Abstract
Endoplasmic reticulum (ER) membrane cholesterol is maintained at an optimal concentration of ∼5 mol % by the net impact of sterol synthesis, modification, and export. Arv1p was first identified in the yeast Saccharomyces cerevisiae as a key component of this homeostasis due to its probable role in intracellular sterol transport. Mammalian ARV1, which can fully complement the yeast lesion, encodes a ubiquitously expressed, resident ER protein. Repeated dosing of specific antisense oligonucleotides to ARV1 produced a marked reduction of ARV1 transcripts in liver, adipose, and to a lesser extent, intestine. This resulted in marked hypercholesterolemia, elevated serum bile acids, and activation of the hepatic farnesoid X receptor (FXR) regulatory pathway. Knockdown of ARV1 in murine liver and HepG2 cells was associated with accumulation of cholesterol in the ER at the expense of the plasma membrane and suppression of sterol regulatory element-binding proteins and their targets. These studies indicate a critical role of mammalian Arv1p in sterol movement from the ER and in the ensuing regulation of hepatic cholesterol and bile acid metabolism.
Keywords: Bile Acid, Cholesterol Metabolism, Dyslipidemia, Genetics, Intracellular Trafficking, Lipid, Membrane Biogenesis
Introduction
The endoplasmic reticulum (ER)4 is the pivotal organelle with regard to cholesterol homeostasis. It is here that cholesterol is synthesized via the mevalonate pathway, sensed by the sterol regulatory element-binding protein (SREBP) cleavage-activating protein system, and neutralized by esterification (1–3). In addition, in certain cells, ER cholesterol is secreted in lipoprotein particles or hydroxylated to form bile acids. Consequently, sterol levels in the ER of all eukaryotic cells are strikingly low relative to the plasma membrane (PM) (4). Movement of sterol between these organelles is rapid; the majority of endogenously synthesized cholesterol is transported from the ER to the PM within 10–20 min by an energy-dependent process (5). Vesicular and nonvesicular cholesterol transport pathways have been described (6, 7); however, the molecular components of these events are surprisingly obscure. The net impact of these processes is striking; any variation around the threshold concentration of ∼5 mol % results in activation or repression of key regulators of lipid homeostasis, including the master regulators, SREBP-1 and -2 (8).
In the yeast Saccharomyces cerevisiae, ARV1 encodes a key component of sterol transport from the ER to the PM and was identified by complementation of yeast mutations that confer viability dependence upon sterol esterification (9). This concept of synthetic lethality was based on the hypothesis that loss of one homeostatic pathway (e.g. sterol esterification) might be tolerated; however, removal of multiple “detoxifying” events would be lethal. Yeast with mutations in ARV1 have striking phenotypes, including an elevated ratio of subcellular sterols relative to the PM and abnormal phospholipid, sphingolipid, and glycosylphosphatidylinositol metabolism (9–11). The yeast ARV1 gene predicts a 322-residue protein with several transmembrane domains. An ARV1 ortholog is present in every eukaryotic genome that has been sequenced. Human ARV1 encodes a 271-amino acid protein, the expression of which complements all known phenotypes associated with mutations in the yeast gene (9, 11).
To address the role of mammalian ARV1 in lipid homeostasis, we used antisense oligonucleotides (ASOs) to “knock down” the expression of the murine ARV1 gene in a subset of tissues. We demonstrate a striking impact of loss of ARV1 on cholesterol and bile acid homeostasis, consistent with impaired anterograde transport of sterol from the ER to the plasma membrane.
EXPERIMENTAL PROCEDURES
Animal Procedures and Knockdown of ARV1 in Mice
Weight-matched, 8–12-week-old C57BL/6 male mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were housed in a specific pathogen-free animal facility in polycarbonate cages (up to 4 animals per cage) in a room with a daylight cycle from 6 a.m. to 6 p.m., temperature of 22 °C ± 1 °C, and humidity of 30–50%. For all experiments, animals were fed with a rodent chow diet prepared by Research Diets, Inc. (New Brunswick, NJ). Mouse ARV1 ASOs were designed by ISIS Pharmaceuticals, Inc., Carlsbad, CA. ASOs were prepared in 0.9% saline (200 μl) and injected intraperitoneally into mice twice a week for 1 or 2 weeks or as a single dose. The ARV1 ASO sequences used in this study were CCTTCTCTTTTGAGGTAGCT and GCACAGTCACTGCTCACATC.
Body weight and food intake were measured once every week. After fasting the animals for 4 h, blood was drawn from the retro-orbital sinus vein. Plasma was obtained from heparinized blood and frozen at −20 °C for future analysis. Samples for real-time PCR were cut and immersed into RNA later (Ambion, Austin, TX) overnight and stored at −80 °C. The left lobe and caudate lobes of mouse livers were snap frozen in liquid nitrogen and stored at −80 °C prior to the measurement of tissue lipids. The University of Pennsylvania Institutional Animal Care and Usage Committee approved the experiments, and all procedures were performed as recommended by the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Laboratory Animal Welfare Act.
Cell Culture and Cell Transfection
Human HepG2 cells were grown in 35-mm culture dishes seeded at 6 × 105 cells/well in Eagle's minimum essential medium (American Type Culture Collection), supplemented with 10% fetal bovine serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin. HepG2 cells were grown on Lipofectin transfection reagents and OptiMEM reduced serum medium, and all cell culture media were purchased from Invitrogen. Cells were transfected by lipid-mediated Lipofectin transfection. Cells were washed once with OptiMEM, then transfection mixes, including 5 μm control ASO or human ARV1 ASO, were incubated with the cells for 6 h. Mnimum essential medium and fetal bovine serum were added back to the cell culture to make a 10% final serum concentration and incubated overnight. After the treatment, the transfection mix was removed, and the culture medium was replaced with complete growth medium and incubated an additional 16–20 h. To assess SREBP-2 processing, cells were incubated in 10% (v/v) newborn calf lipoprotein-deficient serum, 50 μm compactin, 50 μm sodium mevalonate for 16 h.
Plasma Lipid and Lipoprotein Analysis
Plasma total, free, and high density lipoprotein cholesterol; phospholipids, free fatty acids, and triglycerides (TGs), and alanine transaminase levels were measured enzymatically on a Cobas Fara II (Roche Diagnostic Systems) using reagents from Diagnostic Chemicals Limited. Pooled plasma samples (120 μl) from four to six mice were subjected to fast protein liquid chromatography (FPLC) gel filtration (Pharmacia LKB Biotechnology) on two Superose 6 columns at a flow rate of 0.5 ml/min (one run/sample), and fractions of 500 μl each were collected (12). Individual fractions were assayed for cholesterol concentration using an enzymatic assay kit (Wako Chemicals, Richmond, VA).
Hepatic Cholesterol and TG Analysis
PBS-perfused livers were homogenized in PBS and incubated at 37 °C in the presence of deoxycholate for 5 min. Cholesterol and TGs were measured enzymatically using Infinity TG and Infinity cholesterol reagents (Thermo Electron, Melbourne, Australia) (13).
Measurement of Hepatic TG Secretion Rates in Mice
The hepatic cholesterol and TG secretion rates were measured as previously described (14). In brief, mice were injected intraperitoneally with 400 μl of P-407 (1 mg/g) solution in sterile PBS. Blood was collected via retro-orbital plexus prior to injection (0 h) and at 1 h, 2 h, and 4 h after injection. TG was measured enzymatically as described above. The TG secretion rates were calculated as the slope of the plasma concentration versus time after linear regression and expressed in mg/dl per h.
Plasma, Hepatic and Biliary Bile Acids Measurement
Bile acids in plasma, liver, and bile (diluted 1:1000) were measured using a Total Bile Acids kit (Wako Chemicals). For the liver bile acid measurement, total bile acids were extracted by 75% ethanol at 50 °C for 2 h. After clearance by centrifugation, the supernatant was transferred to a new tube, and 20 μl of supernatant was used for measurement. Bile salt compositions of the bile salt pool were determined by HPLC (15). Briefly, mice were anesthetized, and ethanolic extracts of liver, gallbladder, common bile duct, and small intestine were prepared. Prior to extraction, glycocholate was added as an internal standard. The bile salt hydrophobic index, which provides a measure of the overall hydrophobicity of mixed bile salt solutions, was determined according to Heuman (16).
Subcellular Localization of ARV1
CHO K1 cells were plated on glass coverslips in 6-well plates and transfected with either phARV1-EGFP (a PCR-generated fusion of GFP to the 5′ end of human ARV1 in pEGFP-N1 (Clontech)) or pEGFP-N1, using FuGENE 6 reagent. Twenty-four hours after transfection, cells were incubated with 200 nm ER-Tracker at 37 °C for 0.5 h followed by incubation in growth medium for an additional 30 min. Cells were fixed in 3.3% paraformaldehyde before mounting on slides. Laser confocal images were captured using a Zeiss laser scanning microscope LSM 510. For co-localization of hARV1-EGFP and ER-Tracker, samples were viewed using BP 500–550 IR (excitation wavelength at 480 nm, argon laser) and BP435–485 IR (excitation wavelength at 780 nm, multiphoton, Mira 900-F fs-pulsed titanium-sapphire laser) filter sets, respectively.
Subcellular Fractionation and Analysis
The method for fractionation of cells and purification of nuclei is described by Sakai et al. (17). Briefly, the cells were washed with PBS and 1 μl/ml ALLN (N-acetyl-leucinal-leucinal-norleucinal) and swelled in hypotonic buffer A (10 mm HEPES, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 25 μg/ml ALLN, and a mixture of protease inhibitors) for 30 min at 4 °C, passed through a 22.5-gauge needle 30 times, and centrifuged at 1000 × g at 4 °C. The 1000 × g pellet was resuspended in 0.1 ml of buffer B (10 mm HEPES, pH 7.5, 0.42 m NaCl, 2.5% (v/v) glycerol, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 25 μg/ml ALLN, and protease inhibitors) and clarified by centrifugation (“nuclear extract”). The supernatant from the original 1000 × g spin was centrifuged at 10,000 × g for 30 min at 4 °C, and the pellet was dissolved in 0.1 ml of 10 mm Tris-HCl, pH 6.8, 100 mm NaCl, 1% (v/v) SDS, 1 mm EDTA, and 1 mm EGT (“membrane fraction”). Nuclear and membrane fractions were resolved in 10% NuPAGE gels (Invitrogen) and transferred to nitrocellulose membranes. For SREBP-2 detection, a rabbit anti-SREBP-2 polyclonal antibody (Cayman 10007663) was used. The blots were also probed using a rabbit polyclonal antibody against HMG-CoA reductase (a gift of Dr. Ness) (18). A polyclonal anti-β-actin antibody (clone I-19) (Santa Cruz Biotechnology) was used to normalize sample loading.
Measurement of Endogenous Cholesterol and Cholesteryl Ester Synthesis
To measure endogenous cholesterol synthesis, HepG2 cells were labeled with [3H]acetate. After the transfection, cells were incubated with 1 ml of Eagle's minimum essential medium containing 5% fetal bovine serum and 25 μCi of [3H] acetate for 2 and 4 h. At the end of incubation, the cells were washed twice with ice-cold PBS. Lipids were extracted from the cells by adding 2 ml of hexane:isopropyl alcohol (3:2) cholesterol and cholesterol ester separated on TLC plates and quantified (19).
Filipin Staining
Cells were grown on chamber slides and transfected with ASOs as described above. Cells were fixed with 3% paraformaldehyde in PBS for 20 min, washed three times with PBS, and stained with filipin as described (20). Coverslips were mounted with Prolong Antifade reagent (Molecular Probes), and filipin fluorescence was viewed and photographed using a Zeiss Axiovert inverted confocal microscope.
RNA Isolation and Gene Expression Analysis
Total RNA was isolated from indicated tissues using EZ1 RNA Mini or RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. 0.1–1.0 μg of total RNA was reverse transcribed using iScript (Bio-Rad) or SuperScript First-Strand (Invitrogen), according to the manufacturer's directions. Primers were designed using Primer Express 2.0 software (Applied Biosystem; sequences available upon request). For quantitative real-time PCR analysis, the reactions were performed using MicroAmp Optical 96-well plates with optical adhesive covers (Applied Biosystems). The primers were added in appropriate concentrations to a 2 × SYBR Green master mix containing SYBR Green dye, AmpliTaq Gold DNA polymerase, and PCR buffer. In certain cases, real-time PCRs were performed with the MyIQ Single-color Real-time PCR Detection system (Bio-Rad), SYBR Green 2 × Supermix (Bio-Rad), and primers. Expression levels were calculated using ΔΔCt analysis relative to GAPDH, as previously described (21). To determine the mRNA expression of unknown samples, a relative quantification of the mRNA expression was performed using data from pooled hepatic cDNA samples, which were serially diluted. Relative hepatic mRNA expression in the ARV1 ASO-injected mice are reported as a percentage change from mRNA levels of the wild type mice. Human ARV1-specific primers and control glucose-3-phosphate dehydrogenase primers were used in 25-cycle standard PCRs with human multiple-tissue cDNA (panel I and II from Clontech) as templates. 15 μl of each reaction were resolved on 2% agarose gels.
In Vitro Cholesterol Esterification
Cholesterol esterification under conditions where cholesterol is rate-limiting was measured as described (22). Briefly, livers were perfused in situ and homogenized in 0.1 m Tris, pH 7.4, containing protease inhibitors. Microsomes were isolated immediately by differential centrifugation, resuspended in Tris without protease inhibitors, and frozen until use. The assay containing 100 μg of microsomal protein, 1 mg of fatty acid-free BSA, 1 mm glutathione, and 40 μm [14C]oleoyl-CoA (10,000 dpm/nmol) was initiated by the addition of oleoyl CoA and stopped after 20 min with chloroform/methanol (2/1). Lipids were extracted and cholesterol and cholesteryl ester separated by TLC.
Uptake and Esterification of Exogenous Cholesterol
HepG2 cells were transfected with ARV1 or control ASO as described above and incubated in Eagle's minimum essential medium containing 5% lipoprotein-deficient serum and [3H]cholesterol (0.5 μCi). Uptake by the cells and incorporation into esterified cholesterol were determined at 2, 4, and 6 h as described (23).
RESULTS
Expression and Manipulation of ARV1
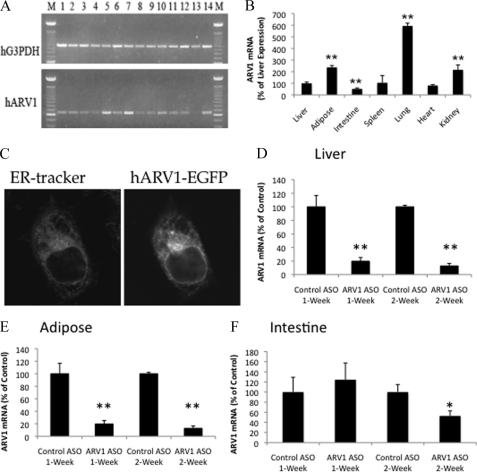
The Arv1 protein is structurally and functionally conserved throughout evolution with representatives in every eukaryotic genome currently available (9). In accordance with a very basic physiological function, the gene encoding this protein is expressed in most human tissues with a peak of expression in liver and adipose (Fig. 1A). Similarly in mice, ARV1 transcripts were detected in all tissues examined, with maximal expression in the lung (Fig. 1B). As might be anticipated for a protein with a key role in ER lipid homeostasis, an Arv1-GFP fusion localized to the ER of transfected Chinese hamster ovary (Fig. 1C) and yeast cells (11). To assess the role of ARV1 in mammalian physiology, C57BL/6 wild type mice were injected with a nontargeting control ASO and two different 2′-O-(2-methoxy)-ethyl-modified ARV1 ASOs corresponding to different regions of the ARV1 gene. Quantitative real-time PCR analysis of total liver RNA from animals treated for 2 weeks indicated that the targeting ASOs reduced hepatic ARV1 mRNA levels to 20 and 50% of control, respectively. Importantly, both ASOs produced qualitatively similar phenotypes with a severity that correlated with repression of the ARV1 gene that was absent from the nontargeting control. Consequently, the more potent ARV1-ASO was used for the remainder of the studies presented here.
FIGURE 1.
ARV1 expression, localization and knockdown. A, ARV1 is expressed prolifically in human tissues. PCRs were used to determine ARV1 expression levels in RNA derived from heart, brain, placenta, lung, liver, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, and colon. Lanes 1–14, respectively, were derived from multiple tissue cDNA panels I and II (Clontech). Reaction products were resolved by agarose gel electrophoresis and ethidium bromide staining. B, ARV1 is expressed in murine tissues. RNA derived from the indicated murine tissues was analyzed by quantitative RT-PCR. C, Arv1p is a resident protein of the endoplasmic reticulum. CHO K1 cells transfected with either phARV1-EGFP (human ARV1 fused at its N terminus with GFP in EGFP-N1 (Clontech)) or pEGFP-N1 were incubated with 200 nm ER-Tracker and visualized by confocal microscopy, as described. D–F, knockdown of ARV1 mRNA levels in mice injected with control or ARV1-specific ASOs is shown. *, p < 0.05; **, p < 0.01.
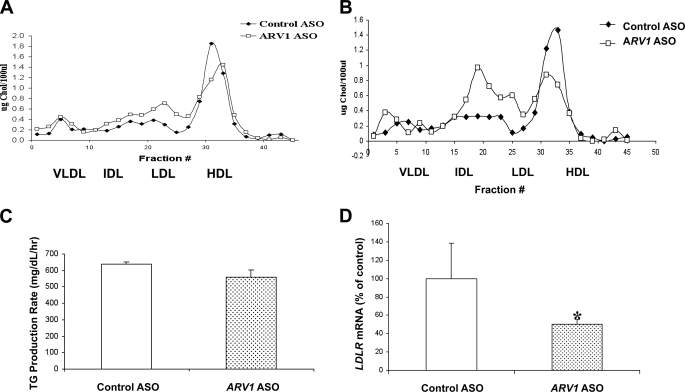
Animals were injected intraperitoneally biweekly with the ARV1 or control ASO at 50 mg/kg body weight over 2 weeks. Animals were killed for analysis at 7 or 14 days. Hepatic ARV1 mRNA levels were decreased to ∼20% of control ASO mice at both time points (Table 1 and Fig. 1D). Similar decreases were observed in terms of repression of ARV1 in adipose and to a much lesser extent, intestine of the same animals (Fig. 1, E and F). By contrast, we did not detect knockdown of ARV1 in the lung, spleen, or kidney of treated animals (data not shown). Plasma alanine transaminase levels were similar in control ASO- and ARV1 ASO-treated mice at 1 week but elevated at 2 weeks in the ARV1 ASO-treated mice (Table 1). Liver TGs were decreased, and liver total cholesterol was unchanged in ARV1 knockdown mice after 2 weeks (Table 1). Plasma cholesterol and phospholipid (Table 1) concentrations in ARV1 ASO-treated mice were significantly elevated at 1 week and further increased after 2 weeks of administration with the ARV1 ASO. Analysis of plasma lipoprotein distribution by FPLC at 1 and weeks (Fig. 2, A and B) showed that the increase was primarily in low density lipoproteins. A decrease in HDL-C was also observed at 1 week that was further decreased at 2 weeks. This decrease is likely due to the increase in plasma bile acids (Fig. 3A). An increase in plasma bile acids has been shown to inhibit LCAT activity and decrease HDL-C (24).
TABLE 1.
Physiological characteristics of C57BL/6 wild type mice treated with ARV1 or control ASO biweekly for either 1 week or 2 weeks
Values are expressed as means ± S.E. ALT, alanine aminotransferase. Representative data are from three separate identical experiments performed with four to six male mice/group per treatment in all experimental groups.
| Characteristic | 1-week treatment |
2-week treatment |
||
|---|---|---|---|---|
| Control ASO | ARV1 ASO | Control ASO | ARV1 ASO | |
| Weight (g) | 28.0 ± 0.5 | 27.9 ± 1.3 | 26.5 ± 1.3 | 23.9 ± 2.5 |
| Liver/body weight | 4.1 ± 0.4% | 5.5 ± 0.18%a | 4.3 ± 0.47% | 6.6 ± 0.37%a |
| Plasma ALT (units/liter) | 25.0 ± 12.2 | 28.0 ± 4.3 | 20.8 ± 1.5 | 583.5 ± 65.8b |
| Plasma cholesterol (mg/dl) | 75.0 ± 1.4 | 121.5 ± 22.4a | 76.8 ± 4.3 | 167.5 ± 40.9b |
| Plasma phospholipids (mg/dl) | 170.5 ± 2.4 | 246.5 ± 37.0a | 204.8 ± 11.4 | 346.0 ± 67.6a |
| Plasma triglycerides (mg/dl) | 47.0 ± 5.0 | 63.0 ± 18.9 | 41.8 ± 7.4 | 23.8 ± 7.5a |
| Plasma free fatty acids (meq/liter) | 1.1 ± 0.1 | 1.1 ± 0.2 | 1.4 ± 0.1 | 1.2 ± 0.1 |
| Hepatic triglycerides (mg/g liver) | 3.7 ± 1.1 | 4.8 ± 0.5 | 5.1 ± 0.8 | 2.7 ± 0.9a |
| Hepatic cholesterol (mg/g liver) | 3.5 ± 0.7 | 3.1 ± 0.4 | 2.5 ± 0.2 | 2.4 ± 0.3 |
| Hepatic ARV1 mRNA | 100 ± 7.9% | 26.9 ± 1.0%a | 100 ± 7.9% | 22.9 ± 1.7%a |
a Significant difference (p < 0.05) between ARV1 ASO-treated mice and control ASO-treated mice.
b Significant difference (p < 0.001) between ARV1 ASO-treated mice or control ASO-treated mice.
FIGURE 2.
Knockdown of hepatic ARV1 affects plasma lipid homeostasis. A and B, FPLC analysis of the plasma lipoproteins of pooled plasma samples derived from four to six mice injected with ASOs biweekly for 1 week (A) or 2 weeks (B). C, measurement of TG secretion rates in mice treated with control ASO or ARV1 ASO. Wild type mice treated with control ASO or ARV1 ASO for a week and mice were injected intraperitoneally with 400 μl of P-407 (1 mg/g) solution at zero time point. Rate of TG production was measured over varying time periods in P-407-treated mice. Two to three separate identical experiments were performed with four to six mice/group per treatment in all experimental groups. Values are expressed as means ± S.E. (error bars) of the mice in the representative experiment. Asterisks denote statistical significant differences from control for ASO-treated animals with p < 0.05 (one asterisk) and p < 0.001 (two asterisks) by unpaired t test. D, hepatic LDLR mRNA levels of mice injected with either control ASO or ARV1 ASO over the time course of 2 weeks.
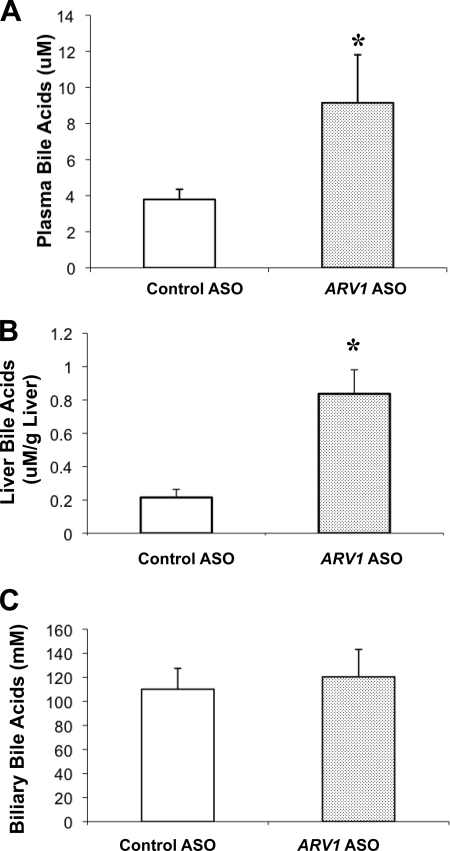
FIGURE 3.
Bile acid quantification in ASO-treated mice. Values are expressed as means ± S.E. (error bars) of the mice in the representative experiment. Values shown are for plasma (A), liver (B), and bile (C). Asterisks denote a statistical significance difference compared with the control with p < 0.05 by unpaired t test. Representative data of at least two separate identical experiments performed with four to six male mice/treatment are shown.
To assess further whether the lipid changes were due to decreased clearance versus elevated synthesis we investigated the ASO-treated animals following intraperitoneal injections with P-407 (1 mg/g). Blood was collected via retro-orbital plexus prior to injection (0 h) and at 1 h, 2 h, and 4 h after injection. TG was measured enzymatically, and secretion rates (mg/dl per h) were calculated as the slope of the plasma concentration versus time after linear regression (Fig. 2C). There was no change in VLDL-TG production with knockdown of ARV1 at 1 week. By contrast, we observed a ∼50% reduction in LDL receptor mRNA abundance in the liver (Fig. 2D), further suggesting that the increased LDL was due to delayed catabolism rather than elevated hepatic VLDL production.
Knockdown of ARV1 Disrupts Bile Acid Metabolism and Results in Farnesoid X Receptor (FXR) Activation
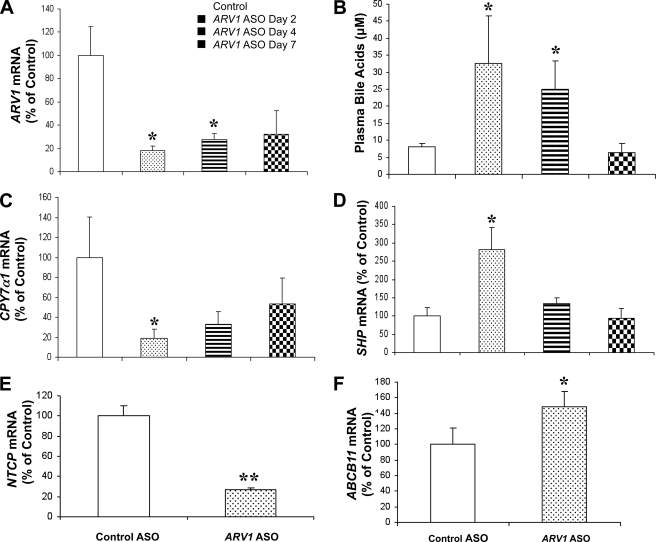
A 2-week administration of the ARV1 ASO was associated with a 2–4-fold increase in plasma bile acids (Fig. 3A) and hepatic bile acids (Fig. 3B), without elevations in biliary bile acids (Fig. 3C). The bile acid pool was not significantly different between the treated (0.36 ± 0.05 μmol/g) and the control group (0.43 ± 0.10 μmol/g). However, in ARV1-ASO treated animals, there was an increase in the hydrophobic index of the total biliary bile acid pool (−0.50 ± 0.04 versus −0.40 ± 0.01 μmol/g, p = 0.001), primarily due to a significant decrease in tauromuricholic acid (66.1 to 53.4 mol%; p < 0.001) and corresponding increase in the more hydrophobic taurocholic acid (26.6 to 40.9 mol%; p < 0.001). In multiple experiments, there was a significant increase in the concentration of cholesterol in gallbladder bile (2.1 ± 0.32 versus 1.38 ± 0.36 μg/μl, p = 0.01) in the absence of changes in biliary PL concentrations in ARV1-ASO animals relative to controls. To establish the primary cause of these observations, we turned to experiments involving a single injection of the ARV1 ASO (Fig. 4). ARV1 gene expression was decreased to 18% of control ASO-treated mice (Fig. 4A) on day 2 and was maintained at 32% of control by day 7. Alanine transaminase levels were not elevated after a single injection, indicating a lack of generalized hepatic injury in the ARV1 ASO-treated animals over this time course. On day 2 after ARV1 ASO injection, the plasma bile acids concentrations were already substantially elevated relative to the control treatment (33 μm compared with 8 μm; Fig. 4B), with a reduction to baseline levels by day 7. The concentrations of hepatic and biliary bile acids did not differ between groups (data not shown).
FIGURE 4.
Knockdown of hepatic ARV1 induces changes in plasma bile acids and activation of FXR regulatory pathways. Wild type mice were injected with a single dose of either control ASO or ARV1 ASO, and subgroups of animals were killed at day 2, day 4, and day 7 after injection. Liver total RNA was isolated, and real-time RT-PCR was performed to assess the hepatic gene expression. A, hepatic expression of ARV1 gene. B, plasma bile acids concentrations at the same time points. of selected genes involved in FXR regulatory pathways. C, hepatic expression of CYP7a1. D, hepatic expression of SHP. In a separate experiment, mice were injected with either control ASO or ARV1 ASO over the time course of 2 weeks, and real-time RT-PCR was performed to assess the hepatic mRNA levels of genes involved in bile acid metabolism including NTCP and ABCB11 (panels E and F, respectively). Asterisks indicate significant differences in ARV1 ASO-treated groups compared with that of the control ASO-treated group with p < 0.05 (one asterisk) and p < 0.001 (two asterisks) by unpaired t test. Error bars, S.E.
To gain further insight into the mechanisms of the disruptions in bile acid and lipid metabolism caused by knockdown of ARV1, hepatic gene expression was analyzed at several time points within 1 week after a single injection of the ARV1 ASO. CYP7α1, encoding the first and rate-limiting enzyme of the neutral bile acid synthetic pathway, was significantly decreased 2 days after a single dose of ARV1 ASO (Fig. 4C). Conversely, the atypical nuclear receptor small heterodimer partner (SHP or NR0B2), a major target of the FXR, was significantly increased in ARV1 ASO-treated mice by 2 days after treatment (Fig. 4D) and may explain the decrease in the mRNA levels of CYP7a1. The mRNA levels of the gene encoding the sinusoidal Na2+-dependent taurocholate co-transport peptide (NTCP) was decreased, and the canalicular bile salt transporter (ABCB11/BSEP) was significantly increased after a 2-week treatment of ARV1 ASO (Fig. 4, E and F). ARV1 was also knocked down in intestine by 50% after 2 weeks of ASO treatment that could affect bile acid homeostasis. We did not, however, detect any transcriptional changes in intestinal components of bile acid homeostasis (e.g. IBABP, OSTα and OSTβ; supplemental Fig. S1). We assume therefore that the major cause of the in vivo bile acid phenotype is hepatic in origin.
Knockdown of ARV1 Results in ER Cholesterol Accumulation and Dysregulation of Lipid Homeostasis in Murine Liver and Human HepG2 Cells
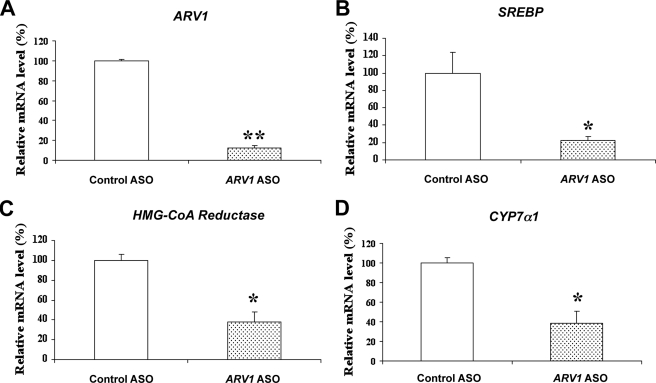
We hypothesized that the knockdown of ARV1 in liver impaired transport of cholesterol from the ER to other cellular compartments such as plasma membrane, resulting in cholesterol accumulation in the ER, and down-regulation of the SREBP pathway. Consequently, the FXR pathways may have become activated due to increases in bile acid synthesis in response to accumulation of unesterified cholesterol in the ER. To test this hypothesis at the cellular level, we treated human hepatoma (HepG2) cells with 5 μm human ARV1 ASO, thereby reducing mRNA levels of ARV1 to 12% of control cells (Fig. 5A). Consistent with ER cholesterol accumulation, mRNA levels of SREBP-1c and HMG-CoA reductase were significantly decreased relative to the control ASO-treated cells (Fig. 5, B and C). Moreover, consistent with the FXR activation observed in the animal studies, CYP7α1 mRNA was also significantly reduced in the HepG2 cells (Fig. 5D).
FIGURE 5.
Regulation of genes involved in lipid and bile acid metabolism in HepG2 cells treated with ARV1 ASO. Total RNA was isolated from HepG2 cells treated with 5 μm control ASO or ARV1 ASO. Real-time RT-PCR was performed to assess the mRNA levels of selected genes involved in lipid and bile acid related pathways. A, ARV1. B, SREBP. C, HMG-CoA reductase. D, CYP7a1. Asterisks indicate significant differences of mRNA levels in ARV1 ASO-treated group compared with that of the control ASO-treated group with p < 0.05 (one asterisk) and p < 0.001(two asterisks) by unpaired t test. Error bars, S.E.
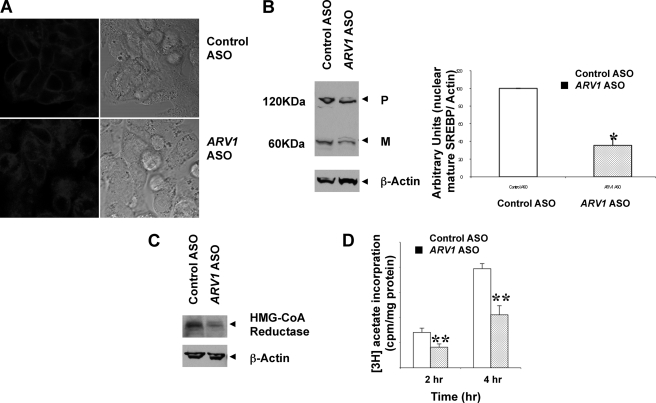
The prior observations may reflect regulatory events downstream of cholesterol accumulation at the ER in ARV1 knockdown cells. To assess this directly, we investigated the sterol status of HepG2 cells after ASO treatment, by staining with filipin which binds unesterified cholesterol, followed by confocal fluorescent microscopy (Fig. 6A). Control ASO-treated cells showed distinct plasma membrane fluorescence, whereas knockdown of ARV1 in HepG2 cells produced an intense intracellular fluorescence accumulation consistent with altered cholesterol trafficking in cells due to decreased ARV1 expression. To corroborate decreased movement of cholesterol out of the ER in the whole animal, we took advantage of the fact that cholesterol availability is rate-limiting in formation of cholesteryl ester. Consequently, the ER substrate pool of cholesterol can be measured by in vitro microsomal acyl-CoA cholesterol acyltransferase assays performed in the absence of exogenous sterol (25, 26). Mice were treated for 2 weeks with or without ARV1 ASO, sacrificed, livers perfused and microsomes isolated and acyl-CoA cholesterol acyltransferase activity determined. Hepatic acyl-CoA cholesterol acyltransferase activity was increased almost 2-fold (123 ± 17 pmol/min/mg versus 66 ± 6 pmol/min/mg; p = 0.01) in mice receiving ARV1 ASO. This is consistent with increased cholesterol in the ER due to decreased transport. The basal esterification of plasma membrane cholesterol can also be determined (27). Equilibration of HepG2 cells with radiolabeled cholesterol in lipoprotein-deficient serum provides an estimate of the amount of cholesterol in the PM. HepG2 cells were treated with or with out ARV1 ASO and then grown in media containing lipoprotein deficient serum for 24 h. Tritiated cholesterol was then added in ethanol and the amount of tracer taken up by the cell and esterified over a 6 h period was determined. There was a 3.08 ± 0.27-fold increase in the amount of [3H]cholesterol/mg protein taken up by the ARV1 ASO treated cells. This is consistent with a depletion of cholesterol at the plasma membrane. There was a similar 2.4 ± 0.06-fold increase in the amount of exogenous cholesterol esterified such that the % of tracer esterified did not change. This would suggest that trafficking of cholesterol from the plasma membrane to the ER is not affected by reduced ARV1 expression.
FIGURE 6.
Impairment of cholesterol trafficking in HepG2 Cells with knockdown of ARV1. HepG2 cells were plated on collagen-coated plate and then treated with 5 μm control ASO or ARV1 ASO. Two days after the transfection, the cells were subjected to the studies. A, free cholesterol accumulation was detected by filipin staining. Cells were stained with 0.05 mg/ml filipin (fluorescence) and examined by confocal microscopy. B, knockdown of ARV1 suppresses the processing of endogenous SREBP-2 in HepG2 cells. Western blot analysis was performed with antibody against SREBP-2 on nuclear fraction of control ASO and ARV1 ASO-treated HepG2 cell. P and M indicate the precursor (125 kDa) and mature (68 kDa) forms of SREBP-2, respectively. Each lane contains 60 μg of nuclear extract and the arbitrary unit of SREBP-2 mature form was normalized to β-actin. Although the precursor form does not change, the mature form was decreased dramatically in ARV1 ASO-treated cell samples. C, knockdown of ARV1 decreases HMG-CoA reductase protein levels in HepG2 cells. Western blot analysis was performed with antibody against human HMG-CoA reductase on membrane fraction of control ASO and ARV1 ASO-treated HepG2 cell. Each lane contains 60 μg of membrane fraction. D, endogenous cholesterol synthesis in HepG2 cells was measured by [3H]acetate incorporation. Cells were incubated and labeled with [3H]acetate for 2 h and 4 h. Cellular lipids were extracted and determined by TLC. Data values represent mean ± S.E. (error bars) of triplicate cultures at each time point. Asterisks denote statistical significance of ARV1-ASO treatment from control cells with p < 0.05 (one asterisk) and p < 0.001(two asterisks) by unpaired t test.
SREBPs reside in the ER and are translocated to the Golgi for proteolytic activation in response to sterol concentration in the ER below a critical threshold of about 5 mol% (8). If knockdown of ARV1 causes cholesterol accumulation in the ER, it should inhibit the proteolytic processing of the SREBPs. HepG2 cells were grown in cholesterol depleted conditions and assessed for SREBP by immunoblots after cell fractionation. The amount of the mature form of SREBP-2 in the nuclear fraction was markedly decreased in the ARV1 ASO-treated cells compared with control ASO-treated cells (Fig. 6B), indicating the inhibition of proteolytic processing of SREBP-2. HMG-CoA reductase is regulated by SREBPs and by proteasomal degradation in response to ER cholesterol. The levels of membrane associated HMG-CoA reductase protein were markedly reduced in HepG2 cells treated with ARV1 ASO relative to control ASO (Fig. 6C), consistent with increased ER cholesterol. Accordingly we observed a significant 40% reduction in the rate of endogenously synthesized cholesterol in ARV1 knockdown cells measured by [3H]acetate incorporation at 2 h and 4 h time points (Fig. 6D). Even though there was a decreased in endogenously synthesized cholesterol, the % of the nascent cholesterol which was esterified increased by 1.5-fold, 14.5 ± 1.8% versus 9.6 ± 0.8% p < 0.01. This is in agreement with a decreased export of nascent cholesterol out of the ER leading to increased esterification by acyl-CoA cholesterol acyltransferase.
DISCUSSION
The molecular mechanisms that maintain mammalian ER cholesterol content, particularly the exit of cholesterol from the ER, remain elusive. Because ARV1 encodes a significant component of subcellular sterol transport in yeast, we hypothesized that it also participates in intracellular transport of cholesterol in mammalian cells. ARV1, a resident ER protein, is expressed ubiquitously in human and murine tissues. Targeting expression of ARV1 by intraperitoneal injection of stable antisense oligonucleotides led to significant and persistent tissue-specific knockdown in liver and adipose and to a lesser extent the intestine. Knockdown of ARV1 in these locations in mice resulted in elevated plasma cholesterol (mainly LDL) and bile acids, accompanied by pronounced activation of FXR, a bile acid-sensing nuclear receptor that controls bile acid homeostasis. We confirmed that knockdown of ARV1 in both the murine liver and in HepG2 cells resulted in intracellular cholesterol accumulation due to impairment of cholesterol removal from the ER, as well as suppression of SREBP-2 processing. We conclude that the ARV1-encoded protein plays a pivotal role in cholesterol metabolism in the hepatocyte as it does in yeast, by facilitating sterol movement out of the ER. Disruption of ARV1 thereby results in an ER accumulation of cholesterol and consequently misregulated cholesterol and bile acid homeostasis.
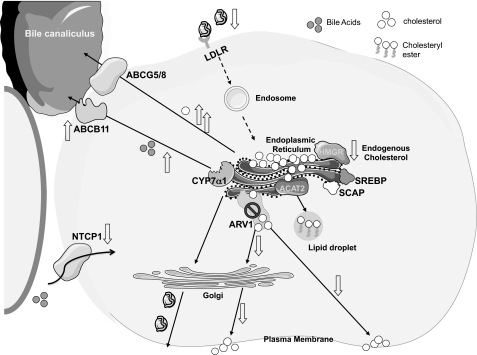
In addition to transport to the PM, ER cholesterol has several possible fates in the liver, including secretion in VLDL, esterification, and hydroxylation to form bile acids. In the current study, knockdown of ARV1 significantly perturbed bile acid metabolism, resulting in a substantial and acute increase in plasma bile acids. ARV1 knockdown was associated with elevated hydrophobic bile acids and cholesterol in bile. Hydrophobic bile acids are better ligands for FXR than their hydrophilic counterparts (28, 29), perhaps accounting for activation of FXR. Consistent with the FXR role in protecting the hepatocyte from bile acid toxicity, we observed rapid and substantial induction of SHP (a key FXR target gene and mediator of many of FXRs transcriptional effects), rapid repression of CYP7α1 (a key enzyme in endogenous bile acid synthesis), up-regulation of ABCB11 (a bile salt export pump involved in transporting bile acids through the hepatocyte canalicular membrane into bile), and repression of NTCP (a bile acid transporter required for hepatic reabsorption of bile acids from plasma). The effects of ARV1 repression on FXR-regulated gene expression and plasma bile acids occurred within 2 days after a single dose of ARV1 ASO and in the absence of any markers of hepatocellular injury. FXR activation, which normally protects the hepatocyte from bile acid toxicity, is thus the most sensitive early marker of acute hepatic ARV1 depletion. However, even though CYP7α1 is down-regulated, the synthesis of bile acids is also dependent upon the availability of cholesterol substrate (30, 31). Because ARV1 depletion increased the supply of substrate, it is feasible that continued bile acid synthesis takes place even though CYP7α1 is decreased. Our data are consistent with a model (Fig. 7) whereby the Arv1 protein plays a pivotal role in lipid metabolism by facilitating cholesterol movement out of the ER. In the setting of reduced hepatic ARV1, elevated ER cholesterol suppresses SREBP processing and down-regulates LDL receptor expression, leading to elevated plasma LDL cholesterol levels. The primary mechanism that results in aberrant bile acid metabolism is less clear. However, accumulation of cholesterol in the ER likely drives bile acid biosynthesis, even while inducing the FXR pathways, by providing more substrate while at the same time affecting the hydrophobic index of newly synthesized bile acids. Moreover, the deprivation of cholesterol at the plasma membrane due to ARV1 knockdown, along with the greater solubilization of PM cholesterol by the more hydrophobic bile acids, significantly alters the properties of this organelle. The increase in hydrophobic bile acids may account for the observed increase of cholesterol in the bile. Of note, the activity of transporters such as ABCB11, ABCC23, and possibly ATP8B1 (32) is reduced when canalicular membrane cholesterol is low. ABCB11 is the major bile salt transporter, thus its inactivation would create a cholestatic situation whereby bile salts are retained in cells. Our studies identify ARV1 as encoding an important new participant in the regulation of cholesterol and bile acid homeostasis in the hepatocytes.
FIGURE 7.
Model of the role of Arv1p in subcellular cholesterol trafficking in mammalian cells. We propose that the ARV1-encoded protein facilitates vesicular and/or nonvesicular cholesterol movement from the ER to the PM. When ARV1 expression is disrupted, cholesterol is cycled inefficiently to the PM and therefore accumulates inside the cells, probably at the ER. Accumulation of intracellular cholesterol causes the following outcomes: down-regulation of SREBP; a concomitant decrease in endogenous cholesterol synthesis and uptake via HMG-CoA reductase and the LDL receptor (LDLR), respectively; an increase in bile acid biosynthesis and elevated cholesterol secretion into bile. ACAT2, acyl-CoA:cholesterol acyltransferase 2; SCAP, SREBP cleavage-activating protein, CYP7α1, cholesterol 7α-hydroxylase; ABCB11 and ABCG5/8, members of the ATP-binding cassette superfamily of membrane transporters. Cholesterol is denoted by the open circles.
Supplementary Material
Acknowledgments
We thank Aisha Wilson and Debbie Cromley for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK48873 and DK56626 (to D. E. C.), Grant DK54320 (to S. L. S.), Grant HL59407 (to D. J. R.), and Grant Training Fellowships in Nutrition T32 DK007647 and Translational Science TL 1RR024158 (to C. F. S.). This work was also supported by the American Diabetes Association (F. T. and S. L. S.), the Ara Parseghian Medical Research Foundation (S. L. S.), and the Bristol Myers Squibb “Freedom to Discover” Unrestricted Cardiovascular Research Grant (to D. J. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ER
- endoplasmic reticulum
- ALLN
- N-acetyl-leucinal-leucinal-norleucinal
- ASO
- antisense oligonucleotide
- FXR
- farnesoid X receptor
- PM
- plasma membrane
- SREBP
- sterol regulatory element-binding protein
- TG
- triacylglycerol.
REFERENCES
- 1.Brown M. S., Goldstein J. L. (1997) Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 3.Sun L. P., Seemann J., Goldstein J. L., Brown M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liscum L., Underwood K. W. (1995) J. Biol. Chem. 270, 15443–15446 [DOI] [PubMed] [Google Scholar]
- 5.Matveev S., Li X., Everson W., Smart E. J. (2001) Adv. Drug Deliv. Rev. 49, 237–250 [DOI] [PubMed] [Google Scholar]
- 6.Prinz W. A. (2007) Prog. Lipid Res. 46, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liscum L., Munn N. J. (1999) Biochim. Biophys. Acta 1438, 19–37 [DOI] [PubMed] [Google Scholar]
- 8.Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S. (2008) Cell Metab. 8, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tinkelenberg A. H., Liu Y., Alcantara F., Khan S., Guo Z., Bard M., Sturley S. L. (2000) J. Biol. Chem. 275, 40667–40670 [DOI] [PubMed] [Google Scholar]
- 10.Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K. (2008) Mol. Biol. Cell 19, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swain E., Stukey J., McDonough V., Germann M., Liu Y., Sturley S. L., Nickels J. T., Jr. (2002) J. Biol. Chem. 277, 36152–36160 [DOI] [PubMed] [Google Scholar]
- 12.Jin W., Millar J. S., Broedl U., Glick J. M., Rader D. J. (2003) J. Clin. Invest. 111, 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao B., Zondlo S., Gibbs S., Cromley D., Hosagrahara V. P., Kirchgessner T. G., Billheimer J., Mukherjee R. (2004) J. Lipid Res. 45, 1410–1417 [DOI] [PubMed] [Google Scholar]
- 14.Millar J. S., Cromley D. A., McCoy M. G., Rader D. J., Billheimer J. T. (2005) J. Lipid Res. 46, 2023–2028 [DOI] [PubMed] [Google Scholar]
- 15.VanPatten S., Ranginani N., Shefer S., Nguyen L. B., Rossetti L., Cohen D. E. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 281, G393–G404 [DOI] [PubMed] [Google Scholar]
- 16.Heuman D. M. (1989) J. Lipid Res. 30, 719–730 [PubMed] [Google Scholar]
- 17.Sakai J., Nohturfft A., Goldstein J. L., Brown M. S. (1998) J. Biol. Chem. 273, 5785–5793 [DOI] [PubMed] [Google Scholar]
- 18.Singer II, Kawka D. W., Kazazis D. M., Alberts A. W., Chen J. S., Huff J. W., Ness G. C. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 5556–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Field F. J., Watt K., Mathur S. N. (2007) J. Lipid Res. 48, 1735–1745 [DOI] [PubMed] [Google Scholar]
- 20.Jacobs N. L., Andemariam B., Underwood K. W., Panchalingam K., Sternberg D., Kielian M., Liscum L. (1997) J. Lipid Res. 38, 1973–1987 [PubMed] [Google Scholar]
- 21.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 22.Tavani D. M., Tanaka T., Strauss J. F., 3rd, Billheimer J. T. (1982) Endocrinology 111, 794–800 [DOI] [PubMed] [Google Scholar]
- 23.Wojtanik K. M., Liscum L. (2003) J. Biol. Chem. 278, 14850–14856 [DOI] [PubMed] [Google Scholar]
- 24.Bravo I., Amigo L., Cohen D. E., Nervi F., Rigotti A., Francone O., Zanlungo S. (2007) Biochim. Biophys. Acta 1770, 979–988 [DOI] [PubMed] [Google Scholar]
- 25.Billheimer J. T., Tavani D., Nes W. R. (1981) Anal. Biochem. 111, 331–335 [DOI] [PubMed] [Google Scholar]
- 26.Lange Y., Steck T. L. (1997) J. Biol. Chem. 272, 13103–13108 [DOI] [PubMed] [Google Scholar]
- 27.Underwood K. W., Jacobs N. L., Howley A., Liscum L. (1998) J. Biol. Chem. 273, 4266–4274 [DOI] [PubMed] [Google Scholar]
- 28.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. (1999) Science 284, 1362–1365 [DOI] [PubMed] [Google Scholar]
- 29.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. (1999) Science 284, 1365–1368 [DOI] [PubMed] [Google Scholar]
- 30.Mitropoulos K. A., Balasubramaniam S., Venkatesan S., Reeves B. E. (1978) Biochim. Biophys. Acta 530, 99–111 [DOI] [PubMed] [Google Scholar]
- 31.Post S. M., Zoeteweij J. P., Bos M. H., de Wit E. C., Havinga R., Kuipers F., Princen H. M. (1999) Hepatology 30, 491–500 [DOI] [PubMed] [Google Scholar]
- 32.Paulusma C. C., de Waart D. R., Kunne C., Mok K. S., Elferink R. P. (2009) J. Biol. Chem. 284, 9947–9954 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.