Abstract
The regulation of synthesis, degradation, and distribution of lipids is crucial for homeostasis of organisms and cells. The sterol regulatory element-binding protein (SREBP) transcription factor family is post-translationally activated in situations of reduced lipid abundance and activates numerous genes involved in cholesterol, fatty acid, and phospholipid synthesis. In this study, we provide evidence that the primary transcript of SREBP2 contains an intronic miRNA (miR-33) that reduces cellular cholesterol export via inhibition of translation of the cholesterol export pump ABCA1. Notably, miR-33 also inhibits translation of several transcripts encoding proteins involved in fatty acid β-oxidation including CPT1A, HADHB, and CROT, thereby reducing fatty acid degradation. The genetic locus encoding SREBP2 and miR-33 therefore contains a protein that increases lipid synthesis and a miRNA that prevents export and degradation of newly synthesized lipids. These results add an additional layer of complexity to our understanding of lipid homeostasis and might open possibilities for future therapeutic intervention.
Keywords: Beta-oxidation, Cholesterol, Cholesterol Metabolism, Lipid, Micro-RNA
Introduction
The expression of enzymes involved in cholesterol, fatty acid, and phospholipid synthesis is coordinated by the family of sterol regulatory element-binding factor (SREBP)3 transcription factors (1, 2). These factors normally reside in an inactive form tethered to the membrane of the endoplasmic reticulum and get activated by proteolytic cleavage when cellular cholesterol and/or fatty acid levels drop (3). Although lower organisms have only one SREBP gene, vertebrates have two, SREBP1 and SREBP2. The locus encoding for SREBP1 gives rise to two distinct mRNAs, SREBP1a and SREBP1c, which are transcribed from two distinct promoters. SREBP1a, SREBP1c, and SREBP2 differ with regard to transcriptional activation capacity, tissue distribution, and mode of regulation (4,5). Overall, SREBP1a contains the strongest transcriptional activity, potently driving expression of the complete set of fatty acid and cholesterol synthesis genes (2). Although a certain preference of SREBP2 for the activation of genes involved in cholesterol synthesis has been suggested it can still activate most of the fatty acid synthesis genes (2).
Micro-RNAs (miRNAs) are short 21–24-nucleotide long, nonprotein-coding RNAs that are increasingly recognized as important regulators of gene expression (6–8). By binding to the 3′ untranslated region of protein-coding mRNA transcripts they can reduce translation from these transcripts and in some cases lead to their degradation (9, 10). Target gene recognition is promiscuous and in some instances pairing of only six to eight nucleotides of the miRNA (the so-called “seed region”) to the 3′ UTR of target transcripts is sufficient for silencing (11). Each miRNA therefore is predicted to target numerous target genes, and in many cases several hundred. Placing individual miRNAs in a functional context, is therefore often not trivial (12).
Herein we present evidence that the genetic locus of SREBP2 not only encodes a sterol sensing transcription factor (2), but also contains a highly conserved miRNA that regulates cholesterol export and fatty acid β-oxidation. miR-33 reduces cellular cholesterol export by directly targeting the transcript of the ATP binding cassette A1 (ABCA1) protein, and reduces β-oxidation by directly targeting transcripts for the β-subunit of the mitochondrial trifunctional protein (hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase β-subunit; HADHB), the liver-specific isoform carnitine palmitoyltransferase 1A (CPT1A) and the carnitine O-octanoyltransferase (CROT). miR-33 therefore cooperates with its embedding gene SREBP2 to maintain cellular lipid levels. We also supply data to suggest that the regulation of β-oxidation could be evolutionarily conserved down to Drosophila melanogaster.
EXPERIMENTAL PROCEDURES
DNA Constructs and Reporter Assays
Genomic regions encompassing the predicted miR-33 binding sites in the 3′ UTR of human ABCA1, HADHB, CROT, ATP8B1, SLC25A25, and CPT1A, as well as the Drosophila CPT1 3′ UTR were amplified by polymerase chain reaction using PfuI polymerase (Fermentas, St. Leon-Rot, Germany) and inserted downstream of the constitutively active luciferase expression cassette of the plasmid pGL3 control (Promega, Madison, WI) (supplemental Fig. S1). Site-directed mutagenesis was performed using a modified QuikChange site-directed mutagenesis system (Stratagene-Agilent, Diegem, Belgium) or fusion PCR. Mutations introduced in reporter constructs are indicated by asterisks. Supplemental Table S1 contains the primers used in this study for cloning.
Synthetic preMIRs and a negative control were obtained from Applied Biosystems (Foster City, CA). Transfections was performed in 24-well plates in 0.5 ml with Lipofectamine 2000 (Invitrogen) using preMIRs at a final concentration of 2 nm (in the final volume of 0.5 ml) as well as 200 ng of firefly luciferase constructs and 50 ng of a constitutively active Renilla luciferase construct driven by the SV40 promoter (Promega, Madison, WI). Relative luciferase activity in all figures refers to firefly luciferase activities normalized to Renilla luciferase. Values are mean ± S.D. of triplicates. Locked nucleic acids and cholesterol-modified 2-O-methyl-oligonucleotides were designed as described previously and obtained from IDT DNA Technologies (Leuven, Belgium) (13). Transfections were performed in 24-well plates using 100 nm antisense oligonucleotide and 1 μl of Lipofectamine 2000 using 100 μl of OptiMEM serum-free medium and 500 μl of total DMEM. miR-33 antagomir denotes a 1:1 mixture of miR-33a and miR-33b antagomir.
Lentiviral Infection
Lentiviral constructs driving expression of miR-33a were generated by introducing a PCR fragment generated from genomic DNA encompassing the human miR-33a into the XhoI-BamHI or a MluI-ClaI of the pGIPZ or pTRIPZ empty vector, respectively (Openbiosystems, Thermofisher, Epsom, UK) (supplemental Fig. S2). miR-33 sponges were generated by ligating six tandem repeats of miR-33 recognition sites in the XhoI-BamHI site of pGIPZ empty vector (14). Packaging was performed using a second generation plasmid system (psPAX2 and pMD2.G; Addgene plasmids 12,259 and 12,260, respectively, Cambridge, MA) by transient transfection with the calcium phosphate coprecipitation method. We used 20 μg of lentiviral vectors, 20 μg of psPAX2, and 10 μg of pMD2.G per 10-cm tissue culture dish. Cells were infected 24 to 48 h after transfection in the presence of 4 mg/ml of Polybrene (Sigma). Infected cells were selected for 4 days with 1.5 (HepG2), 0.5 (THP1), and 2 μg/ml (Y1 cells) of puromycin (Merck, Darmstadt, Germany). HepG2 cells were kind gifts of Miikka Vikkula (Université Catholique de Louvain), THP1 cells were from the laboratory of Paul Tulkens (Université Catholique de Louvain), Y1 cells from Tobias Else (University of Michigan), and 293T cells from Eric Fearon (University of Michigan). Where indicated, cells were treated with 500 ng/ml of doxycyclin (Sigma) or 5 mm T0901317 (Cayman, Ann Arbor, MI).
Quantitative PCR
RNA was isolated using TRIzol (Invitrogen) and reverse transcription was performed using RevertAidTM H Minus reverse transcriptase (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's protocols using 2 μg in a total volume of 20 μl. We used 5 μl of these diluted cDNAs in subsequent quantitative PCR in MyiQTM thermal cycler using SYBR Green as a fluorescent dye (Bio-Rad) and hotstart Taq polymerase (Eurogentec, Seraing, Belgium). PCR programs were 95 °C for 3 min, 40 cycles at 95 °C for 10 s and 60 °C for 1 min with acquisition of fluorescent information, followed by a melting curve. Experiments shown represent averages and standard deviations for three to six biological replicates. Primers for amplification can be obtained upon request. Human tissue RNA was a kind gift from Eric Fearon (University of Michigan) and originally obtained from Ambion (Ambion-Invitrogen, Lennik, Belgium).
Cholesterol Export
Apolipoprotein AI was overexpressed, delipidized, and purified as described previously (15). Cells were plated at high density (2.5 × 105 cells per well) in 24-well plates and loaded for 24 h with 0.5 mCi of [1,2-3H]cholesterol (PerkinElmer Life Sciences) per well containing 500 μl of medium. After overnight equilibration in DMEM containing 0.2% BSA, cells were incubated in the presence or absence of 5 mg of apoAI for 7 h. Radioactivity was measured in the cell culture medium and normalized to the radioactivity remaining in the cells. Experiments were performed using 5 to 8 replicates per experimental condition and values shown represent mean ± S.D.
Thin Layer Chromatography (TLC)
Cellular lipids were extracted essentially by the Folch method (16) and subsequently dried under N2, and the pellet was resuspended in a volume of chloroform/methanol (2:1) proportional to the protein concentration assessed in 5% of the cells. Equal amounts of lipids were separated by TLC with diethyl ether/hexane/glacial acetic acid (35:65:1, v/v/v) as solvent on high performance TLC silica plates (Merck, Darmstadt, Germany). Plates were dried and evenly sprayed with 0.05% of Primulin in acetone/water (4:1, v/v) (17). Visualization was performed using 340 nm excitation and a digital camera, or, for quantification, using the Cy2 channel of a fluorescent imager (GE Healthcare, Diegem, Belgium). Lipid species were identified by comparison of Rf values with lipid standards.
β-Oxidation Assays
Cellular β-oxidation was assessed as described previously (18). Briefly, for each well in a 24-well plate 1.25 ml of 22 mm palmitic acid and 2 mCi of [9,10-3H]palmitic acid (PerkinElmer Life Sciences) were dried under N2, resuspended in 12.5 ml of phosphate-buffered saline (PBS) containing 10 mg/ml of fatty acid-free bovine serum albumin (BSA, Sigma), and then further diluted with 37.5 ml of PBS. Cells were washed once with Hanks' balanced salt solution (Invitrogen) and then preincubated in 200 μl of PBS containing 0.5 mg/ml of BSA in the presence or absence of 100 μm Etomoxir (Sigma) for 1 h. After addition of 50 μl of the radioactive palmitic acid mixture, cells were incubated for 2 h. Anion exchange columns were used to separate radioactivity incorporated into water from palmitic acid. Radioactive counts were normalized to the amount of protein per well as determined by bicinchoninic acid assay (ThermoFisher Belgium, Tournai, Belgium).
Northern Blot Analysis
We separated 10 μg of total RNA on denaturing 15% polyacrylamide/urea mini-gels (acrylamide/bisacrylamide = 19:1) using Tris borate-EDTA (TBE) buffer (19). Transfer was performed in 0.5× TBE buffer using a semi-dry transfer system (Trans-Blot SD, Bio-Rad) at 100 mA/gel (10 × 7.5 cm) for 90 min onto a positively charged nylon membrane (Hybond N+, GE Healthcare). RNA was cross-linked using ultraviolet light (Stratalinker 1800, Agilent-Stratagene, Diegem, Belgium). To label probes, 50 nmol of oligonucleotides were radioactively labeled using 30 units of T4-polynucleotide kinase (Fermentas) in a total volume of 50 μl for 60 min in the presence of 50 μCi of [γ-32P]ATP. Unincorporated nucleotides were separated using G25-Sepharose spin columns (GE Healthcare) according to the manufacturer's instructions. Membranes were prehybridized for 1 h and then hybridized overnight in rapid hybridization buffer at 106 cpm/ml (RapidHyb, GE Healthcare) at 42 °C. Three stringent washes were performed and the signals were revealed either by exposure to autoradiography film (U6) or by use of a storage phosphorscreen (Pharos FX, Bio-Rad).
Western Blot Analysis
Proteins were extracted using radioimmunoprecipitation buffer (RIPA) and genomic DNA was sheared by brief sonification. Equal amounts of clear lysates were resolved on 10% Tris glycine polyacrylamide mini-gels and transferred to PVDF membranes (Immobilon P, Millipore) using a tank-transfer system. Equal transfer was validated by staining with Ponceau Red. Membranes were blocked with 10% skimmed milk in Tris-buffered saline (TBS) and incubated with primary antibodies in TBS containing 0.05% Tween 20, 2% BSA, and 0.05% sodium azide overnight at 4 °C. Antibodies were used at the following dilutions: ABCA1 at 1:1,000 (AB.H10, Santa Cruz Biotechnologies), HADHB at 1:1,000 (ARP48133, Aviva Systems Biology, San Diego, CA), HADHA at 1:1,000 (HPA015536, Sigma), CPT1 1:1000 (Proteintech, Manchester, UK), GAPDH at 1:2,000 (6C5, Pierce-ThermoFisher), and β-actin at 1:10,000 (AC-40, Sigma). Secondary horseradish peroxidase-coupled antibodies (rabbit, GE Healthcare; mouse, Sigma) were used at 1:10,000 in 10% skimmed milk in TBS containing 0.05% Tween 20. Signals were revealed using an enhanced chemiluminescence reagent (SuperSignal West Pico chemiluminescent substrate, Pierce-ThermoFisher), and subsequent exposure to autoradiography film (Super RX film, Fujifilm Medical Systems, Sint-Niklaas, Belgium).
Animals
All mice were raised under standard husbandry conditions and housed in colony cages with a 12-h light/dark cycle. They had free access to water and food, unless otherwise specified. Male C57/BL6 mice (Janvier, Le Genest-St-Isle, France) were fed regular chow containing 10% of calories as fat (Carfill, Oud-Turnhout, Belgium) or a high fat diet containing 60% of calories derived from fat (Diet 12492, Research diets, New Brunswick, CT) for 16 weeks starting either at 4 or 8 weeks of age. The experiments were performed with the approval of the ethical committee of the host institution.
RESULTS
miR-33a Is Produced from an Intron of the SREBP2 Gene and Targets the Transcript of ABCA1
miR-33a and miR-33b are localized in corresponding introns of the human SREBP2 and SREBP1 genes, respectively (Fig. 1A and supplemental Fig. S3). Localization and sequence of this miRNA family is highly conserved down to insects and chordates, but not in worms. Most species (including the mouse), however, only possess miR-33a in the SREBP2 gene and have lost the sequence of miR-33b within the SREBP1 gene (Fig. 1A and supplemental Fig. S3). With regard to their sequence, miR-33a and miR-33b differ only by two nucleotides outside the seed sequence (Fig. 1B).
FIGURE 1.
miR-33a localization within SREBP2 and sequences are evolutionary conserved. A, schematic representation of the human SREBP2 locus with embedded miR-33a sequence including evolutionary conservation of the latter. B, alignment of human miR-33a and miR-33b.
According to several target gene prediction algorithms, the mRNA transcript of the cellular cholesterol export pump ABCA1 contains putative binding sites miR-33a and miR-33b in its 3′ UTR (Pictar, Targetscan, Diana micro-T 3.0) (20–23)(Fig. 2A). Given the large overlap in predicted target genes for miR-33a and miR-33b, and the fact that miR-33a is completely conserved in most animal species, we focused our work on miR-33a (Fig. 1A). The predicted binding site in ABCA1 is highly conserved during evolution and present in mammals, birds, fish, and reptiles (supplemental Fig. S4A). To test whether it is functional, we cloned the region of the 3′ UTR of the ABCA1 gene surrounding the predicted miR-33a binding site downstream of a constitutively active firefly luciferase cassette. Cotransfection of preMIR-33a significantly reduced the activity of the reporter construct containing the 3′ UTR of ABCA1 (Fig. 2B). Introduction of site-directed mutations in the region of perfect complementarity to the seed region of miR-33 abolished this response, demonstrating that miR-33a can directly target this transcript (Fig. 2, B, asterisks, in A shown as sites of mutation, and supplemental Fig. S4B).
FIGURE 2.
miR-33 targets the ABCA1 3′ UTR for translational inhibition. A, predicted annealing of human miR-33a to the ABCA1 3′ UTR. Asterisks denote the bases changed in the mutant constructs. B, reporter assay to assess the influence of miR-33 on the ABCA1 3′ UTR. 293T cells were transfected with a construct carrying part of the ABCA1 3′ UTR (ABCA1), a mutant of the ABCA1 3′ UTR with site-directed mutagenesis of the sequence complementary to the miR-33 seed (ABCA1 MUT), or the parental pGL3 control plasmid (control) in the presence of a synthetic pre-miR-33 or a negative control pre-miRNA. Values are normalized to the activity of a constitutively active Renilla luciferase expression construct and represented as mean ± S.D. of triplicates. Asterisk denotes p < 0.05 in Student's t test (n = 3).
miR-33 Inhibits Endogenous ABCA1 Levels and Inhibits Cellular Cholesterol Export
To determine whether expression of miR-33a regulates endogenous levels of ABCA1, we transduced HepG2 cells with a lentiviral expression vector constitutively driving expression of a 500-bp genomic region surrounding the miR-33a sequence. We observed that miR-33a causes a marked decrease of ABCA1 protein levels (Fig. 3A, lanes 1 and 2) as assessed by immunoblot analysis. This effect was maintained when cells were treated with the liver X receptor (LXR)-agonist T0901317 (Fig. 3A, lanes 3 and 4), which has previously been shown to activate transcription of ABCA1 (24, 25).
FIGURE 3.
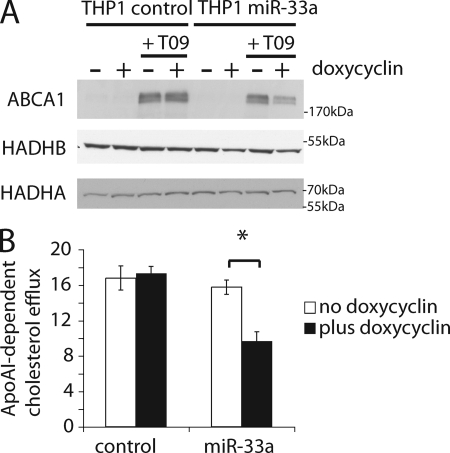
miR-33 reduces endogenous ABCA1 protein levels and modulates cellular cholesterol export in HepG2 cells. A and B, HepG2 cells were transduced with lentiviral constructs driving constitutive expression of miR-33a (miR-33) or a control construct (control). Western blot analysis (A) and apoAI-dependent cholesterol export (B) were performed after incubation for 16 h in the presence or absence of the LXR agonist T0901317. Values represent mean ± S.E. of 6 replicates. Asterisks denote p < 0.05 in Student's t test (n = 6). HepG2 cells were transduced with miRNA sponges targeting miR-33 or a control construct. Western blot analysis (two replicates are shown) (C) and apoAI-dependent cholesterol export (D) were performed in the absence of T0901317 and are represented as in A and B. HepG2 were transfected with cholesterol-coupled antisense oligonucleotides (antagomir) targeting miR-33a and miR-33b (equal amounts) or a negative control (i.e. the sense sequence of Arabidopsis thaliana miRNA159a). Western blot analysis (E) and apoAI-dependent cholesterol export (F) were performed 48 h after transfection and are represented as above (n = 6).
ABCA1 acts as a cellular cholesterol export pump. Although there are a number of cholesterol efflux pumps, ABCA1 has the unique capability to export cholesterol to lipid-free apolipoprotein AI (apoAI) and is therefore involved in the initial step of high-density lipoprotein particle formation in liver, as well as reverse cholesterol transport (26). To test whether the observed reduction of ABCA1 upon activation of miR-33 expression might be sufficient to reduce cellular cholesterol export, we loaded cells with radioactively labeled cholesterol, and observed significantly reduced apoAI-dependent cholesterol export in the HepG2 liver cancer cell line (Fig. 3B) and THP1 monocytic leukemia cell line derived from macrophages (Fig. 4B). Given rather low baseline levels of ABCA1 in THP1 cells, this experiment was performed only in the presence of the LXR agonist T0901317 (24, 25).
FIGURE 4.
miR-33 reduces endogenous ABCA1 protein levels and modulates cellular cholesterol export in THP1 macrophages. A, Western blot analysis of THP1 cells engineered to express miR-33a under the control of a doxycyclin-dependent promoter (THP1 miR-33) or control cells (THP1 control). Cells were differentiated into macrophages by treatment with PMA for 3 days. Protein lysates were analyzed upon treatment with or without doxycyclin for 24 h and the LXR agonist T0901317 for 16 h. B, apoAI-dependent cellular cholesterol export was measured in the absence or presence of doxycyclin for 24 h upon treatment for 16 h with the LXR agonist T0901317 (n = 7). Error bars denote ±S.D.
Notably, overexpression of competitive antagonists of miR-33 action (i.e. miRNA sponges) in HepG2 cells led to an up-regulation of ABCA1 protein levels, which was accompanied by a significant increase in apoAI-dependent cellular cholesterol export (Fig. 3, C and D). Because the observed effect was rather small, we sought to confirm these findings with an independent method of inhibiting miR-33. To this end, we used cholesterol-coupled 2-O-methyl-oligonucleotides (i.e. antagomirs) (13). When antagomir 33 was transfected into HepG2 cells, an even more dramatic stimulation of apoAI-dependent cholesterol export was observed (Fig. 3, E and F). In parallel experiments we established a human macrophage cell line, THP1, where miR-33a is expressed under a doxycyclin responsive promoter. Treatment of a control cell line with doxycyclin does not alter expression of ABCA1 protein levels in the absence or presence of T0901317 (Fig. 4A, first to fourth lanes). However, the cell line transduced with the conditional miR-33a lentivirus shows strong reduction of ABCA1 levels upon treatment with doxycyclin (Fig. 4A, fifth to eighth lanes). Similar results were also obtained in the human adrenocortical carcinoma cell line Y1 (supplemental Fig. S5). Taken together, our data suggest that endogenous miR-33 targets and represses translation of ABCA1, and reduces cellular cholesterol export.
miR-33 Targets Components of Cellular β-Oxidation
Evolution has conserved a protein coding for a master regulator of cholesterol synthesis (SREBP-2) and a miRNA that prevents the cellular export of cholesterol in the same primary transcript (Figs. 1–4). However, SREBPs not only activate cholesterol synthesis but also fatty acid and phospholipid synthesis (2). We therefore wondered whether other pathways in lipid metabolism might also be regulated by miR-33. Our attention was immediately drawn to fatty acid β-oxidation, because three distinct proteins involved in this pathway are predicted targets for miR-33 in mammals and birds (HADHB, CPT1A, and CROT) (Fig. 5, A, C, and E, and supplemental Figs. S6–S8 for evolutionary conservation). CPT1A is the liver-specific isoform of the enzyme that converts acyl-CoA to acylcarnitine and thereby allows fatty acids to enter the mitochondrion where fatty acid β-oxidation takes place (27). Although not strictly an enzyme of the β-oxidation, the activity of CPT1A is rate-limiting for β-oxidation, and is the most highly regulated element in this metabolic pathway (28). CROT, sometimes abbreviated COT, is a peroxisomal enzyme that couples short chain fatty acids to carnitine and thereby allows their entry in the mitochondrial matrix. Peroxisomal β-oxidation is limited to the degradation of long- and medium-chain fatty acids as it is not able to oxidize short chain fatty acids (29). Coupling to carnitine via CROT thereby allows mitochondrial entry of short chain fatty acids in a pathway that potentially bypasses CPTIA. The gene HADHB encodes for the β-subunit of the mitochondrial trifunctional protein that carries out the last three steps of β-oxidation (30).
FIGURE 5.
miR-33 directly inhibits translation of the transcripts of several components of cellular β-oxidation. A, predicted binding of miR-33a to the 3′ UTR of the human HADHB transcript. B, reporter gene experiment using the 3′ UTR of the human HADHB transcript (HADHB) or a mutant containing a site-directed mutation in the miR-33 seed binding region (HADHB MUT) were cotransfected with pre-miR-33 or a pre-miR control, identically to the experiment described in the legend to Fig. 2B. C, predicted binding of miR-33a to the 3′ UTR of the human CROT transcript. D, reporter gene experiment using the 3′ UTR of the human CROT transcript (CROT) or a mutant containing site-directed mutations of one and/or the other predicted miR-33 seed binding region (CROT MUT1/2/1 + 2) were cotransfected with pre-miR-33 or a pre-miR control, and evaluated identically to the experiment described in Fig. 2B. E, predicted binding of miR-33a to the 3′ UTR of the human CPT1A transcript encoding the rate-limiting step of cellular β-oxidation. F, reporter gene experiment using the 3′ UTR of the human CPT1A transcript (CPT1A) or a mutant containing site-directed mutations of one and/or the other predicted miR-33 seed binding region (CPT1A MUT1/2/1 + 2) were cotransfected with pre-miR-33 or a pre-miR control, and evaluated identically to the experiment described in the legend to Fig. 2B. Asterisks denote p < 0.05 in Student's t test (n = 3). Error bars denote ±S.D.
We first attempted to validate that the predicted target sites in the transcripts of CPT1A, CROT, and HADHB are targeted by miR-33. To this end, we cloned the regions encompassing the predicted miR-33 binding site behind a constitutively active firefly luciferase expression cassette (Fig. 5, A, C, and E). Cotransfection of these constructs with preMIR-33 significantly reduced activation of constructs containing the wild type 3′ UTR when compared with transfection with a preMIR control (Fig. 5, B, D, and F). Site-directed mutagenesis of one or two of the predicted seed binding sites completely abolished this response (mutated nucleotides indicated by asterisks). Not all predicted binding sites seemed to contribute to the response of miR-33 to the same extent. For example, deletion of the non-conserved predicted binding site 1 of the CPT1A transcript did not affect regulation by miR-33. It should be noted that several prediction algorithms do not recognize CPT1A as a miR-33 target gene. This is due to the fact that these algorithms utilize a short 3′ UTR generated from an alternative exon (supplemental Fig. S9A). Quantitative RT-PCR in different human organs, however, suggests that the transcript lacking the predicted miR-33 binding site is expressed at very low concentrations (supplemental Fig. S9B). It should be noted that several additional genes involved in other aspects of lipid metabolism or mitochondrial energy metabolism such as the NPC1 gene (Niemann-Pick disease, type C1) involved in intracellular cholesterol distribution, the phospholipid flippase ATP8B1, or the mitochondrial phosphate carrier SLC25A25 also contain predicted miR-33 binding sites in their 3′ UTR. In reporter assays, these 3′ UTRs convey a varying degree of responsiveness to miR-33 (supplemental Fig. S10).
miR-33 Inhibits Cellular Fatty Acid β-Oxidation
To ascertain effects of miR-33 on endogenous target genes, we performed a Western blot analysis for HADHB and CPT1A on lysates from HepG2 cells that had been transduced with a lentivirus driving expression of miR-33. As shown in Fig. 6A, miR-33 suppresses HADHB and CPT1A protein levels. Of note, a similar effect on HADHB levels, albeit to a smaller extent, can be observed in THP1 cells and Y1 cells (Fig. 4A and supplemental Fig. S5). The mitochondrial trifunctional protein consists of an α subunit (encoded by HADHA) and a β subunit (encoded by HADHB) (31). Previous studies have shown that reduction of one or the other component in heterozygote knock-out mice leads to a concomitant reduction of the other subunit (31–33). In line with these observations, the strong reduction of HADHB in HepG2 was accompanied by a reduction in HADHA. Unfortunately we were not able to obtain specific Western blot results using commercially available and custom-made antibodies against CROT. In addition to their function as inhibitors of protein translation, miRNAs often lead to a small reduction of target transcript abundance. We therefore were curious to see whether the mRNA transcript abundance of CROT was altered upon expression of miR-33. As shown in Fig. 6B, CROT mRNA levels were significantly reduced in cell lines overexpressing miR-33 to a degree that is similar to the reduction of ABCA1 and CPT1A, consistent with the idea that this transcript is indeed targeted by miR-33 and on the protein level might even be more strongly decreased.
FIGURE 6.
miR-33 reduces β-oxidation and alters cellular fatty acid metabolism. A, Western blot analysis of HepG2 cells transduced with lentiviral constructs driving expression of miR-33a (miR-33) or a control plasmid. Two biological replicates are shown. Loading control denotes an unspecific band in recognized by the HADHB antibody to demonstrate even loading of samples. B, quantitative RT-PCR for ABCA1, CROT, and CPT1A was performed in the same cells as in A. Expression was normalized to the abundance of U6 expression using the 2−dCt method. Note that these data are derived from six biological replicates and standard deviations are depicted to facilitate interpretation. The asterisk denotes p < 0.05 in Student's t test (n = 6). C, cellular β-oxidation was assessed by measuring conversion of [9,10-3H]palmitic acid into water in the cells represented in A. Values represent counts per protein amount in each well and are mean ± S.E. The asterisk denotes p < 0.05 in Student's t test (n = 6). D, quantification of lipid distribution in the cells described in A by TLC. Lipids were stained with the fluorescent dye Primulin and quantified using a fluorescent image scanner. Representative TLC are shown in supplemental Fig. S11 and values are shown as mean ± S.D. (n = 3).
To evaluate effects of miR-33 on fatty acid β-oxidation, we measured incorporation of radioactivity from [9,10-3H]palmitic acid into water. To measure the mitochondrial contribution to β-oxidation, we measured β-oxidation in the presence or absence of etomoxir, an irreversible inhibitor of CPT1A and CROT (34, 35). As shown in Fig. 6C, overexpression of miR-33 is associated with a 20% reduction of mitochondrial β-oxidation. When analyzing cellular extracts by TLC we observed a significant increase in cellular free fatty acid levels and triacylglycerides (Fig. 6D and supplemental Fig. S11). Inhibition of miR-33 with miRNA sponges or antisense oligonucleotides did not cause a reproducible increase in fatty acid β-oxidation or alteration of HADHB and CPT1A protein levels in HepG2 cells (data not shown).
Given the fact that CROT serves to channel medium chain fatty acids, the end products of peroxisomal β-oxidation, into the mitchondrion it seemed conceivable that miR-33 might alter the chain lengths of cellular fatty acids (29). Preliminary studies using separation by TLC, and gas chromatography of fatty acid chain lengths in triacylglycerides, phospholipids, and cholesterol esters do not appear to support this hypothesis (data not shown).
It has been previously noted that cellular miR-33 levels are dependent on culture conditions and exogenous lipid supply (36). Culturing cells at high density and/or in lipid-free medium leads to an increase of miR-33 (supplemental Fig. S12A). Given the influence of miR-33 on cholesterol export and β-oxidation, we investigated whether miR-33 are altered under conditions of fasting or high fat diet. In the livers of fasting mice and in two separate cohorts of mice exposed to a high fat diet for 16 weeks, we did not observe any significant alteration of miR-33 expression levels, albeit there was a small but non-significant trend for the high-fat diet group toward lower miR-33 levels (supplemental Fig. S12, B—D). Further studies are required to establish how miR-33 is regulated under varying physiological conditions, including dietary content and fasting/refeeding. In these studies, we are relying on Northern blot analysis, because miR-33 shows tissue-dependent differences in 3′ end length (Isomirs), which might confound quantification of miRNA levels by methods that rely on reverse transcription using hairpin primers (supplemental Fig. S13) (37, 38).
CPT1 in Cholesterol Auxotroph Drosophila Contains a Functional miR-33 Binding Site
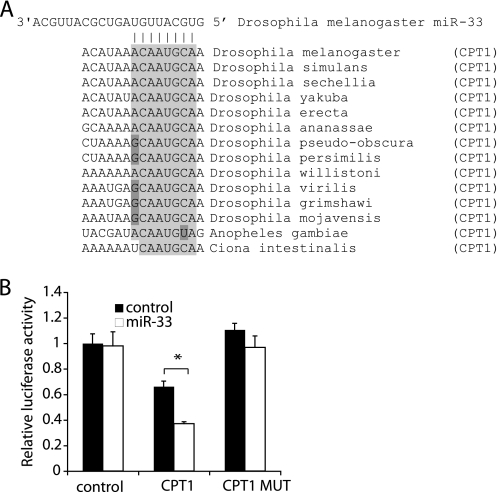
As mentioned above, SREBPs of higher organisms help to coordinately induce genes involved in cholesterol, fatty acid, and phospholipid synthesis. Notably, miR-33 is highly conserved in D. melanogaster (and other Drosophilids), which are auxotrophs for cholesterol, i.e. they cannot synthesize cholesterol. It has been previously shown that the homologs of SREBPs do not respond to changes in cholesterol, but rather changes in palmitate and/or phospholipids (39, 40). Notably, the transcript of Drosophila CPT1 contains a predicted miR-33 binding site in its 3′ UTR that is completely conserved in all Drosophilids (Fig. 7A). When cloned behind a constitutively active firefly luciferase expression cassette, this element confers responsiveness to miR-33 in reporter assays (Fig. 7B). Mutation of the seed region completely abolishes this response suggesting that this element mediates the response to miR-33.
FIGURE 7.
Conserved targeting of the β-oxidation gene CPTI in the cholesterol auxotrophs Drosophila. A, evolutionary conservation of the predicted miR-33 binding site in the 3′ UTR of the Drosophila, Anopheles, and C. intestinalis CPT1 transcripts. B, reporter gene experiment using the 3′ UTR of the CPT1 transcript (CPT1) or mutants containing site-directed mutations of the predicted miR-33 seed binding region (CPT1 MUT) were cotransfected with pre-miR-33 or a pre-miR control, and evaluated identically to the experiment described in the legend to Fig. 2B. The asterisk denotes p < 0.05 in Student's t test (n = 3). Error bars denote S.D.
DISCUSSION
Cholesterol is an essential component of cell membranes and serves as the starting point for the synthesis of steroid hormones and bile acids. Genes involved in its synthesis are coordinately up-regulated by the action of SREBP transcription factors (2). The discovery that miR-33 inhibits ABCA1 expression and consequentially cellular cholesterol export suggests that one genetic locus increases synthesis and limits export of cholesterol through two distinct mechanisms (Fig. 8A). When decreases in cellular cholesterol levels trigger the activation of SREBP transcription factors, the presence of miR-33 prevents cholesterol from leaving the cell by suppressing expression of ABCA1, thus allowing cellular cholesterol levels to be restored (Fig. 8A).
FIGURE 8.
miR-33 cooperates with SREBPs to maintain cellular lipid levels. A, schematic representation of the regulatory network in mammals. B, schematic representation of the regulatory network in the cholesterol auxotroph D. melanogaster.
Although cholesterol is required for most cells, its inappropriate accumulation in the wall of blood vessels is a hallmark of arteriosclerosis. High levels of low density lipoprotein cholesterol are a major risk factor for the development of arteriosclerosis and death from cardiovascular causes (41). On the other hand, elevated levels of high density lipoprotein cholesterol reduces the risk of arteriosclerosis (42, 43). Agents that alter HDL cholesterol levels therefore carry the potential to reduce cardiovascular diseases.
In a process termed reverse cholesterol transport, peripheral tissues export their excess cholesterol to HDL using the cholesterol export pumps ABCA1 and ABCG1 (44). ABCA1 is unique in the way that it can export cholesterol to nascent HDL particle or naked ApoAI. This process seems to be essential for the formation of HDL particles because liver- and intestine-specific knock-out mice for ABCA1 show strong reductions in HDL levels (45–47). The reciprocal relationship also appears to be true in that increased gene dosage of ABCA1 increases plasma HDL levels and protects from arteriosclerosis in animal models of hyperlipidemia (48).
Inactivating mutations in ABCA1 lead to the development of Tangier disease, which is characterized by the absence of HDL and the accumulation of cholesterol in peripheral tissues (42). Loss-of-function alleles of ABCA1 are associated with a decrease in HDL levels and might be associated with an increase in cardiovascular mortality (42, 49). Given its preventive effect in mice, increasing ABCA1 protein levels by inhibiting miR-33 might therefore prove useful in the prevention of cardiovascular diseases. Future in vivo experiments will be required to evaluate this possibility.
miR-33 and the Inhibition of β-Oxidation
Activation of SREBPs stimulates transcription of several genes involved in fatty acid synthesis. Inhibition of fatty acid degradation by miR-33 therefore is an elegant way to prevent degradation of newly synthesized lipids (Fig. 8A). D. melanogaster is a cholesterol auxotroph and activation of SREBP is regulated by reduced levels of palmitate and/or phospholipids. Interestingly, a conserved miR-33 binding site is present in the CPT1 transcript of all Drosophila species. No predicted binding sites exist in the closest homolog of ABCA1 (CG1718). This suggests, that the bifunctional SREBP-miR-33 locus in Drosophila is concentrated on the maintenance of cellular fatty acid levels (Fig. 8B). Surprisingly, based on miRNA binding site prediction algorithms, binding sites for miR-33 in β-oxidation genes are absent in lower vertebrates. At first glance this might suggest that the modulation of β-oxidation by miR-33 is of lesser importance than its effects on cholesterol export. However, the CPT1 genes in fish have undergone additional duplications leading to a total of five CPT1 homologs (localized in Zebrafish chr25, 24637246–24664477; chr24, 35296460–35323563; chr3, 29494843–29531322; chr7, 49453562–49466880; and chr18, 9105052–9120233) (50). It is therefore likely that the selective pressure for the maintenance of a functional miR-33 binding site in these genes is less pronounced. It should also be noted that annotation of 3′ UTRs in lower animals is not complete. Extending annotated gene transcripts with expressed sequence tags reveals that CPT1A in the frog Xenopus tropicalis and even the primitive chordate Ciona intestinalis (sea squirt) contain miR-33 seed binding sites (supplemental Fig. S7, C and D). Taken together with conservation of the binding site in Drosophila CPT1, this reinforces the notion that the influence of miR-33 on β-oxidation is of ancient origin.
Several explanations seem possible to explain why we did not observe an increase in β-oxidation and HADHB levels upon inhibition of miR-33. First, our inhibition is likely not complete. Second, it is possible that these transcripts might be under parallel repression by other miRNAs, so that reduction of miR-33 does not matter. Third, in the case of CPT1A, it should also be noted that the miR-33 binding site is localized quite distant from the open reading frame. It has recently been noted that cancer cell lines and proliferative cells often express transcripts with shorter 3′ UTRs (51). Therefore it seems possible that a significant portion of CPT1A in the tissue culture cell lines escapes repression by miR-33. Fourth, alterations in enzyme concentration in situations where the other regulatory mechanisms are working undisturbed might not necessarily be expected to alter flux through β-oxidation. Because we were unable to assess CROT protein abundance with commercial antibodies, it remains to be determined whether this protein might in fact increase upon miR-33 inhibition.
CROT and CPT1A are enzymes that couple fatty acids to carnitine. Although CROT is localized to the peroxisomes, it serves to transfer remnants from peroxisomal β-oxidation to the mitochondria and therefore fuels mitochondria with fatty acids independently of CPT1A (29). As mentioned before, CPT1A is the rate-limiting and most highly regulated step in β-oxidation. Interestingly, the dominant allosteric inhibitor malonyl-CoA regulates CROT similarly to CPT1A (28, 52). In an elegant twist, evolution has maintained the regulation by miR-33 of two enzymes that are sensitive to malonyl-CoA by miR-33. Increases in β-oxidation in situations of low malonyl-CoA levels might therefore be limited by miR-33.
In a more restricted number of species, SREBP1 contains the intronic miR-33b, which differs by two nucleotides in the center from miR-33a. Notably, these two changes lead to a perfect complementarity of miR-33b with the CROT 3′ UTR over a total of 18 nucleotides, which could allow miR-33b to induce Ago2-dependent cleavage of the CROT transcript (supplemental Fig. S8, B versus C). Because the embedding gene SREBP1 is more potent at activating genes involved in fatty acid synthesis than SREBP2, it makes sense teleologically that the embedded miRNA would more efficiently inhibit β-oxidation.
The relevance of targeting HADHB is not quite as apparent, although reduced HADHB concentrations might help maintain adequate intermediary product levels in a situation where the enzyme that supplies the substrates for β-oxidation (CPT1A and CROT) is inhibited. The situation is also a bit reminiscent of the situation of the SREBP proteins themselves. High levels of SREBP proteins do not necessarily mean high transcriptional activity, because these proteins are maintained in an inactive form attached to the membrane of the endoplasmic reticulum and need to be activated by proteolytic cleavage when lipid levels fall. Protein amounts of SREBPs therefore represent the maximum potential to respond to changes in lipid levels. Likewise, altering the levels of HADHB might change the potential maximum flux through β-oxidation, but might have little influence under most conditions where fatty acid supply in the mitochondria is rate-limiting for β-oxidation.
The protein product of the HADHB transcript forms the β-subunit of the mitochondrial trifunctional protein, which carries out three steps of β-oxidation. Surprisingly, there is evidence that reduced gene dosage or activity of this enzyme complex leads to the accumulation of lipids in the liver (hepatic steatosis) (32, 33). This was observed both in mice that carry a hypomorphic allele of the HADHB gene and, notably, in mice heterozygote for a null mutation of the HADHA gene (encoding the α-subunit) (32, 33). These observations are consistent with the idea that HADHA and HADHB levels limit capacity for β-oxidation in certain settings.
Clinically, regulation of β-oxidation might be of significant importance. Presently, non-alcoholic fatty liver disease (NAFLD) is by far the most common liver ailment (53). With the increasing prevalence of obesity, it affects 10–25% of the general population and up to 75% of obese type II diabetic patients (54). Although generally benign, about 10% of the cases progress to non-alcoholic steatohepatitis (NASH), which severely affects liver function in up to 25% of cases. A recent study of Cabellero and co-workers (55) suggests that SREBP2 transcript levels in NAFLD and NASH are elevated 3- and 7-fold, respectively. Given the fact that SREBP2 transcript levels correlate well with miR-33a levels in most tissues and cell lines (data not shown), it seems likely that miR-33a in these diseases would be elevated as well. Even small reductions of β-oxidation by miR-33 in a situation of hyperalimentation might contribute to an accumulation of lipids in the liver of patients with NAFLD or NASH. Interfering with miR-33 function in vivo therefore not only holds promise in increasing HDL cholesterol levels but may also prove beneficial for treatment of NAFLD and NASH.
Unfortunately, no single mouse model or feeding regimen reliably reproduces the features of NAFLD and NASH. In our two cohorts of mice on the high fat diet we did not observe an increase in miR-33 levels. In fact, there was a small, but not significant, trend for reduced miR-33 levels, as one might intuitively expect in a situation of hyperalimentation. Future studies on samples from the patient with NAFLD and NASH will be warranted.
Supplementary Material
Acknowledgments
We are grateful to Eric Fearon, Miikka Vikkula, Paul Tulkens, and Tobias Else for cell lines, Hassan Bassem and Natalie de Geest for Drosophila genomic DNA, and Emile Van Schaftingen for continuous intellectual and material support. The apoAI bacterial expression plasmid was a generous gift from W. Sean Davidson. We thank Michel Ghislain and Agnieszka Goj for help with the Phosphorimager, Hervé Degand for help with the fluorescent scanner, and Nicolas Poncelet for technical assistance in the early stages of this project.
Addendum
While this manuscript was in preparation and under review, two articles were presented online describing the effect of miR-33 on ABCA1 and cholesterol metabolism (56–58).
This work was supported by a Brains-Back-to-Brussels grant of the Region-Bruxelles-Capitales (to G. T. B.), by a fellowship of the Fonds Spéciaux de Recherche – Université Catholique de Louvain (FRS-UCL) and the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture (FRIA) (to L.-A. C.), by a return grant of the Belgian Science Policy (to I. G.), by a grant of the Fonds de la Recherche Scientifique (to I. A. L.), by funds from the de Duve Institute (to G. T. B.), by the National Institute of Health Grant DK51563 (to O. A. M.), and the Michigan Nutrition Obesity Research Center Grant 5P30DK089503 (to C. F. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S13.
- SREBP
- sterol regulatory element-binding factor
- CPT1A
- carnitine palmitoyltransferase 1A
- CROT
- carnitine O-octanoyltransferase
- HADHB
- hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-coenzyme A hydratase β-subunit
- miRNA
- micro-RNA
- LXR
- liver X receptor
- NAFLD
- non-alcoholic fatty liver disease
- NASH
- non-alcoholic steatohepatitis.
REFERENCES
- 1.Horton J. D., Goldstein J. L., Brown M. S. (2002) Cold Spring Harbor Symp. Quant. Biol. 67, 491–498 [DOI] [PubMed] [Google Scholar]
- 2.Horton J. D., Goldstein J. L., Brown M. S. (2002) J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 4.Shimano H., Horton J. D., Shimomura I., Hammer R. E., Brown M. S., Goldstein J. L. (1997) J. Clin. Invest. 99, 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. (1997) J. Clin. Invest. 99, 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 7.Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farh K. K., Grimson A., Jan C., Lewis B. P., Johnston W. K., Lim L. P., Burge C. B., Bartel D. P. (2005) Science 310, 1817–1821 [DOI] [PubMed] [Google Scholar]
- 9.Behm-Ansmant I., Rehwinkel J., Izaurralde E. (2006) Cold Spring Harbor Symp. Quant. Biol. 71, 523–530 [DOI] [PubMed] [Google Scholar]
- 10.Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. (2009) RNA 15, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajewsky N. (2006) Nat. Genet. 38, (suppl.), S8–13 [DOI] [PubMed] [Google Scholar]
- 12.Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 14.Ebert M. S., Neilson J. R., Sharp P. A. (2007) Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tubb M. R., Smith L. E., Davidson W. S. (2009) J. Lipid Res. 50, 1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 17.White T., Bursten S., Federighi D., Lewis R. A., Nudelman E. (1998) Anal. Biochem. 258, 109–117 [DOI] [PubMed] [Google Scholar]
- 18.Keller P., Petrie J. T., De Rose P., Gerin I., Wright W. S., Chiang S. H., Nielsen A. R., Fischer C. P., Pedersen B. K., MacDougald O. A. (2008) J. Biol. Chem. 283, 14355–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 22.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 23.Maragkakis M., Alexiou P., Papadopoulos G. L., Reczko M., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., Simossis V. A., Sethupathy P., Vergoulis T., Koziris N., Sellis T., Tsanakas P., Hatzigeorgiou A. G. (2009) BMC Bioinformatics 10, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Repa J. J., Turley S. D., Lobaccaro J. A., Medina J., Li L., Lustig K., Shan B., Heyman R. A., Dietschy J. M., Mangelsdorf D. J. (2000) Science 289, 1524–1529 [DOI] [PubMed] [Google Scholar]
- 25.Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., Tontonoz P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12097–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tall A. R., Yvan-Charvet L., Terasaka N., Pagler T., Wang N. (2008) Cell Metab. 7, 365–375 [DOI] [PubMed] [Google Scholar]
- 27.Bonnefont J. P., Djouadi F., Prip-Buus C., Gobin S., Munnich A., Bastin J. (2004) Mol. Aspects Med. 25, 495–520 [DOI] [PubMed] [Google Scholar]
- 28.Saggerson D. (2008) Annu. Rev. Nutr. 28, 253–272 [DOI] [PubMed] [Google Scholar]
- 29.Wanders R. J., Ferdinandusse S., Brites P., Kemp S. (2010) Biochim. Biophys. Acta 1801, 272–280 [DOI] [PubMed] [Google Scholar]
- 30.Eaton S., Bursby T., Middleton B., Pourfarzam M., Mills K., Johnson A. W., Bartlett K. (2000) Biochem. Soc. Trans. 28, 177–182 [DOI] [PubMed] [Google Scholar]
- 31.Ibdah J. A., Paul H., Zhao Y., Binford S., Salleng K., Cline M., Matern D., Bennett M. J., Rinaldo P., Strauss A. W. (2001) J. Clin. Invest. 107, 1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibdah J. A., Perlegas P., Zhao Y., Angdisen J., Borgerink H., Shadoan M. K., Wagner J. D., Matern D., Rinaldo P., Cline J. M. (2005) Gastroenterology 128, 1381–1390 [DOI] [PubMed] [Google Scholar]
- 33.Kao H. J., Cheng C. F., Chen Y. H., Hung S. I., Huang C. C., Millington D., Kikuchi T., Wu J. Y., Chen Y. T. (2006) Hum. Mol. Genet. 15, 3569–3577 [DOI] [PubMed] [Google Scholar]
- 34.Lopaschuk G. D., Wall S. R., Olley P. M., Davies N. J. (1988) Circ. Res. 63, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 35.Morillas M., Clotet J., Rubí B., Serra D., Ariño J., Hegardt F. G., Asins G. (2000) Biochem. J. 351, 495–502 [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuchiya S., Oku M., Imanaka Y., Kunimoto R., Okuno Y., Terasawa K., Sato F., Tsujimoto G., Shimizu K. (2009) Nucleic Acids Res. 37, 3821–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benes V., Castoldi M. (2010) Methods 50, 244–249 [DOI] [PubMed] [Google Scholar]
- 38.Marti E., Pantano L., Banez-Coronel M., Llorens F., Minones-Moyano E., Porta S., Sumoy L., Ferrer I., Estivill X. (2010) Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B. (2002) Science 296, 879–883 [DOI] [PubMed] [Google Scholar]
- 40.Seegmiller A. C., Dobrosotskaya I., Goldstein J. L., Ho Y. K., Brown M. S., Rawson R. B. (2002) Dev. Cell 2, 229–238 [DOI] [PubMed] [Google Scholar]
- 41.Nabel E. G. (2003) N. Engl. J. Med. 349, 60–72 [DOI] [PubMed] [Google Scholar]
- 42.Brunham L. R., Kastelein J. J., Hayden M. R. (2008) JAMA 300, 1997–1998; author reply 1998 [DOI] [PubMed] [Google Scholar]
- 43.Kannel W. B. (1983) Am. J. Cardiol. 52, 9B–12B [DOI] [PubMed] [Google Scholar]
- 44.Yvan-Charvet L., Ranalletta M., Wang N., Han S., Terasaka N., Li R., Welch C., Tall A. R. (2007) J. Clin. Invest. 117, 3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunham L. R., Singaraja R. R., Duong M., Timmins J. M., Fievet C., Bissada N., Kang M. H., Samra A., Fruchart J. C., McManus B., Staels B., Parks J. S., Hayden M. R. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 548–554 [DOI] [PubMed] [Google Scholar]
- 46.Singaraja R. R., Stahmer B., Brundert M., Merkel M., Heeren J., Bissada N., Kang M., Timmins J. M., Ramakrishnan R., Parks J. S., Hayden M. R., Rinninger F. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1821–1827 [DOI] [PubMed] [Google Scholar]
- 47.Brunham L. R., Kruit J. K., Iqbal J., Fievet C., Timmins J. M., Pape T. D., Coburn B. A., Bissada N., Staels B., Groen A. K., Hussain M. M., Parks J. S., Kuipers F., Hayden M. R. (2006) J. Clin. Invest. 116, 1052–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singaraja R. R., Bocher V., James E. R., Clee S. M., Zhang L. H., Leavitt B. R., Tan B., Brooks-Wilson A., Kwok A., Bissada N., Yang Y. Z., Liu G., Tafuri S. R., Fievet C., Wellington C. L., Staels B., Hayden M. R. (2001) J. Biol. Chem. 276, 33969–33979 [DOI] [PubMed] [Google Scholar]
- 49.Frikke-Schmidt R., Nordestgaard B. G., Stene M. C., Sethi A. A., Remaley A. T., Schnohr P., Grande P., Tybjaerg-Hansen A. (2008) JAMA 299, 2524–2532 [DOI] [PubMed] [Google Scholar]
- 50.Morash A. J., Le Moine C. M., McClelland G. B. (2010) Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R579–589 [DOI] [PubMed] [Google Scholar]
- 51.Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008) Science 320, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A'Bháird N. N., Ramsay R. R. (1992) Biochem. J. 286, 637–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Angulo P. (2002) N. Engl. J. Med. 346, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 54.Marchesini G., Moscatiello S., Di Domizio S., Forlani G. (2008) J. Clin. Endocrinol. Metab. 93, S74–80 [DOI] [PubMed] [Google Scholar]
- 55.Caballero F., Fernández A., De Lacy A. M., Fernández-Checa J. C., Caballería J., García-Ruiz C. (2009) J. Hepatol. 50, 789–796 [DOI] [PubMed] [Google Scholar]
- 56.Rayner K. J., Suraez Y., Davalos A., Parathath S., Fitzgerald M. L., Tamehiro N., Fisher E. A., Moore K. J., Fernandez-Hernando C. (2010) Science 328, 1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Najafi-Shoushtari S. H., Kristo F., Li Y., Shioda T., Cohen D. E., Gerszten R. E., Naar A. M. (2010) Science 328, 1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marquart T. J., Allen R. M., Ory D. S., Baldán A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12228–12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.