, 18, and 27, respectively). In contrast, a control antibody to the multidrug resistance efflux transporter ABCG2 failed to yield any supershift of complex a (Fig. 2A, compare lanes 1, 11, and 21 to 9, 19, and 28, respectively). These data demonstrate that NRF-1 binds to the minimal PCFT promoter region at three different NRF-1 consensus binding sites.
FIGURE 1.
Identification of three putative NRF-1 binding sites in the human PCFT promoter. Alignment of the 5′ region of the human PCFT promoter containing 296 bp upstream to the ATG (23, 24) with that of the following mammals: monkey, cow, mouse, and rat. Putative NRF-1 binding sites termed NRF-1 A, B, and C are denoted by open squares (the dark box in NRF-1A represents the highest bioinformatic score). The beginning of the 5′-UTR is marked by the arrow. Nucleotide conservation is indicated by the asterisks in all mammalian organisms studied.
FIGURE 2.
NRF-1 binds and activates the human PCFT promoter. A, EMSA was performed using nuclear extract from HeLa cells and oligonucleotides containing the authentic sequence of NRF-1 sites from the minimal PCFT promoter region. DNA-protein complex a and the high molecular weight complex formed using antibody-dependent supershift analysis (complex b) were resolved by electrophoresis on non-denaturing polyacrylamide gels and examined by a phosphorimager as detailed under “Experimental Procedures.” Molar excess competitions (250-fold) were performed with nonradioactive oligonucleotides. The sequence of the consensus and mutant NRF-1 oligonucleotides is indicated in the lower left side of the panel. The core NRF-1 binding site is underlined, and inactivating mutations in the core consensus are depicted in bold. B, chromatin immunoprecipitation was performed using mouse IgG, rabbit IgG as well as polymerase II (PolII) and NRF-1-specific antibodies. Immunoprecipitation (Ip) in the upper panel corresponds to PCR targeting the PCFT promoter region −130 to +90 (220 bp). Ip in the lower panel corresponds to PCR targeting a 174-bp region of an intergenic genomic DNA sequence from chromosome 12 (negative control). C, luciferase reporter gene assay in HeLa cells following transient co-transfection with pRL-O Renilla plasmid and the various pGL3-271 bp PCFT promoter constructs harboring point mutations (indicated by X) of the designated NRF-1 sites (black rectangles). Results are presented as a luciferase/Renilla ratio (%), normalized to the pGL3-271-bp vector assigned 100% activity. Results are the mean ± S.D of three independent experiments performed in duplicates.
To provide further corroboration of the physical association of NRF-1 to its binding sites in the PCFT promoter region, we performed a ChIP assay in HeLa cells (Fig. 2B). The anti-NRF-1 antibody specifically co-precipitated the PCFT promoter fragment, although failing to do so with the unrelated intergenic region from chromosome 12 (Fig. 2B). Collectively, results obtained with EMSA and the ChIP assays establish that NRF-1 binds to the minimal PCFT promoter region.
All Three NRF-1 Binding Sites Are Essential for the Activity of the PCFT Promoter
After corroborating the actual binding of NRF-1 to its binding sites in the minimal PCFT promoter, we next determined whether or not this NRF-1 binding can functionally transactivate PCFT gene expression. To this end, we used the luciferase reporter assay in HeLa cells by first introducing inactivating mutations in individual or all three NRF-1 binding sites; this was undertaken using a construct harboring a 271-bp fragment of the PCFT promoter (24) cloned upstream to a luciferase reporter gene (Fig. 2C). When compared with the luciferase activity of the control 271-bp construct, which was assigned a 100% reporter activity, a mutation introduced in NRF-1A site resulted in a ∼56% decrease in promoter activity (Fig. 2C, 271-A). Likewise, an inactivating mutation in NRF-1B also resulted in a similar decrease of ∼63% in luciferase reporter activity (Fig. 2C, 271-B), whereas inactivation of the NRF-1C site revealed a lesser decrease of 33% (Fig. 2C, 271-C). Based on the functionality of each of the three NRF-1 binding sites, we now generated constructs harboring inactivating mutations in two or all three NRF-1 binding sites and examined their luciferase reporter activity. Both the double NRF-1 A+B mutant as well as the triple NRF-1 A+B+C mutant displayed essentially the same residual luciferase activities of ∼40% (Fig. 2C, 271 A+B) and 38% (Fig. 2C, 271 A+B+C), respectively. This site-directed inactivation of single or multiple NRF-1 binding sites illustrates that all three NRF-1 binding sites are essential for PCFT promoter activity.
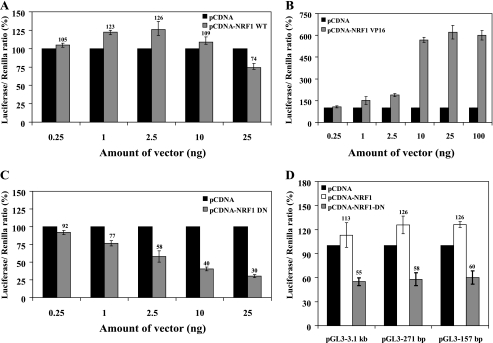
PCFT Promoter Activity Is Modulated by Alterations in Cellular NRF-1 Levels
It was previously shown that overexpression of NRF-1 does not significantly elevate the expression of target genes both in vivo (35) and in vitro with cultured mammalian cells (27). To explore the impact of cellular NRF-1 levels on PCFT promoter activity, we transiently transfected into HeLa cells various NRF-1 constructs including wild type (WT) NRF-1, constitutively active NRF-1 VP16 fusion protein as well as a dominant negative NRF-1 DN. Then, PCFT promoter activity was determined using the 271-bp construct upon a luciferase reporter assay (Fig. 3). Transient overexpression of NRF-1 WT vector harboring the full-length human NRF-1, resulted in a relatively low, yet significant increase in PCFT promoter activity, when compared with empty vector, with the highest increase of ∼30% at 2.5 ng of DNA of NRF-1 WT construct (p = 0.0001) (Fig. 3A); increasing the amount of NRF-1 WT resulted in an inhibitory effect, which was also previously reported in other studies (36). To overcome this obstacle we employed the NRF-1 VP16 vector in which NRF-1 is constitutively active due to its fusion to the potent viral transactivation domain VP16, and examined its ability to transactivate luciferase gene expression. As evident from Fig. 3B, transient introduction of NRF-1 VP16 markedly enhanced PCFT promoter activity in a dose-dependent manner with as much as 6-fold induction of luciferase reporter activity at 25 ng of DNA of the NRF-1 VP-16 construct (p < 0.0001) (Fig. 3B). In contrast, introduction of the NRF-1 DN, which contains amino acids 1–342 but is devoid of the transactivation domain, showed a remarkable dose-dependent decrease in PCFT promoter activity with ∼70% repression observed at 25 ng of DNA (p < 0.0001) (Fig. 3C). Consistent with our findings above, these results suggest that alterations in the functional status of cellular NRF-1 modulate the transcriptional activity of the PCFT promoter.
FIGURE 3.
Luciferase reporter assay reveals that PCFT promoter activity is modulated by NRF-1 levels. A–C, HeLa cells were transiently co-transfected with plasmids: 1) pRL-O Renilla; 2) pGL3-271 bp PCFT promoter construct; and 3) various amounts (ng) of NRF-1 WT (A), NRF-1 VP16 (B), or NRF-1 DN (C). Luciferase activity was then determined as detailed under “Experimental Procedures.” Results are presented as a luciferase/Renilla ratio (%), normalized to the pGL3-271-bp construct along with the corresponding amount of pCDNA empty vector assigned 100% activity. D, luciferase reporter assay in HeLa cells following co-transfection with plasmids: 1) pRL-O Renilla plasmid; 2) 2.5 ng of NRF-1 WT or NRF-1 DN; 3) PCFT promoter constructs pGL3-3.1 kb, -271 bp, or -157 bp. Results are presented as a luciferase/Renilla ratio (%), normalized to the corresponding PCFT promoter construct along with the 2.5 ng of pCDNA empty vector assigned 100% activity. Results shown are the mean ± S.D of three independent experiments performed in duplicates.
NRF-1 Activity within the Minimal PCFT Promoter Is Not Required for Upstream Elements
Our previous study demonstrated that the GC-box sites within the minimal PCFT promoter have a dual role in regulation of PCFT gene expression (24). Mutational inactivation of these GC-box sites in the context of the minimal PCFT promoter resulted in ∼60% decrease in promoter activity (24). In contrast, mutational inactivation of the same GC-box sites within the context of the larger 3.1-kb construct nearly abolished PCFT promoter activity, hence resulting in only 6% residual activity. Hence, one possibility was that members of the Sp1 family that bind to their proximal GC-box binding sites may potentially be essential for the transactivation of factors that may bind to remote upstream enhancer elements. To test the viability of this possibility in the context of NRF-1, we transiently co-transfected various NRF-1 constructs including NRF-1 WT and NRF-1 DN along with different PCFT reporter constructs including the full-length 3.1-kb plasmid as well as the 271- and 157-bp constructs containing only the minimal PCFT promoter region (Fig. 3D). The results obtained suggest that NRF-1 displays the same stimulatory effect on both 3.1-kb and 271- and 157-bp PCFT constructs when using NRF-1 WT. Conversely, transient introduction of the NRF-1 DN vector equally repressed PCFT reporter activity in all three constructs by ∼40%. These data suggest that NRF-1 activity in the minimal PCFT promoter is not essential for the transactivation capacity of upstream enhancer elements in the PCFT promoter.
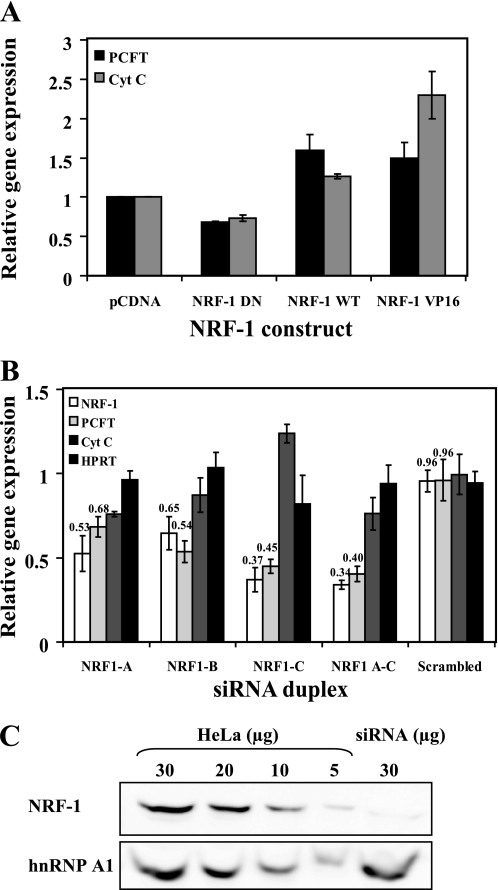
Stable Transfection with Various NRF-1 Constructs Alters PCFT mRNA Levels
To examine the impact of NRF-1 on PCFT mRNA levels, we stably transfected HeLa cells with various NRF-1 constructs and determined their impact on the transcript levels of PCFT and cytochrome c, an established NRF-1-responsive gene, using quantitative real time PCR analysis. Prior to real time analysis, we verified that all stable transfectant populations overexpressed the transfected gene by performing RT-PCR (data not shown). Stable introduction of the NRF-1 DN construct resulted in a consistent 30% decrease in both PCFT and cytochrome c mRNA, when compared with mock transfection (Fig. 4A). Furthermore, overexpression of the NRF-1 WT construct resulted in 60 and 25% increases in PCFT and cytochrome c mRNA levels, respectively (Fig. 4A). Finally, introduction of the constitutively active NRF-1 VP16 construct showed a 2.3-fold increase in cytochrome c transcript levels; whereas PCFT mRNA levels increased by 50% (Fig. 4A). Taken in toto, consistent with the above findings with transient transfections, the present results with stable transfections demonstrate that alterations in the functional levels of NRF-1 induce marked changes in PCFT mRNA levels. These data strongly support the conclusion that PCFT is a target gene of NRF-1.
FIGURE 4.
NRF-1 regulates PCFT mRNA levels. A, HeLa cells were stably transfected with one of the following expression vectors: pCDNA3, NRF-1 DN, NRF-1 WT, or NRF-1 VP16 and a stable population was established by drug selection using G418 or hygromycin. PCFT and cytochrome c (CytC) mRNA levels were determined using real time PCR. Gene expression was normalized to β2-microglobulin mRNA levels. Results are presented as relative expression levels of PCFT or cytochrome c mRNA when compared with cells transfected with the empty pCDNA3 vector, which was assigned a value of 1. B, NRF-1 RNA interference leads to down-regulation of PCFT mRNA; HeLa cells were transfected with each of three different siRNA duplexes that targeted NRF-1 or a combination of all three. For a negative control, HeLa cells were transfected with scrambled siRNA that was not directed to any gene. The mRNA expression level of PCFT, cytochrome c, NRF-1, and hypoxanthine-guanine phosphoribosyltransferase (HPRT) were determined using real time PCR. Gene expression was normalized to β2-microglobulin mRNA. Results are presented relative to the expression of the above genes in a mock transfection, assigned a value of 1. Results are the means of three independent experiments ± S.D. C, Western blot analysis of NRF-1 protein levels following NRF-1 silencing via siRNA. HeLa cells were transfected with NRF-1 C siRNA duplex and nuclear proteins were isolated. Different amounts of nuclear proteins from HeLa cells (5, 10, 20, and 30 μg) as well as 30 μg of nuclear proteins from NRF-1-silenced cells were resolved by electrophoresis on 10% polyacrylamide gels under denaturing conditions. NRF-1 protein levels were determined using an anti-NRF-1 antibody. The protein level of the nuclear splicing factor heterogeneous nuclear ribonucleoprotein (hnRNP) A1 was used for estimation of equal loading.
Knockdown of NRF-1 Decreases Cellular PCFT mRNA Levels
To date, there is little published information regarding the successful down-regulation of NRF-1 via siRNA. Moreover, most successful studies in NRF-1 silencing employed mouse cell lines (37–39). Using human cell lines, Asangani et al. (33) reported on the silencing of NRF-1, however, the latter brought about only 30% reduction in the studied NRF-1 target gene, calpain small subunit 1 (CAPNS1). To provide further confirmation to our findings that NRF-1 is capable of up-regulating PCFT gene expression by regulating PCFT mRNA levels, we employed siRNA constructs targeted to NRF-1 mRNA in HeLa cells. Using real time PCR analysis, mRNA levels of NRF-1, PCFT, cytochrome c, and a control gene hypoxanthine-guanine phosphoribosyltransferase (HPRT) were determined (Fig. 4B). HeLa cells were transfected either with one of three siRNA duplexes targeted to NRF-1 (i.e. NRF1 A, NRF1 B, or NRF1 C) or with a combination of all three siRNA duplexes. Control cells received siRNA that targets GAPDH or a scrambled siRNA that fails to bind any known mRNA. Additional control was obtained through a mock transfection where cells received no dsRNA. Validated primers against NRF-1, PCFT, GAPDH, cytochrome c, and hypoxanthine-guanine phosphoribosyltransferase were used in real time PCR and expression levels were normalized to β2-microglobulin, a well established control housekeeping gene. As evident from Fig. 4B, each of the siRNA duplexes brought about a 35–66% decrease in NRF-1 mRNA levels, a decrease that was accompanied by a parallel decrease of 32–60% in PCFT mRNA levels (p < 0.0001) (Fig. 4B), whereas cytochrome c mRNA was decreased by up to 25% (p < 0.01) (Fig. 4B). In contrast, the mRNA levels of the control hypoxanthine-guanine phosphoribosyltransferase gene remained unchanged (Fig. 4B). To determine the extent of the decrease in NRF-1 protein levels following siRNA-dependent NRF-1 knockdown, Western blot analysis was performed with nuclear proteins isolated from HeLa cells after transient transfection with the NRF-1 C siRNA duplex. NRF-1 protein levels were decreased by 80% upon NRF-1 knockdown, when compared with untreated control HeLa cells (Fig. 4C). It is noteworthy that the extent of the decrease (∼60%) in PCFT gene expression upon NRF-1 silencing is consistent with the extent of decrease in established NRF-1 target genes including respiratory genes (Table 1). Although the extent of decrease in these NRF-1-responsive genes is 30–75% (Table 1), the major physiological impact of NRF-1 regulation on these genes is well established.
TABLE 1.
The extent of decrease in NRF-1 target genes following NRF-1 knockdown
| Gene | Gene function | mRNA decreasea | Transfected cell line | Ref. |
|---|---|---|---|---|
| CAPNS1 | Ca2+-dependent intracellular cysteine protease | 30% | HeLa | 33 |
| Cyctochrome c | Oxidative phosphorylation | 75% | C2C12 | 39 |
| COX5b | Oxidative phosphorylation | 70% | C2C12 | 39 |
| MEF2A | Transcription factor | 67% | C2C12 | 39 |
| COX mRNAs: 4il, 5a, 5b, 6a1, 6b, 6c, 7a2, 7b 7c, 8a | Oxidative phosphorylation | 40–75% | N2a | 38 |
| TFAM, TFB1M, TFB2M | Transcription of mitochondrial genes | 35–70% | N2a | 38 |
| SURF1 | Surfeit1- involved in the COX complex | 35–70% | N2a | 38 |
| VDAC1 | Voltage-dependent anion channel | 35–70% | N2a | 38 |
| TOM20 | Mitochondrial protein transporter | 35–70% | N2a | 38 |
a Decrease in mRNA levels following NRF-1 knockdown.
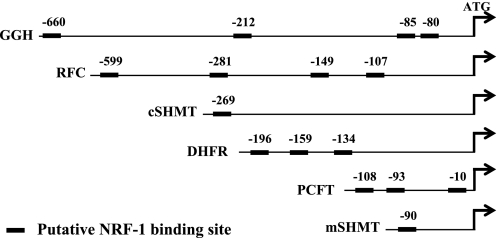
To expand the scope of the impact of NRF-1 on the gene expression status of folate transport and folate-dependent enzymes in the folate metabolic pathway, we further studied a large panel of folate transporters and folate-dependent enzymes. To this end, NRF-1 was silenced using the NRF-1 C siRNA duplex and followed by real time PCR quantification of the expression status of various folate transporters and key enzymes in folate metabolism (Table 2). We first studied all known folate transporters including PCFT, the reduced folate carrier (RFC/SLC19A1), folate receptor α (FRα), as well as the mitochondrial folate transporter (MFT/SLC25A32). NRF-1 silencing brought about a similar and significant decrease of 25% in both RFC and FRα mRNA levels (Table 2). Interestingly, NRF-1 silencing had no effect on MFT gene expression, although MFT is a nuclear encoded gene required for folate transport from the cytoplasm to the mitochondria (Table 2). We next examined the impact of NRF-1 knockdown on the expression status of genes encoding for key folate-dependent enzymes that are central to, and crucial for the folate metabolism including: dihydrofolate reductase (DHFR), thymidylate synthase (TS), glycinamide ribonucleotide transformylase (GARTF), 5-aminoimidazole-4-carboxamide ribonucleotide transformylase (AICARTF), 5,10-methylenetetrahydrofolate reductase (MTHFR), folylpoly-γ-glutamate synthetase (FPGS), and γ-glutmayl hydrolase (GGH). NRF-1 silencing resulted in significant decreases in dihydrofolate reductase (27%), 5,10-methylenetetrahydrofolate reductase (14%), and γ-glutmayl hydrolase (15%), whereas no significant effect was observed with other folate-dependent genes (Table 2). Finally, the impact of NRF-1 silencing on two folate-dependent enzymes involved in glycine biosynthesis, cytosolic and mitochondrial serine transhydroxymethylase (cSHMT and mSHMT, respectively) was also examined. Knockdown of NRF-1 resulted in a 20% decrease in transcript levels of both cSHMT and mSHMT (Table 2). In summary, these results suggest that some of the folate-dependent enzymes in the folate metabolic pathway appear to be regulated to some extent, by cellular levels of NRF-1. It is likely that the lower impact of NRF-1 silencing observed with these folate-dependent genes when compared with PCFT may presumably occur via secondary regulatory mechanisms and mediators that are downstream to NRF-1, thus relying on cellular folate pool sensing. This presumption is further substantiated by the fact that the promoters of some of these genes encoding for folate transporters and folate-dependent enzymes lacked consensus NRF-1 binding sites (Fig. 5); specifically, upon bioinformatic examination of the presence of putative NRF-1 binding sites in the promoters of all studied genes encoding for folate transporters and folate-dependent enzymes that were down-regulated by NRF-1 knockdown, only RFC, dihydrofolate reductase, γ-glutmayl hydrolase, cSHMT, and mSHMT had one or more consensus NRF-1 binding site(s), whereas FRα and 5,10-methylenetetrahydrofolate reductase were devoid of NRF-1 binding sites in their 2-kb promoter region (Fig. 5).
TABLE 2.
Changes in mRNA levels of genes involved in folate transport and metabolism following NRF-1 silencing
Gene expression is presented relative to mock transfection assigned as 1. Results are depicted as the mean ± S.D. of 3–4 independent experiments.
|
NRF1 C siRNA |
Scrambled siRNA |
|||
|---|---|---|---|---|
| Gene expression | p value | Gene expression | p value | |
| NRF-1 | 0.37 ± 0.07 | 0.001 | 0.96 ± 0.06 | 0.06 |
| PCFT | 0.45 ± 0.07 | 0.002 | 0.98 ± 0.11 | 0.70 |
| RFC | 0.75 ± 0.07 | 0.003 | 0.94 ± 0.12 | 0.22 |
| FRα | 0.77 ± 0.11 | 0.012 | 0.91 ± 0.1 | 0.09 |
| MFT | 1.04 ± 0.18 | 0.348 | 1.05 ± 0.12 | 0.26 |
| DHFR | 0.73 ± 0.07 | 0.003 | 1.01 ± 0.18 | 0.46 |
| TS | 0.83 ± 0.18 | 0.078 | 0.89 ± 0.16 | 0.14 |
| GARTF | 1.06 ± 0.09 | 0.169 | 0.97 ± 0.18 | 0.40 |
| AICARTF | 1.12 ± 0.09 | 0.049 | 1.24 ± 0.03 | 0.07 |
| MTHFR | 0.86 ± 0.04 | 0.002 | 0.99 ± 0.09 | 0.45 |
| FPGS | 0.99 ± 0.08 | 0.419 | 1.06 ± 0.06 | 0.10 |
| GGH | 0.85 ± 0.03 | 0.001 | 0.98 ± 0.04 | 0.29 |
| cSHMT | 0.79 ± 0.13 | 0.022 | 1.09 ± 0.12 | 0.41 |
| mSHMT | 0.80 ± 0.07 | 0.004 | 0.96 ± 0.25 | 0.12 |
FIGURE 5.
Bioinformatic analysis of consensus NRF-1 binding sites in the promoters of genes encoding for folate transporters and enzymes in the folate metabolic pathway. The bioinformatic program Genomatics was used to predict the presence of potential NRF-1 binding sites in the 2-kb promoter region as well as the 5′ UTRs of the above genes, the expression of which was decreased following NRF-1 knockdown. The putative NRF-1 binding sites are denoted by solid rectangles. The position of the 5′-most nucleotide of each putative NRF-1 binding site is shown above the solid rectangle.
PGC-1α Does Not Collaborate with NRF-1 in Regulation of PCFT Gene Expression
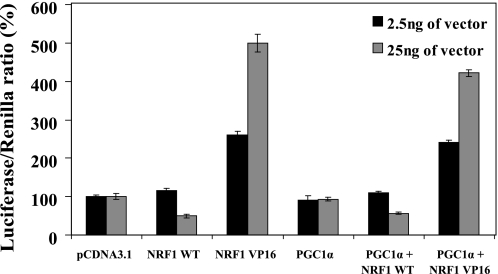
The peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α) was first discovered as a coactivator protein that although lacking a DNA-binding domain, possesses the ability to induce gene expression by directly interacting with NRF-1 (40). There is ample evidence demonstrating that NRF-1 collaborates with PGC-1α, thereby up-regulating expression of various genes, in particular those that encode for proteins involved in mitochondrial respiration (26). After corroborating the role that NRF-1 plays in up-regulation of PCFT gene expression, we aimed at exploring whether or not PGC-1α also participates in transactivation of the PCFT promoter. We hence used the pGL3-271bp reporter construct upon co-transfection either with PGC-1α alone or PGC-1α in combination with NRF-1 WT or NRF-1 VP16. As shown in Fig. 6, not only did PGC-1α fail to induce PCFT promoter activity, it in fact displayed a slight repression of PCFT promoter activity, achieved upon individual transfection of NRF-1 WT or NRF-1 VP16 (Fig. 6). These results suggest that PGC-1α does not collaborate with NRF-1 in the up-regulation of the PCFT gene expression.
FIGURE 6.
PGC-1α does not collaborate with NRF-1 in the regulation of PCFT gene expression. HeLa cells were co-transfected with plasmids: 1) pRL-O Renilla; 2) pGL3-271 bp PCFT promoter construct; 3) 2.5 or 25 ng of one of the following plasmids: NRF-1 WT, NRF-1 VP16, PGC-1α, or a combination of them, as indicated in the figure. Luciferase reporter activity was then determined. Results are presented as a luciferase/Renilla ratio (%), normalized to the pGL3-271 bp construct along with the appropriate amount of pCDNA empty vector assigned 100% activity. Results are the mean ± S.D of three independent experiments performed in duplicates.
DISCUSSION
In the present study we discovered that NRF-1, the dominant transcription factor regulating mitochondrial biogenesis and respiration, binds to, and transactivates the human PCFT promoter; this conclusion is supported by several lines of evidence. First, bioinformatic analysis identified three adjacent putative NRF-1 binding sites residing within the minimal promoter region of PCFT, the characterization of which we have recently described (23, 24). Moreover, evolutionary conservation analysis of the putative NRF-1 binding sites in the PCFT promoter region of various mammalian organisms revealed that the NRF-1A was the most conserved site. Second, the actual binding of NRF-1 to these individual sites was established using several approaches; (a) EMSA performed with the authentic oligonucleotides representing the putative NRF-1 binding sites in the PCFT promoter, confirmed the formation of nuclear factor (i.e. NRF-1)-DNA complexes. (b) Competition EMSA experiments with established consensus NRF-1 oligonucleotides but not with known mutant consensus NRF-1 oligonucleotides, abolished the formation of nuclear factor-DNA complexes. (c) NRF-1 antibody-mediated supershift EMSA experiments demonstrated that NRF-1 was present in the nuclear factor-DNA complexes. (d) ChIP experiments confirmed the specific binding of NRF-1 to its binding sites in the PCFT promoter, but not to an unrelated intergenic region from chromosome 12. Third, the functional contribution of NRF-1 to transcriptional transactivation of the PCFT promoter was achieved using various approaches. Luciferase reporter constructs harboring the minimal as well as the full PCFT promoter regions revealed transactivation, whereas mutational inactivation of individual or all three NRF-1 binding sites resulted in a marked loss (∼60%) of luciferase reporter activity. Consistently, transient introduction of WT NRF-1 or the constitutively active fusion protein NRF-1 VP16 into HeLa cells resulted in a prominent increase in luciferase reporter activity (30% and 6-fold, respectively) using the above PCFT promoter constructs. Moreover, transient introduction of NRF-1 DN, a dominant-negative form of NRF-1, resulted in a significant decrease of 70% in luciferase reporter activity; a value that was similar to the decrease in promoter activity observed when disrupting all three NRF-1 binding sites in the PCFT promoter. Fourth, the ultimate impact of alterations in cellular NRF-1 levels, as well as changes in its functional status, on PCFT mRNA levels were also explored. Stable transfection of an NRF-1 DN construct resulted in a 30% decrease in the mRNA levels of both PCFT and cytochrome c, the latter of which is a well established NRF-1-resposnive respiratory gene. Likewise, stable overexpression of the WT NRF-1 construct brought about 60% and 25% increases in PCFT and cytochrome c mRNA levels, respectively. Moreover, introduction of the constitutively active NRF-1 VP16 construct also resulted in a 2.3-fold increase in cytochrome c mRNA levels, whereas PCFT mRNA levels were increased by 50%. One possible mechanism to account for the fact that overexpression of WT NRF-1 did not result in a major increase in PCFT promoter activity as well as mRNA levels may be associated with the phosphorylation state of NRF-1 (41). In this respect, because phosphorylation enhances the transcriptional activity of NRF-1 (41), it is possible that the rate-limiting step for NRF-1 activity in transfectant cells is its phosphorylation status. Finally, silencing NRF-1 using siRNA technology with several different siRNA duplexes resulted in ∼60% diminished PCFT mRNA levels. Table 2 presents the extent of decrease in gene expression of well established NRF-1 targets following NRF-1 knockdown. Upon NRF-1 silencing, the expression levels of these NRF-1-responsive genes, most of which are respiratory genes, was decreased by 30–75%. The ∼60% reduction in PCFT mRNA levels is highly consistent with the extent of decrease in expression levels of NRF-1-responsive genes. Although the extent of decrease in these established NRF-1-responsive genes after NRF-1 silencing may seem modest, the major impact that NRF-1 has on mitochondrial biogenesis and respiratory physiology is very well documented (26–28). Analysis of the mRNA levels of all known folate transporters revealed that NRF-1 highly regulates PCFT and to a lesser extent the RFC and FRα genes. We hence propose that NRF-1 may play an important role in maintaining sufficient levels of PCFT in the upper small intestine where PCFT functions as the obligatory proton gradient-dependent influx transporter responsible for folate uptake into the blood. NRF-1 may also contribute to the regulation of folate transport from the blood to various organs and tissues via up-regulation of the folate transport systems including RFC and FRα (3, 7, 42). Moreover, as opposed to PCFT, which was shown here to be a bona fide NRF-1 responsive gene via direct binding of NRF-1 to the PCFT promoter, the somewhat lower impact of NRF-1 silencing on the decrease in the expression of various folate-dependent genes than that observed with PCFT may presumably occur via indirect secondary regulatory mechanisms and mediators that are downstream to NRF-1, thus relying on molecular sensing of cellular folate pools. In addition, one should bear in mind that although NRF-1 is a major regulator of mitochondria biogenesis, it is not the sole transcriptional regulator. Thus, alternative transcription factors including NRF-2, ERRα, as well as peroxisome proliferator-activated receptor α also play important roles as regulators of mitochondria biogenesis (28) and may therefore possibly contribute to the regulation of expression of genes in folate transport and the folate metabolic pathway. Consistently, analysis of the promoters of the folate-dependent genes studied here that were down-regulated by NRF-1 silencing revealed that some promoters contained a single or multiple NRF-1-binding sites; others were completely devoid of NRF-1-binding sites. This supports the possibility that the impact of NRF-1 on these genes may not be a direct effect via NRF-1 binding to their promoters but rather through an intermediary NRF-1-regulated gene, the product of which may promote the expression of some folate-dependent genes. It is noteworthy that the bioinformatic analysis of the 2-kb promoter region of the various folate-dependent genes revealed that all consensus NRF-1 binding sites mapped to the transcriptionally active core promoter region, thus supporting the likelihood of the functionality of these putative NRF-1 binding sites.
There is ample evidence establishing that NRF-1 physically associates with PGC-1α, thereby achieving transcriptional transactivation of various NRF-1-responsive genes, in particular those that encode for proteins involved in mitochondria biogenesis and the respiratory chain (26). However, in the current study we found that PGC-1α does not collaborate with NRF-1 in the up-regulation of PCFT gene expression. Yet, one should bear in mind that the PGC-1 family of transcriptional coactivators is comprised of PGC-1α, PGC-1β, and PGC-1-related coactivator. All three coactivators are known to physically interact with NRF-1 and collaborate in multiple nucleo-mitochondrial interactions at the transcriptional level (28). Hence, it is possible that other members of the PGC-1 family including PGC-1β and PGC-1-related coactivator as well as yet unknown nuclear factors may participate in regulation of expression of genes including PCFT, other folate transporters, as well as folate-dependent enzymes; however, this hypothesis must await experimental corroboration.
The current finding that PCFT is an NRF-1-responsive gene, although it is not a bona fide mitochondrial respiration or biogenesis gene, raises the question as to the possible link between folate transport via PCFT, folate metabolism, and mitochondrial biogenesis-respiration. A close examination of this important question uncovers multiple functional metabolic linkages between PCFT-dependent folate transport, folate metabolism, and mitochondrial biogenesis-respiration functions. First, PCFT was initially described as HCP1, a low-affinity heme carrier protein (15). Recent studies suggest that some heme transport may possibly occur via HCP1/PCFT (SLC46A1) (43, 44). Interestingly, heme biosynthesis involves interlinked reactions occurring both in the mitochondria and cytosol (45, 46), primarily of erythrocytes that by themselves are extremely dependent on folate cofactors for erythropoiesis (47). In this respect, the first and dedicated step in heme biosynthesis occurs in mitochondria and involves the conversion of glycine and succinyl-CoA to δ-aminolevulinic acid (45, 46). Remarkably, the major source of cellular glycine is entirely dependent on tetrahydrofolate cofactors, as loss of function mutations in the MFT result in cellular glycine auxotrophy (48, 49). Moreover, recent studies have established that mitochondria are the predominant source of cellular glycine through the mitochondrial enzyme serine transhydroxymethylase (mSHMT), which converts serine to glycine via a tetrahydrofolate-dependent transmethylation reaction (50). Interestingly, targeted disruption of mSHMT, despite the presence of cytosolic SHMT (cSHMT), results in cellular glycine auxotrophy (51, 52). Moreover, mice lacking cSHMT are viable and fertile, hence demonstrating that cSHMT is not an essential source of tetrahydrofolate-activated one-carbon units (51). Therefore, mSHMT-derived one-carbon units are essential for folate-mediated one-carbon metabolism in the cytoplasm. The above findings therefore provide a strong metabolic link between mitochondrial heme biosynthesis and folate-dependent mitochondrial provision of glycine. Consistently, it appears that NRF-1 contributes to the transcriptional regulation of both cSHMT and mSHMT as both decreased upon siRNA-dependent NRF-1 knockdown. Second, it has been recently shown that a 4-week folate deprivation of Wistar rats induces liver mitochondrial oxidative decay including large mitochondrial DNA deletions, cytochrome c dysfunction, membrane depolarization, and superoxide overproduction (53). Hence, absorption of folates from the upper intestine to the blood via PCFT and further from the circulation to tissues and then from the cytosol into mitochondria through MFT is crucial for mitochondrial biogenesis and respiratory function. NRF-1 may therefore be crucial for regulation of intestinal folate transport, particularly under states of folate deprivation (47). Third, ATP, the principal bioenergy currency of the cell, fuels most biosynthetic reactions in the cytoplasm through its hydrolysis into ADP and inorganic phosphate. Because re-synthesis of ATP occurs in the mitochondrial matrix, ATP is exported into the cytoplasm, whereas ADP is imported into the matrix; this exchange is accomplished by a single exchanger, the ADP/ATP carrier (54). The generation of this key bioenergetic molecule, ATP, through mitochondrial oxidative phosphorylation, completely relies on the import of the purine ADP from the cytosol. Importantly, de novo purine biosynthesis including ADP is absolutely dependent on the availability of the reduced folate 5,10-formyltetrahydrofolate, a requirement that must be met by intestinal absorption of folates via PCFT (3, 42, 47). Fourth, one key metabolite that links intestinal folate absorption, folate metabolism in the cytosol and mitochondria is S-adenosylmethionine (AdoMet). AdoMet is the methyl group donor for a multitude of biological methylation reactions. In mitochondria, AdoMet is required for the methylation of DNA, RNA, and proteins, as an intermediate in the biosynthesis of lipoic acid and ubiquinone (55). AdoMet is synthesized from ATP and methionine, the latter of which is synthesized in the cytosol from homocysteine and 5-methylterahydrofolate in a B12 vitamin-dependent reaction catalyzed by methionine synthase. Hence, lack of folates will result in impaired AdoMet biosynthesis and hence mitochondrial deprivation of AdoMet with severe consequences on a multitude of transmethylation reactions of DNA, RNA proteins, lipoic acid, and ubiquinone.
Another important support for the apparent linkage between NRF-1, folate transport, and folate metabolism as well as mitochondria biogenesis-respiration relates to the finding that NRF-1 has been previously shown to transactivate gene expression of folate-dependent enzymes in de novo purine nucleotide biosynthesis (56). Specifically, NRF-1 was shown to up-regulate the bidirectional transcription of two adjacent genes glutamine phosphoribosylpyrophosphate amidotransferase (GPAT) and 5-aminoimidazole ribonucleotide carboxylase (AIRC). The former gene encodes for GPAT, the key regulatory enzyme of the entire purine biosynthetic pathway that catalyzes the first step in this biosynthesis, was found to be closely linked to the AIRC gene on human chromosome 4 (57). The AIRC gene encodes the enzyme AIRC, which is, like GPAT, a 10-formyltetrahydrofolate-dependent enzyme that catalyzes steps six and seven in this purine biosynthetic pathway. Hence, NRF-1 binding to the common promoter of the GPAT and AIRC genes was found to be required for stable binding of Sp1. Deletion of a 54-bp promoter region resulted in decreased expression of GPAT and AIRC in transfected HepG2 cells; this decreased expression was accounted for by point mutations in an NRF-1-binding site and either of two flanking Sp1 sites. Thus, NRF-1 markedly contributes to the coordinated expression of both human GPAT and AIRC.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- RFC
- reduced folate carrier
- PCFT
- proton-coupled folate transporter
- NRF-1
- nuclear respiratory factor-1
- HFM
- hereditary folate malabsorption
- DN
- dominant-negative
- MFT
- mitochondrial folate transporter
- FR
- folate receptor
- SHMT
- serine transhydroxymethylase
- AdoMet
- S-adenosylmethionine
- GPAT
- glutamine phosphoribosylpyrophosphate amidotransferase
- AIRC
- 5-aminoimidazole ribonucleotide carboxylase
- HCP-1
- heme carrier protein-1
- PGC-1α
- peroxisome proliferator-activated receptor γ co-activator 1α
- EMSA
- electrophoretic mobility shift assay
- DHFR
- dihydrofolate reductase
- TS
- thymidylate synthase
- GARTF
- glycinamide ribonucleotide transformylase
- AICARTF
- 5-aminoimidazole-4-carboxamide ribonucleotide transformylase
- MTHFR
- methylenetetrahydrofolate reductase
- FPGS
- folylpoly-γ-glutamate synthetase
- GGH
- γ-glutmayl hydrolase.
REFERENCES
- 1.Appling D. R. (1991) FASEB J. 5, 2645–2651 [DOI] [PubMed] [Google Scholar]
- 2.Stockstad E. L. R. (1990) Folic Acid Metabolism in Health and Disease, pp. 1–21, Wiley-Liss, New York [Google Scholar]
- 3.Assaraf Y. G. (2007) Cancer Metastasis Rev. 26, 153–181 [DOI] [PubMed] [Google Scholar]
- 4.Dixon K. H., Lanpher B. C., Chiu J., Kelley K., Cowan K. H. (1994) J. Biol. Chem. 269, 17–20 [PubMed] [Google Scholar]
- 5.Matherly L. H., Hou Z., Deng Y. (2007) Cancer Metastasis Rev. 26, 111–128 [DOI] [PubMed] [Google Scholar]
- 6.Wong S. C., Proefke S. A., Bhushan A., Matherly L. H. (1995) J. Biol. Chem. 270, 17468–17475 [DOI] [PubMed] [Google Scholar]
- 7.Zhao R., Matherly L. H., Goldman I. D. (2009) Expert Rev. Mol. Med. 11, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elnakat H., Ratnam M. (2004) Adv. Drug Deliv. Rev. 56, 1067–1084 [DOI] [PubMed] [Google Scholar]
- 9.Jansen G. (1999) in Receptor- and Carrier-mediated Transport System for Folates and Antifolates (Jackman A. L. ed.) pp. 293–321, Humana, Totowa, NJ [Google Scholar]
- 10.Kamen B. A., Smith A. K. (2004) Adv. Drug Deliv. Rev. 56, 1085–1097 [DOI] [PubMed] [Google Scholar]
- 11.Kamen B. A., Wang M. T., Streckfuss A. J., Peryea X., Anderson R. G. (1988) J. Biol. Chem. 263, 13602–13609 [PubMed] [Google Scholar]
- 12.Lu Y., Low P. S. (2002) Adv. Drug Deliv. Rev. 54, 675–693 [DOI] [PubMed] [Google Scholar]
- 13.Rothberg K. G., Ying Y. S., Kolhouse J. F., Kamen B. A., Anderson R. G. (1990) J. Cell Biol. 110, 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar M. D., Ratnam M. (2007) Cancer Metastasis Rev. 26, 141–152 [DOI] [PubMed] [Google Scholar]
- 15.Shayeghi M., Latunde-Dada G. O., Oakhill J. S., Laftah A. H., Takeuchi K., Halliday N., Khan Y., Warley A., McCann F. E., Hider R. C., Frazer D. M., Anderson G. J., Vulpe C. D., Simpson R. J., McKie A. T. (2005) Cell 122, 789–801 [DOI] [PubMed] [Google Scholar]
- 16.Qiu A., Jansen M., Sakaris A., Min S. H., Chattopadhyay S., Tsai E., Sandoval C., Zhao R., Akabas M. H., Goldman I. D. (2006) Cell 127, 917–928 [DOI] [PubMed] [Google Scholar]
- 17.Lasry I., Berman B., Straussberg R., Sofer Y., Bessler H., Sharkia M., Glaser F., Jansen G., Drori S., Assaraf Y. G. (2008) Blood 112, 2055–2061 [DOI] [PubMed] [Google Scholar]
- 18.Min S. H., Oh S. Y., Karp G. I., Poncz M., Zhao R., Goldman I. D. (2008) J. Pediatr. 153, 435–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R., Min S. H., Qiu A., Sakaris A., Goldberg G. L., Sandoval C., Malatack J. J., Rosenblatt D. S., Goldman I. D. (2007) Blood 110, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geller J., Kronn D., Jayabose S., Sandoval C. (2002) Medicine 81, 51–68 [DOI] [PubMed] [Google Scholar]
- 21.Sofer Y., Harel L., Sharkia M., Amir J., Schoenfeld T., Straussberg R. (2007) J. Child Neurol. 22, 783–786 [DOI] [PubMed] [Google Scholar]
- 22.Zhao R., Goldman I. D. (2007) Cancer Metastasis Rev. 26, 129–139 [DOI] [PubMed] [Google Scholar]
- 23.Gonen N., Bram E. E., Assaraf Y. G. (2008) Biochem. Biophys. Res. Commun. 376, 787–792 [DOI] [PubMed] [Google Scholar]
- 24.Stark M., Gonen N., Assaraf Y. G. (2009) Biochem. Biophys. Res. Commun. 388, 79–85 [DOI] [PubMed] [Google Scholar]
- 25.Scarpulla R. C., Agne K. M., Wu R. (1981) J. Biol. Chem. 256, 6480–6486 [PubMed] [Google Scholar]
- 26.Scarpulla R. C. (2002) Biochim. Biophys. Acta 1576, 1–14 [DOI] [PubMed] [Google Scholar]
- 27.Scarpulla R. C. (2006) J. Cell Biochem. 97, 673–683 [DOI] [PubMed] [Google Scholar]
- 28.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 29.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31.Ohlsson H., Edlund T. (1986) Cell 45, 35–44 [DOI] [PubMed] [Google Scholar]
- 32.Fan X., Lamarre-Vincent N., Wang Q., Struhl K. (2008) Nucleic Acids Res. 36, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asangani I. A., Rasheed S. A., Leupold J. H., Post S., Allgayer H. (2008) Gene 410, 197–206 [DOI] [PubMed] [Google Scholar]
- 34.Evans M. J., Scarpulla R. C. (1990) Genes Dev. 4, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 35.Baar K., Song Z., Semenkovich C. F., Jones T. E., Han D. H., Nolte L. A., Ojuka E. O., Chen M., Holloszy J. O. (2003) FASEB J. 17, 1666–1673 [DOI] [PubMed] [Google Scholar]
- 36.Kherrouche Z., De Launoit Y., Monte D. (2004) Biochem. J. 383, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhar S. S., Liang H. L., Wong-Riley M. T. (2009) Biochim. Biophys. Acta 1793, 1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhar S. S., Ongwijitwat S., Wong-Riley M. T. (2008) J. Biol. Chem. 283, 3120–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramachandran B., Yu G., Gulick T. (2008) J. Biol. Chem. 283, 11935–11946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 41.Herzig R. P., Scacco S., Scarpulla R. C. (2000) J. Biol. Chem. 275, 13134–13141 [DOI] [PubMed] [Google Scholar]
- 42.Assaraf Y. G. (2006) Drug Resist. Updat. 9, 227–246 [DOI] [PubMed] [Google Scholar]
- 43.Dang T. N., Bishop G. M., Dringen R., Robinson S. R. (2010) Glia 58, 55–65 [DOI] [PubMed] [Google Scholar]
- 44.Laftah A. H., Latunde-Dada G. O., Fakih S., Hider R. C., Simpson R. J., McKie A. T. (2009) Br. J. Nutr. 101, 1150–1156 [DOI] [PubMed] [Google Scholar]
- 45.Bonkovsky H. (1990) Hepatology: A Textbook of Liver Disease, W. B. Saunders, Philadelphia, PA [Google Scholar]
- 46.Ferreira G. C., Gong J. (1995) J. Bioenerg. Biomembr. 27, 151–159 [DOI] [PubMed] [Google Scholar]
- 47.Ifergan I., Assaraf Y. G. (2008) Vitam. Horm. 79, 99–143 [DOI] [PubMed] [Google Scholar]
- 48.McCarthy E. A., Titus S. A., Taylor S. M., Jackson-Cook C., Moran R. G. (2004) J. Biol. Chem. 279, 33829–33836 [DOI] [PubMed] [Google Scholar]
- 49.Titus S. A., Moran R. G. (2000) J. Biol. Chem. 275, 36811–36817 [DOI] [PubMed] [Google Scholar]
- 50.Wright J. B., Brown S. J., Cole M. D. (2010) Mol. Cell Biol. 30, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacFarlane A. J., Liu X., Perry C. A., Flodby P., Allen R. H., Stabler S. P., Stover P. J. (2008) J. Biol. Chem. 283, 25846–25853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stover P. J., Chen L. H., Suh J. R., Stover D. M., Keyomarsi K., Shane B. (1997) J. Biol. Chem. 272, 1842–1848 [DOI] [PubMed] [Google Scholar]
- 53.Chang C. M., Yu C. C., Lu H. T., Chou Y. F., Huang R. F. (2007) Br. J. Nutr. 97, 855–863 [DOI] [PubMed] [Google Scholar]
- 54.Klingenberg M. (2008) Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 55.Roje S. (2006) Phytochemistry 67, 1686–1698 [DOI] [PubMed] [Google Scholar]
- 56.Chen S., Nagy P. L., Zalkin H. (1997) Nucleic Acids Res. 25, 1809–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brayton K. A., Chen Z., Zhou G., Nagy P. L., Gavalas A., Trent J. M., Deaven L. L., Dixon J. E., Zalkin H. (1994) J. Biol. Chem. 269, 5313–5321 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.