Abstract
Our previous studies have shown an association between Helicobacter pylori infection, the strong up-regulation of cathepsin X (CTSX, also called cathepsin Z/P), and the development of gastric cancer. In the present study, we analyzed primary and conventional gastric epithelial cell lines to establish an optimal in vitro mouse model system for the examination of H. pylori-induced overexpression of Ctsx in a functional way. Gastric epithelial cells were isolated from stomachs of wild-type C57BL6/N and Ctsx−/− mice and compared with the gastric cancer cell line CLS103. Indirect co-cultures of epithelial cells and macrophages were infected with H. pylori strain SS1 and analyzed for the expression of cathepsins, cytokines, and adhesion factors. Cellular interactions, migration capability, and adherence of H. pylori were assessed using time-lapse video microscopy and colony-forming assays. Isolated primary cells from wild-type and transgenic mice revealed qualities and expression profiles similar to those of corresponding tissue samples. Adherence of H. pylori was significantly higher in primary compared with commercially cells. Thus, induction of cathepsins, cytokines, and adhesion proteins was detected solely in primary cells and co-cultured macrophages. Microarray and migration experiments indicated that Ctsx is involved in B/T-cell proliferation/migration and adhesion of macrophages. Primary epithelial cells from stomach of Ctsx−/− mice represent an excellent model of H. pylori gastritis to elaborate the special functions of Ctsx in regulating the immune response to H. pylori.
Keywords: Cell Motility, Cell-Cell Interaction, Epithelial Cell, Inflammation, Macrophage, Helicobacter Pylori, Cathepsin X, Gastritis, Mouse Model, Primary Epithelial Cells
Introduction
Helicobacter pylori is a Gram-negative bacterium, which colonizes the stomach (1–3), principally in the region of the antrum and corpus. H. pylori is widespread throughout the world and is colonized in more than half of the world's population (4). For the majority of infected persons, however, the H. pylori-induced gastric inflammation remains undiscovered (5). Otherwise, infection with H. pylori can be associated with the development of chronic gastritis, gastric adenocarcinoma (6), peptic ulcers, and gastric MALT lymphoma (7, 8). Moreover, H. pylori induces direct DNA damage (9), tissue inflammation, cell apoptosis, and proliferation (10, 11).
Cathepsins X, B, L, and K (CTSX/B/L/K) are differentially expressed in normal and chronically inflamed gastric mucosa. In contrast to CTSB, -L, and -K, only CTSX is strongly expressed in H. pylori-infected gastric mucosa and has also been found to be up-regulated in intestinal-type gastric cancer (12–14). Its expression is restricted primarily to cells of the immune system, such as monocytes, macrophages, or dentritic cells. Up-regulation in gastric cancer was also due to different expressions in neoplastic epithelium. The physiological function of CTSX still needs to be clarified. Obermajer et al. (15) focused on a potential function of CTSX in regulating immune response by activation of Mac-1 and lymphocyte function-associated antigen 1 (LFA-1) β2 integrins, resulting in increased adhesion and phagocytosis of macrophages, as well as in maturation of dendritic cells. Furthermore, CTSX has been shown to bind to cell surface heparin sulfate proteoglycans (16) and integrin αVβ3 (17), indicating once more a major role of CTSX in cellular adhesion, phagocytosis, and lymphocyte proliferation. CTSX expression was found to be regulated by different cell-type-dependent pathways and stimulants. Up-regulation of CTSX expression in NCI-N87 epithelial cells has been found to be independent of infections with cagA+ H. pylori strains in vitro, but fine membrane localization in surface epithelial cells could be recognized only in CagA+ biopsies in vivo. Furthermore, induction involves activation of JNK and p38 signaling pathways, as well as IL-1β and IL-6. In contrast, up-regulation of CTSX in monocytes has been shown to be dependent on the translocation of CagA in corresponding epithelial cells in vitro and was regulated through activation of MEK1/2 signaling, as well as TNF-α and IL-8 (12). Sivaraman et al. (18) demonstrated that cathepsin L plays an active part in activating CTSX, and Vasiljeva et al. (19) described the compensatory function of CTSX in CTSB-deficient tumor cells, making the analysis of the function of CTSX detached from other cathepsins almost impossible.
All experiments conducted by us so far analyzed CTSX expression and regulation in H. pylori-infected gastric mucosa, were done on gastric cancer cell lines (AGS, NCI-N87) and/or commercially available monocytic cells (THP-1) of human origin. Sequencially Krueger et al. (20) isolated normal human gastric epithelial cells from gastric biopsies, which is the ideal in vitro model system for investigation of the gastric epithelium, similar to the in vivo situation. In addition to cell culture studies, several groups investigate H. pylori infections in mouse or gerbil models (21), but the expression or functional studies of cathepsins have not yet been subject in this research. Sevenich et al. (22) provided us with Ctsx and Ctsb transgenic mice, which naturally represent the best system for the analysis of Ctsx functions not only in H. pylori infections.
In the present study, we used primary mouse gastric epithelial cells from WT and Ctsx−/− mice, as well as the murine gastric carcinoma cell line CLS 103 and a mouse macrophage cell line J-774A.1, to analyze the influence of loss of Ctsx during H. pylori infection on the expression of cytokines, chemokines, and genes involved in T- and B-cell activation as well as proliferation and differentiation. Moreover, we compared Ctsx-expressing and -nonexpressing epithelial cells for their ability to interact with macrophages and to initiate their migration.
EXPERIMENTAL PROCEDURES
Animals
Mice with deficiencies in cathepsin B (mutant allele Ctsbtm1Jde, designated here as Ctsb−/−) and cathepsin X (mutant allele Ctsztm1Thre; designated here as Ctsx−/−) were generated by gene targeting in mouse embryonic stem cells as described (22, 23). Both mouse lines have been backcrossed for 10 generations to the C57BL6/N genetic background. The animals were housed in a 12:12 light/dark cycle with unconditional access to water and food. They are kept in specific pathogen-free facilities. In addition, stomachs from specific pathogen-free WT, Ctsx−/−, and Ctsb−/− mice were analyzed to be negative for H. pylori in routine tests.
Cultivation of H. pylori
The H. pylori strain SS1 (Sydney strain 1), a mouse-adapted strain, was cultured in thin layers on 10% horse serum agar plates supplemented with vancomycin (10 μg/ml), trimethoprim (5 μg/ml), and nystatin (1 μg/ml) and incubated for 48 h at 37 °C in an anaerobic jar containing a CampyGen gas mix of 5% O2, 10% CO2, and 85% N2 (Oxoid, Wesel, Germany) as reported previously (20). All antibiotics were obtained from Sigma-Aldrich (Deisenhofen, Germany). The H. pylori strain SS1 was provided by Professor Steffen Backert (School of Biomolecular & Biomedical Science, University College Dublin). For the infection, bacteria were harvested in PBS, pH 7.4, and added to the serum-starved cells at a multiplicity of infection of 50.
Tissue Preparation, Cell Culture, and H. pylori Infection
For the isolation of primary gastric epithelial cells, uninfected WT, Ctsx−/−, and Ctsb−/− mice were sacrificed at 12 to 20 weeks, stomachs were removed and immediately cleaned in RPMI 1640 medium (Invitrogen). The stomachs were cut into small pieces (1–2 mm2) and incubated by stirring constantly in 25 ml collagenase (Sigma)/dispase (Invitrogen) solution (12,000 units of collagenase I, 125 units of dispase, 125 mg BSA per 100 ml Quantum 286) for 2 h at 37 °C. Six-well plates were precoated with 1 ml MatrigelTM (5 μl/ml; BD Biosciences) and incubated at 37 °C for 2 h to allow polymerization of MatrigelTM. After pelleting dispersed cells and washing with PBS (PAA, Linz, Austria), the cells were resuspended in epithelial cell culture medium Quantum 286 (PAA) supplemented with gentamycin (10 μg/ml), penicillin/streptomycin (5 μg/ml, Invitrogen) and seeded into Matrigel-coated six-well-plates (Biochrom, Berlin, Germany) containing Quantum 286 medium (PAA) with antibiotics penicillin/streptomycin/gentamycin (Invitrogen) and incubated at 37 °C for at least 24 h. Cell purity was examined using light microscopy. Cells were classified as epithelial cells (cobblestone morphology), fibroblasts (spindle-shaped morphology), or unclassified (rounded cells of undetermined origin). Due to the Matrigel coating, the percentage of fibroblast and unclassified cells could be reduced to 2.7 + 0.7% and 0.2 + 0.1%, respectively. 97.1 + 8.2% of cells exhibited cobblestone morphology and grew in clusters.
The murine gastric carcinoma cell line CLS 103 was obtained from CLS (Heidelberg, Germany). The cell line was grown in six-well plates (Biochrom, Berlin, Germany) for 2 days. Each well contained 2 ml of RPMI 1640 medium (Invitrogen) supplemented with antibiotics (penicillin/streptomycin) and 10% FCS (Invitrogen, Karlsruhe, Germany).
The mouse macrophage cell line J-774A.1 is active in antibody-dependent phagocytosis and was also received from CLS. The cell line was cultured in high glucose DMEM supplemented with 10% fetal calf serum and 1% antibiotics (penicillin/streptomycin, 10 mg/ml) and incubated at 37 °C under standard conditions of 5% CO2.
For co-culture experiments, mouse gastric epithelial cells were seeded on the upper part of 0.4 μm trans-well chambers (Corning-Costar), infected with the H. pylori strain SS1 and co-cultured with J-774A.1 in the lower compartment of chambers. For infection, incubation medium was removed and replaced by medium without antibiotics. Uninfected cells were used as controls.
Bacterial Colonization Assay
Epithelial cells were plated in 6-cm tissue culture plates at 5 × 105 cells per plate and allowed to grow for 24 h to subconfluency. H. pylori SS1 was added to cells at an multiplicity of infection of 50. Infected cultures were washed after 4 h three times with PBS (pH 7.6) to remove nonadherent bacteria, and total cell lysates were harvested after 15 min of incubation with PBS/0.2% saponin. Serial 10-fold dilutions of cell extracts were cultured on 10% horse serum agar plates as described above and were incubated for 3 to 5 days under microaerobic conditions.
Quantitative Real-time PCR Analysis
Assays were performed on the LightCyclerTM (Roche Diagnostics). Several dilutions of plasmids containing the cDNA fragments of the different genes were used as internal controls. For plasmid construction, the cDNA fragments were amplified using the following primers: mCtsx, mCtsb, mCtsl, mIL-1β, mCxcl1, mMCP-1, and mIL-6. The sequences of all PCR primers with their optimal annealing temperature are listed in Table 1. The same primers were used for qPCR reactions. PCR products were inserted into the pCR2.1-TOPO vector (Invitrogen). The copy number of the resulting plasmids was calculated after DNA quantification.
TABLE 1.
Sequences of PCR primers, length of PCR products, and optimal annealing temperature for the indicated mouse genes
| Primer | Sense | Antisense | Template | Annealing |
|---|---|---|---|---|
| bp | ° C | |||
| Actin | GTGCTGTCCCTGTATGCCTCTG | AACCGCTCGTTGCCAATAGTG | 349 | 55 |
| Ctsx | CCTGTCCGGGAGGGAGAA | TGGTTGATAACGGCCTGGTC | 137 | 55 |
| Ctsb | TGCGTTCGGTGAGGACATAGA | GGACGGGAGCCATTGACAT | 371 | 54 |
| Ctsl | GGAGATGAACGCCTTTGGTG | TTACAGCCCTGATTGCCTTGA | 311 | 54 |
| IL-1β | CAACCAACAAGTGATATTCTCCATG | GATCCAACATCTCCAGCTGCA | 446 | 57 |
| Cxcl1 | GCACCCAAACCGAAGTCATAGC | TTGTCAGAAGCCAGCGTTCACC | 174 | 60 |
| MCP-1 | GCTCTCTCTTCCTCCACCACCAT | GCTCTCCAGCCTACTCATTGGGAT | 170 | 60 |
| IL-6 | CACAAAGCCAGAGTCCTTCAGAGA | CTAGGTTTGCCGAGTAGATCTC | 226 | 52 |
| CD81 | AATCCAAGCZCCGCAGGCCG | GGCCCCATAGCACCCCAGGA | 278 | 60 |
| Cdkn1a | AACGCGCTCCCAGACGAAGT | GCGATATCCAGACATTCAGAGCCACA | 268 | 60 |
| Prkcd | TGCCCTGGTTTGCACCGCAT | GGTCAACACATCACCAGTCTCCTACA | 294 | 60 |
| Glmn | GTGCTGGAAGCCCGGGTACTT | GGAGGCATCGAACAACTGGACCAA | 298 | 60 |
| IL-18 | TGGCCCAGGAACAATGGCTGC | TGGTCTGGGGTTCACTGGCACT | 251 | 60 |
| Spp1 | GCCACATGGCTGGTGCCTGA | CGGCCGTTGGGGACATCGAC | 314 | 60 |
| Ap3b1 | TGAGCCTGCGCCCAGAAACG | GTGGGCGGCCAGTCCTTTCC | 286 | 60 |
| Gadd45g | GCGTCAGGATCGCCTCACCG | GTGCAGGTCTCGGGCTTCGG | 262 | 60 |
Twenty-four hours after infection, cells were harvested, and total RNA was isolated using a High Pure RNA isolation kit (Roche Diagnostics) according to the manufacturer's instructions. For cDNA synthesis, total mRNA was reverse-transcribed using a Transcriptor High Fidelity cDNA kit (Roche Diagnostics). PCR reactions were done using the quantitative PCR Mastermix kit (Quantace, London) according to the manufacturer‘s instructions. The standard temperature profile included initial denaturation for 10 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 15s, annealing at 52 °C to 60 °C (primer-dependent) for 15 s, and extension at 72 °C for 10 s. Mouse actin served as a housekeeping gene.
All oligonucleotide primers were purchased from BioTez (Berlin, Germany). The PCR products were analyzed on a 1.8% agarose gel and visualized by ethidium bromide staining.
Cellular Interaction in Culture Inserts
Mouse gastric epithelial cells (WT and Ctsx−/−) and mouse macrophage cells (J-774A.1) were isolated and seeded in six-well plates as described above. After 48 h, a modified scratch-assay was performed. We used culture inserts from Ibidi (Munich, Germany), which created a cell-free gap of ∼500–600 μm after removing the culture insert. At first, all cells were counted in a Coulter Counter ZII (Coulter Immunotech, Marseille, France) and diluted to a cell suspension of 5 × 105 cells/ml. Subsequently, the epithelial cells (WT and Ctsx−/−) were transferred into the left well of the culture insert and the macrophage cell suspension in the right one. Mouse gastric epithelial cells (WT, Ctsx−/−) were infected with H. pylori SS1 as described above. The culture insert was removed, medium was changed, and the cellular interaction between the infected or noninfected mouse gastric epithelial cells (WT, Ctsx−/−) and mouse macrophages (J-774A.1) was monitored by time-lapse video microscopy (LEICA TCS SP2 microscope) with time intervals of 15 min. To quantify the migration, a scale bar was photographed using the same magnification, and images were combined to determine the mean distance traveled by 10 cells at the leading edge over the course of the experiment. Data of three separate experiments were presented as μm/h (migration speed).
cDNA Array Analysis
The Oligo GEArray® Mouse T-cell and B-cell Activation Microarray was purchased from SABiosciences Corp. (Biomol, Hamburg, Germany). The Oligo GEArray® is designed to profile expression of 113 focused genes representing T- and B-cell activation, proliferation, and differentiation, which are key steps of adaptive immunity. The complete lists of genes are listed and available on the SABiosciences Web site. In addition, genes that activate macrophages, neutrophils, and natural killer cells are involved.
Total RNA from primary gastric epithelial cells (infected with H. pylori and uninfected WT and Ctsx−/− mice) was transcribed to labeled cRNA according to the manufacturer‘s instructions (TrueLabelingAMPTM 2.0 and ArrayGradeTM cRNA Cleanup kit, SABiosciences Corp.). Prehybridization, washing steps, hybridization, and chemiluminescence detection were carried out following the protocol of the manufacturer. For signal development, we used the GEArray® chemiluminescent detection system (SABiosciences Corp.). Images were quantified using the GeneGnome and GeneTools image scanning and analysis package (Syngene BioImaging Systems, Synoptics Ltd.).
Statistical Analysis
Data are expressed as mean ± S.D. Statistical analyses were performed using Student's t test. Differences or significance between the groups was assumed for p ≤ 0.05 (*).
RESULTS
Expression of and Ratio among Cathepsins (Ctsx/b/l) Is Comparable with Isolated Epithelial Cells and Corresponding Tissue
We tested the expression of relevant cathepsins (Ctsx/b/l) in the complete mouse stomach and extracted gastric epithelial cells (WT, Ctsx−/−, and Ctsb−/−), all of C57BL/6 background, to clarify the general expression pattern of cathepsins in the WT compared with the gene-manipulated mice. RNA was isolated from gastric tissues and primary gastric epithelial cells. Expression of the cathepsins was studied by quantitative RT-PCRs.
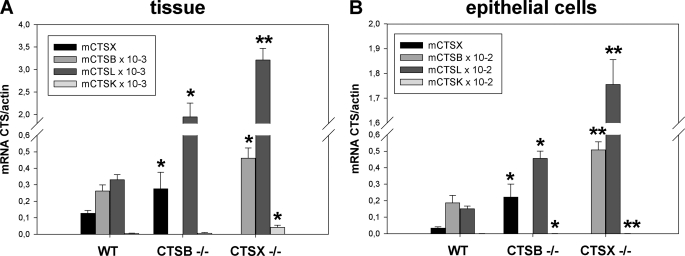
Ctsx expression was increased 2-fold in Ctsb−/− tissue compared with WT stomachs, whereas in extracted epithelial cells of similar origins, the increase was 6.5-fold. Ctsb expression was increased 1.7-fold in Ctsx−/− mice compared with WT tissue and 2.7-fold in isolated cells. Compared with WT mice, Ctsl is also overexpressed (up to 100-fold) in Ctsb−/− and in Ctsx−/− mice. The absolute expression in the epithelial cells is ∼2-fold lower than in tissue because of other Ctsx-expressing cells. Taken together, the expression and the increase of the expression are comparable in tissue and corresponding epithelial cells. Gene-manipulated mice generally expressed significantly higher levels of the cathepsins not depleted. Statistical data (t test) are shown in bar graphs in (Fig. 1, A and B).
FIGURE 1.
Expression of cathepsins (Ctsx/b/l) in gastric tissue and isolated epithelial cells. Gastric tissue (A) and primary gastric epithelial cells (B) from wild-type C57BL6/N, Ctsb, and Ctsx knock-out mice (WT, Ctsb−/−, and Ctsx−/−) were isolated and prepared for quantitative RT-PCRs to analyze and compare the expression levels of cathepsins in tissue and epithelial cells. The absolute expression in the epithelial cells is ∼2-fold lower than in tissue. Ctsx expression was increased 2-fold in Ctsb−/− tissue compared with WT stomachs, whereas the increase was 6.5-times stronger in extracted epithelial cells of similar origin. Cathepsin B expression was increased 1.7-fold in Ctsx−/− mice compared with WT tissue and increased 2.7-fold in isolated cells. Compared with WT mice, cathepsins L and K are also overexpressed (up to 1000-fold) in Ctsb−/− and Ctsx−/− mice. Gene-manipulated mice generally expressed significantly higher levels of the cathepsins not depleted. The t test, performed among these data sets compared with WT tissue or cells, revealed significance (*, p ≤ 0.05; **, p ≤ 0.01) and the results are presented in the bar graphs figure as means ± S.E.
H. pylori Induces Ctsx Overexpression in Primary Gastric Epithelial cells ex Vivo
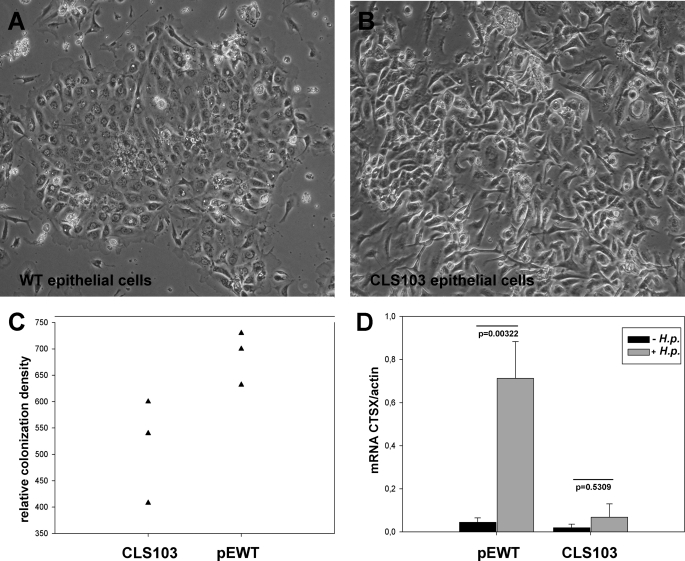
To compare Ctsx expression in wild-type primary gastric epithelial cells with the murine gastric carcinoma cell line CLS 103, we infected the different cell types with H. pylori and performed quantitative RT-PCR and bacterial adhesion assay. Our previous studies have shown that of all the cathepsins in gastric mucosa, only Ctsx was up-regulated by H. pylori (12, 13). Therefore, we have concentrated only on Ctsx expression. The wild-type primary gastric epithelial cells (Fig. 2A) and the murine gastric carcinoma cell line CLS 103 (Fig. 2B) were grown on six-well plates for 2 days. The epithelial cells were infected with H. pylori strain SS1. Uninfected cells served as controls. The density of SS1 colonization was markedly higher in primary cells compared with CLS 103 cells (Fig. 2C). Ctsx was detected by quantitative real-time RT-PCR. Mouse actin mRNA was used as a housekeeping gene. As shown in Fig. 2D, Ctsx is significantly up-regulated (17-fold) at the mRNA level in H. pylori-infected epithelial cells of WT mice (p = 0.00322). H. pylori infection of the CLS 103 cell line resulted in a 3-fold increase of Ctsx, which was not statistically significant. The Ctsx expression after H. pylori infection is generally 13-fold higher in WT cells than in CLS 103 cells.
FIGURE 2.
Effect of H. pylori infection on the expression of Ctsx in primary gastric epithelial cells ex vivo. Wild-type primary gastric epithelial cells (A) and the murine gastric epithelial carcinoma cell line CLS 103 (B) were co-cultured with and without H. pylori (H.p.) strain SS1 (100×). The adhesion of the H. pylori SS1 strain was significantly more pronounced on WT primary cells compared with CLS 130 cells (C). Total RNA was extracted and analyzed by RT-PCR for mRNA expression of Ctsx. Uninfected WT and CLS 103 primary gastric epithelial cells were compared regarding their Ctsx expression after H. pylori infection (D). Ctsx expression after H. pylori infection is generally 13-fold stronger in WT cells than in CLS 103 cells. Bar graphs represent the expression of Ctsx and its significance. Colonization density was measured by colony-forming assay.
Induction of Proinflammatory Cytokines in Conjunction with Ctsx Expression in Mono-cultures and Co-cultures after H. pylori Infection
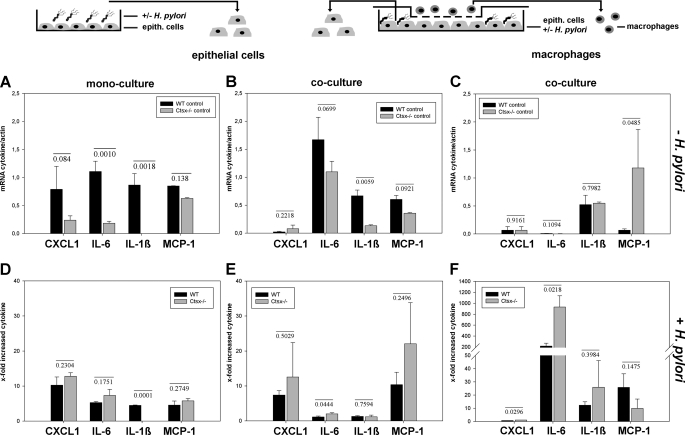
A characteristic feature of H. pylori is its ability to stimulate the production of proinflammatory cytokines. To analyze the influence of H. pylori infection on the expression of cytokines in conjunction with the Ctsx level, we used primary mouse gastric epithelial cells from wild-type and Ctsx−/− mice, as well as the mouse macrophage cell line J-774A.1. The wild-type and Ctsx−/− primary cells were co-cultured with J-774A.1 cells, infected with H. pylori SS1, and tested for the expression of mouse chemo- and cytokines (CXCL1, IL-6, IL-1β, and MCP-1). Mono-cultures with the same treatment served as controls.
Ctsx expression is significantly increased after H. pylori infection, whereas Ctsb showed no significant increase in WT and Ctsx−/− mice after infection (Figs. 1 and 2). The results obtained with co-cultured epithelial cells were similar. The macrophages in co-culture with Ctsx−/− epithelial cells induced Ctsx more strongly (2-fold) than macrophages co-cultured with WT epithelial cells (0.2-fold; data not shown).
We then focused on the analysis of the expression of cytokines in mono- and co-cultures after H. pylori infection. In co-cultured epithelial WT cells, CXCL1 was 35-fold less expressed than in mono-cultured epithelial WT cells (Fig. 3, A and B), with a 7.4-fold increase in co-culture and a 10.2-fold elevation in mono-cultured WT epithelial cells after H. pylori infection (Fig. 3, E and D). Ctsx−/− cells showed a comparable increase of CXCL1 after H. pylori infection (Fig. 3D), unless CXCL1 was less strongly expressed in Ctsx−/− mono-cultured epithelial cells than in WT mono-cultured epithelial cells (Fig. 3A). As in WT, the CXCL1 expression in co-cultured Ctsx−/− epithelial cells was further reduced (Fig. 3B), but the increase is similar after H. pylori infection, with a tendency to express CXCL1 in Ctsx−/− more strongly than WT cells (Fig. 3E).
FIGURE 3.
Induction of proinflammatory cytokines in mono-cultures and co-cultures after H. pylori induction. Primary gastric epithelial cells of WT and Ctsx−/− mice were isolated, mono- or co-cultured with J-774A.1, infected with H. pylori strain SS1 (see schematic illustration) and tested for expression of mouse cytokines CXCL1, IL-6, IL-1β, and MCP-1. infection. A–C, the basal cytokine expression was analyzed in uninfected WT, Ctsx−/−, and J-774A.1 cells using quantitative RT-PCR. D–F, the x-fold increased cytokine expression after H. pylori infection formed from a quotient of mRNA cytokine after H. pylori infection and without H. pylori infection. Bar graphs are presented as means ± S.E. The p values from t test analysis are shown on top of the bars. epith., epithelial.
Compared with WT, IL-6 expression is markedly reduced in Ctsx−/− mono-cultured (p ≤ 0.001) and co-cultured (p ≤ 0.06) epithelial cells (Fig. 3, A and B). After H. pylori infection, the expression of IL-6 was significantly more increased (p ≤ 0.044) only in Ctsx−/− co-cultured epithelial cells compared with WT (Fig. 3E).
IL-1β behaved like IL-6 in WT mono- and co-cultured epithelial cells (Fig. 3, A–E), with a 4.5-fold increase after H. pylori infection in mono-cultures (Fig. 3D). In Ctsx−/− mono-cultured epithelial cells, IL-1β was expressed only at the detection limit (Fig. 3A). The level of IL-1β was slightly increased in co-cultured Ctsx−/− cells but was nonetheless significantly reduced (p ≤ 0.0059) compared with WT co-cultures (Fig. 3B). After infection, there was no induction detectable as in WT cells (Fig. 3E).
MCP-1 is less expressed in Ctsx−/− mono- and co-cultured cells compared with the corresponding WT cells (Fig. 3, A and B). No significant differences in H. pylori-dependent induction of MCP-1 were found for Ctsx−/− and WT cells (Fig. 3, D and E).
The expression of MCP-1 is significantly weaker in macrophages (J774A.1) co-cultured with WT cells (p ≤ 0.048) compared with Ctsx−/− co-cultured macrophages (Fig. 3C); however, the induction of MCP-1 after H. pylori infection is comparable (Fig. 3F). J774A.1 generally expresses CXCL1 at the detection limit without progression after H. pylori infection of the epithelial cells (Fig. 3, C and F). IL-6 was even expressed at the detection limit (Fig. 3C) but was markedly increased by H. pylori infection in both WT and Ctsx−/− co-cultured cells with a significant higher induction (p ≤ 0.0218) in Ctsx−/− co-cultured J774A.1 (Fig. 3F). Cytokine IL-1β, which indicated no induction in co-cultured epithelial cells after H. pylori infection, improved a massive induction (12- to 25-fold) in macrophages co-cultured with WT and Ctsx−/− cells (Fig. 3F), with no significant difference in induction between the two co-cultures.
In summary, Ctsx−/− epithelial cells show weaker expression of CXCL1, IL-6, IL-1β, and MCP-1, but display a tendency for higher cytokine induction after H. pylori infection. Furthermore, macrophages in co-culture with Ctsx−/− epithelial cells express up to 18-fold more MCP-1 and show higher induction of all the other cytokines compared with J774A.1 in co-culture with WT cells. Fig. 3 shows all statistical data, presented in bar graphs for each cytokine.
Analysis of mRNA Expression in WT and Ctsx−/− Epithelial Cells for Genes Known to Be Involved in T-cell and B-cell Regulation after H. pylori Infection
To analyze changes in the gene expression of primary gastric epithelial cells in response to H. pylori SS1, we performed cDNA microarray analysis using the Oligo GEArray® Mouse T-cell and B-cell activation microarray. This array contains 113 genes involved in T-cell and B-cell activation, proliferation, and differentiation. We used mRNA from primary gastric WT and Ctsx−/− epithelial cells with and without H. pylori SS1 infection. All images were quantified, and the final results are expressed as -fold changes given as a relative gene expression (Table 2). The majority of genes showed no differences in expression between WT and Ctsx−/− epithelial cells by infection with H. pylori. But there were 13 differently expressed genes in Ctsx−/− epithelial cells, the expression of which was >1.5-fold up-regulated after H. pylori infection. Some of these genes, for example Shb, Ccnd3, Ap3b1, and Gadd45g, were not influenced in WT epithelial cells after H. pylori infection (indicated by a dash in Table 2). Interestingly, 9 of these 13 genes are involved in T-cell or B-cell proliferation, whereas only four genes were up-regulated in T-cell and B-cell differentiation. Results were confirmed by quantitative RT-PCR. Primer sequences are listed in Table 1. The most abundant gene found to be up-regulated 3.03-fold was Spp1 in Ctsx−/− epithelial cells, followed by CD81 (2.01-fold). Induction of Hmgb3, Inpp5d, and Ccnd3, as shown by microarray data, could not be validated using quantitative RT-PCR. Furthermore, Glmn was found to be the only gene more strongly up-regulated in WT cells. Nonetheless, the majority of genes were more strongly up-regulated in Ctsx−/− infected cells than in WT H. pylori-infected cells.
TABLE 2.
Expression profiles of genes known to be involved in T- and B-cell activation after H. pylori infection in WT and Ctsx−/− epithelial cells
For the complete gene list, see the Oligo GEArray® Mouse T- and B-cell activation microarray (SABiosciences Corp.). Total RNA from primary gastric epithelial cells (uninfected and infected with H. pylori) were treated under the same conditions. All images were quantified using the GeneGnome and GeneTools image scanning and analysis software (Syngene BioImaging Systems). Common gene names are indicated. Expression ratios (before/after H. pylori infection) were calculated per blot or quantitative RT-PCR, and final results are expressed as -fold changes given as a relative gene expression (according to the formula: ratio treatment/ratio control).
| Microarray |
Quantitative RT-PCR (± S.D.) |
|||
|---|---|---|---|---|
| WT | Ctsx−/− | WT | Ctsx−/− | |
| -fold | -fold | |||
| B-cell proliferation | ||||
| CD81 | 1.40 | 2.12 | 1.43 ± 0.4 | 2.01 ± 0.6 |
| Cdkn1a | 1.06 | 1.76 | 1.07 ± 0.4 | 1.60 ± 1.2 |
| Prkcd | 1.52 | 1.90 | 1.05 ± 1.0 | 1.20 ± 0.8 |
| Shb | – | 1.25 | – | – |
| B-cell differentiation | ||||
| Hmgb3 | 1.42 | 2.19 | – | – |
| Inpp5d | 1.61 | 1.70 | – | – |
| T-cell proliferation | ||||
| Ccnd3 | – | 1.90 | – | – |
| Glmn | 1.23 | 1.97 | 2.01 ± 1.2 | 1.70 ± 0.1 |
| IL-18 | 1.50 | 1.75 | 1.32 ± 0.1 | 1.59 ± 0.1 |
| IL-10 | 1.55 | 1.65 | – | – |
| Spp1 | 1.02 | 2.62 | 1.05 ± 0.9 | 3.03 ± 0.1 |
| T-cell differentiation | ||||
| Ap3b1 | – | 1.85 | 1.09 ± 0.1 | 1.52 ± 0.2 |
| Gadd45g | – | 1.55 | 0.51 ± 0.2 | 1.38 ± 0.1 |
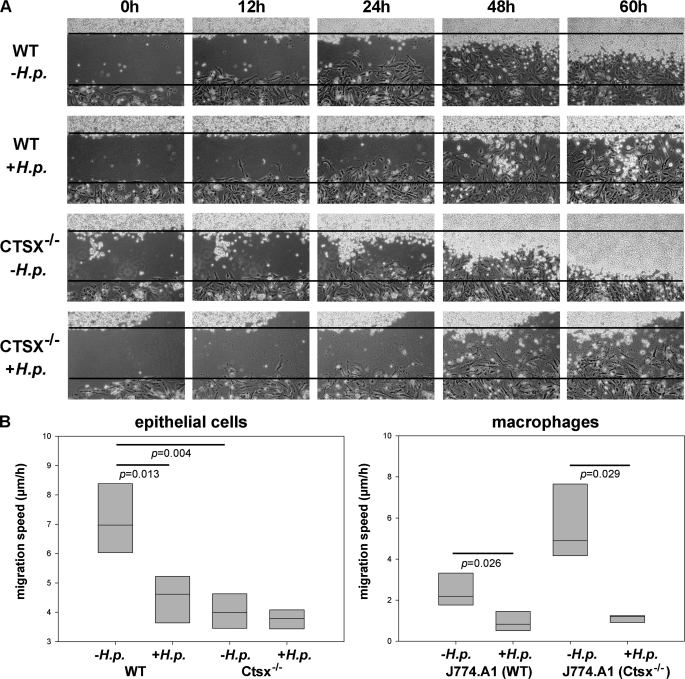
Morphological Changes and Cellular Interaction of Mouse Gastric Epithelial Cells and Mouse Macrophage Cells in Confrontation Culture
Mouse gastric epithelial cells (WT and Ctsx−/−) and mouse macrophages (J-774A.1) were seeded overnight in a culture insert. The epithelial cells (± H. pylori) were filled in the lower well, and the macrophages were filled in the upper well and incubated for 24 h. The culture insert was deleted, and the changes in cell morphology and their interaction were monitored for 60 h. Fig. 4A shows the interaction of H. pylori SS1-infected WT (second line) and Ctsx−/− (fourth line) epithelial cells with macrophages over a time period of 60 h. The time when the inserts were removed to create a wound was defined as point 0 h. In general, the Ctsx−/− epithelial cells grow and migrate significantly slower (p < 0.05) than the WT cells irrespective of the infection status. Furthermore, migration speed of infected cells and corresponding macrophages is significantly lower (p < 0.05) compared with uninfected controls (Fig. 4B). Within 24 to 48 h, the epithelial cells reached the borderline of the macrophages, whereas the macrophages did not migrate within the first 48 h. Interestingly, after this incubation, the control epithelial cells were rapidly overgrown from the macrophages. This observation was neither reflected in infected WT cells nor in Ctsx−/−-infected epithelial cells. Macrophages co-cultured with Ctsx−/− cells started earlier and migrate faster than in co-culture with WT-cells.
FIGURE 4.
Cellular interaction in confrontation cultures. A wound was set between uninfected (first and third lines) and H. pylori (H.p.)SS1-infected (second and fourth lines) WT and Ctsx−/− epithelial cells and mouse macrophage cells (J-774A.1). After 6 h of infection, the inserts were removed (defined as point 0 h). Time-dependent changes in cell morphology and their interaction were monitored by time-lapse video microscopy (A). After ∼48 h, the epithelial cells reached the borderline of macrophages. The Ctsx−/− epithelial cells grow and migrate significantly slower (p < 0.05) than the WT cells irrespective of the infection status. Furthermore, migration speed of infected cells and corresponding macrophages is significantly lower (p < 0.05) compared with uninfected controls (B). Until 60 h, the control epithelial cells were rapidly overgrown from the macrophages. This observation was reflected neither in infected WT cells nor in Ctsx−/−-infected epithelial cells. Bar graphs are presented as means ± S.E. The p values from t test analysis are shown on top of the bars.
Expression of MIF-1 in WT and Ctsx−/− Epithelial Cells Co-cultured with Macrophages
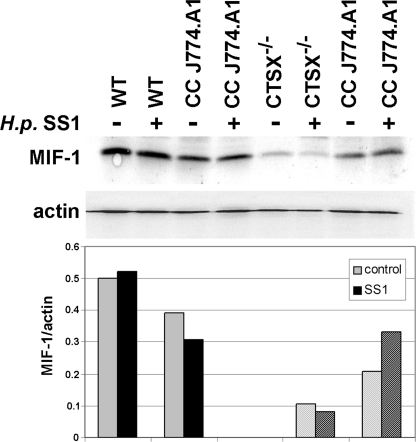
Wong et al. demonstrated that the macrophage migration inhibitory factor (MIF)2 is a critical mediator of both the innate and the Th1 immune response in gastritis induced by H. pylori infection in a transgenic mouse model (32). For this reason, wild-type and Ctsx−/− primary gastric epithelial cell lines were co-cultured with J-774A.1 cells infected with H. pylori SS1 and tested for the protein expression of mouse MIF-1 using Western blot analysis. MIF-1 expression was not influenced by H. pylori infection in WT and Ctsx−/− cells. However, MIF-1 was significantly reduced (five times) in Ctsx−/−cells compared with WT cells. The co-cultured macrophages showed no marked differences in MIF-1 expression after H. pylori infection (Fig. 5). These results could explain the tendency seen in confrontation cultures where macrophages migrate faster in juxtapositioning to Ctsx−/− cells.
FIGURE 5.
Expression of MIF-1 in WT and Ctsx−/− epithelial cells. Total protein was extracted from H. pylori SS1-infected and uninfected co-cultures of WT and Ctsx−/− cells and analyzed for MIF-1 expression by Western blotting. MIF-1 expression is five times weaker in Ctsx−/− cells as in WT cells independent of infection status. Even if the macrophages from Ctsx−/− co-cultures (CC) showed reduced MIF-1 expression in controls, no marked differences were seen after H. pylori SS1 infection.
DISCUSSION
H. pylori induces a local immune response in gastric mucosa that can be manifested in the form of a chronic active gastritis. CTSB/L/K and X are differentially expressed in normal and chronically inflamed gastric mucosa, but only CTSX shows a significant induction in H. pylori-infected gastric epithelial cells and macrophages (12–14). In the present study, we used primary mouse gastric epithelial cells in a comparison with murine gastric carcinoma cell line CLS 103 and a mouse macrophage cell line J-774A.1 to analyze the influence of Ctsx deficiency on H. pylori-induced expression of cytokines, chemokines, and other cathepsins. At first, we demonstrated the expression of different cathepsins (Ctsx/b/l) in the complete mouse gastric system and mouse gastric epithelial cells (WT, Ctsx−/−, and Ctsb−/−) to clarify the general expression of cathepsins. Compared with WT mice, all other cathepsins are significantly up-regulated in Ctsx−/− and Ctsb−/− knock-out mice, and their increased expression levels are similar in primary gastric epithelial cells and in gastric tissue samples. WT cells express low levels of Ctsx, whereas the expression in Ctsb−/− knock-out mice and cells is two times higher. Present data indicate a compensatory function of Ctsx for Ctsb. This was already shown in Ctsb-deficient mice, where an increase in extracellular Ctsx was detected (19), indicating that the cathepsin network counterbalanced deficits in one cathepsin by others.
In addition, we demonstrated that compared with extracted tissues isolated primary cell cultures are particularly qualified for analysis of differentially expressed cathepsins and the corresponding consequences. A further advantage of the primary cells is their better response to and the higher colonization density of H. pylori compared with commonly used cancer cells. Therefore, the following experiments were proceeded with primary gastric epithelial cells combined with macrophages to copy an in vivo-like situation to further investigate the effects on immune cells. The murine gastric carcinoma cell line CLS 103 has not been very well studied so far, but this cell line is established from the primary squamous cell carcinoma of NMRI mice. It has, therefore, impairments similar to the human cell lines AGS and NCI-N87, which we described as unsuitable for H. pylori infection studies (20).
In contrast to other cathepsins and their regulation, little is known about the specific physiological functions and the mechanisms by which H. pylori induces the expression of Ctsx in epithelial cells and especially macrophages. Until now, CTSX was reported to increase the adhesion and phagocytosis of macrophages, as well as maturation of dendritic cells. Furthermore, it regulates the proliferation and migration of T-lymphocytes (15). It is a fact that H. pylori activates signal transduction pathways and therewith the production of various cytokines by epithelial cells that attract for example macrophages (24). Macrophages were generally known as a key source of CTSX expression, and it is well established that cytokines, like IL-6 and IL-1β, are responsible for the increased expression of cathepsins (25). To analyze the expression of several cytokines (mCXCL1, mIL-6, mIL-1β, and mMCP-1), we compared infected and uninfected mono-cultured, as well as co-cultured gastric epithelial cells and mouse macrophage cell line J-774A.1. In our mono-cultured and co-cultured WT and Ctsx−/− cells, as well as in macrophages, all analyzed cytokines were increased to a greater or lesser extent after H. pylori infection. Whereas all cytokines (except of MCP-1) showed clearly reduced expression levels with undetectable mIL-1β in Ctsx−/− mono-cultures, important differences must be further examined in co-cultures. The most striking difference was observed for mIL-1β, which is decreased 5-fold in Ctsx-deficient cells compared with WT cells. This interaction of IL-1β and Ctsx needs to be further investigated and indicates a new field of action for this protease possibly in signal transduction events. Otherwise, CXCL1 and IL-6 were three and two times increased in co-cultured H. pylori-infected Ctsx−/− cells compared with WT. MCP-1 is well established as a monocyte chemoattractant, and it seems that knockdown of Ctsx in epithelial cells resulted also in MCP-1 stimulation in corresponding macrophages. There remains the question why especially co-cultured macrophages of Ctsx−/− cells showed a strengthened MCP-1 expression and induction. The use of J-774A.1 cells, which are isolated from ascites fluid of BALB/c mice exhibiting a reticulum cell sarcoma (ATCC, TIB-67TM) is questionable, as already described above for the epithelial cells. Continuing investigations will be conducted with primary macrophages from the spleen of our WT and Ctsx−/− mice. However, the results achieved so far suggest that Ctsx could influence the macrophage migration, indicating a well coordinated and distinct action of Ctsx in the inflammatory process.
In addition to macrophages, T- and B-lymphocytes were recruited in the wake of inflammatory response (26). Our gene expression microarray analysis of H. pylori-infected WT and Ctsx−/− epithelial cells revealed B- and T-cell specific gene expression signatures that clearly discriminated from uninfected cells. There were 13 different genes in Ctsx−/− epithelial cells and 9 different genes in WT epithelial cells up-regulated after H. pylori infection. Four genes were down-regulated (Shb, Ccnd3, Ap3b1, and Gadd45g) in WT epithelial cells after H. pylori infection. The most up-regulated genes in our analysis were Spp1 (3.03-fold), followed by CD81 (2.01-fold), which correlate well with the previous study conducted by Junnila et al. (27). SPP1 (also known as ETA-1 (early T-lymphocyte activation 1) or osteopontin) is primarily expressed during tissue renewal and tissue remodeling and is involved in stress response, cell adhesion, inflammation, wound healing, prevention of apoptosis, and immune response (28, 29). Also, CD81 is a widely expressed cell-surface protein and is reported to influence adhesion, morphology, activation, proliferation, and differentiation of B-, T-, and other cells (30). In summary, knockdown of Ctsx resulted in an increase of distinct B- and T-cell activators, indicating that Ctsx prevents lymphocyte differentiation and/or activation. These results are in line with studies analyzing the interaction of CTSX with β2 integrin receptors. CTSX activates the Mac-1 receptor, leading to suppression of lymphocyte proliferation and cluster formation. Otherwise, via activation of LFA, CTSX promotes the immune response (31). Together with these findings, our data support a function of CTSX in controlling the immune answer.
The confrontation cultures reflected our findings from expression studies and confirmed the data of Sevenich et al. (22). Knockdown of Ctsx resulted in a trend toward decreased epithelial motility and increased macrophage migration, but no convincing difference could be observed in an in vitro functional assay. This could be due to the compensation of the loss of Ctsx by Ctsb (19) and, obviously, to the self-regulating opposite functions of Ctsx. In vitro systems will not be able to clarify in vivo functions of CTSX in the gastric carcinogenesis to the full extent. Further studies of our group are being performed using Ctsx−/− transgenic mice in a gastritis model and their crossings with hypergastrinemic INS-GAS mice to create a corresponding gastric cancer model.
Acknowledgments
We greatly appreciate the gift of knock-out mice from Dr. Thomas Reinheckel (Institute of Molecular Medicine and Cell Research, Freiburg, Germany) and the SS1 H. pylori strain from Dr. S. Backert (University College Dublin, Dublin, Ireland). We thank Dr. Sabine Brandt, Dr. Roland Hartig, Doreen Medau, and Kirsten Herrmanns for technical assistance.
This work was supported by Grant KR 1901/4-1 from the Deutsche Forschungsgemeinschaft.
- MIF
- macrophage migration inhibitory factor.
REFERENCES
- 1.Blaser M. J., Atherton J. C. (2004) J. Clin. Investig. 113, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall B. J., Warren J. R. (1984) Lancet 1, 1311–1315 [DOI] [PubMed] [Google Scholar]
- 3.Montecucco C., Rappuoli R. (2001) Nat. Rev. Mol. Cell Biol. 2, 457–466 [DOI] [PubMed] [Google Scholar]
- 4.Graham D. Y., Adam E., Reddy G. T., Agarwal J. P., Agarwal R., Evans D. J., Jr., Malaty H. M., Evans D. G. (1991) Dig. Dis. Sci. 36, 1084–1088 [DOI] [PubMed] [Google Scholar]
- 5.Dooley C. P., Cohen H., Fitzgibbons P. L., Bauer M., Appleman M. D., Perez-Perez G. I., Blaser M. J. (1989) N. Engl. J. Med. 321, 1562–1566 [DOI] [PubMed] [Google Scholar]
- 6.Forman D., Newell D. G., Fullerton F., Yarnell J. W. G., Stacey A. R., Wald N., Sitas F. (1991) Br. Med. J. 302, 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wotherspoon A. C., Ortiz-Hidalgo C., Falzon M. R., Isaacson P. G. (1991) Lancet 338, 1175–1176 [DOI] [PubMed] [Google Scholar]
- 8.Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R. J. (2001) N. Engl. J. Med. 345, 784–789 [DOI] [PubMed] [Google Scholar]
- 9.Schmausser B., Mueller S. O., Eck M., Möller M., Müller-Hermelink H., Stopper H. (2000) Cancer Lett. 152, 145–149 [DOI] [PubMed] [Google Scholar]
- 10.Peek R. M., Jr., Wirth H. P., Moss S. F., Yang M., Abdalla A. M., Tham K. T., Zhang T., Tang L. H., Modlin I. M., Blaser M. J. (2000) Gastroenterology 118, 48–59 [DOI] [PubMed] [Google Scholar]
- 11.Xia H. H., Talley N. J. (2001) Am. J. Gastroenterol. 96, 16–26 [DOI] [PubMed] [Google Scholar]
- 12.Krueger S., Kuester D., Bernhardt A., Wex T., Roessner A. (2009) J. Pathol. 217, 581–588 [DOI] [PubMed] [Google Scholar]
- 13.Krueger S., Kalinski T., Hundertmark T., Wex T., Küster D., Peitz U., Ebert M., Nägler D. K., Kellner U., Malfertheiner P., Naumann M., Röcken C., Roessner A. (2005) J. Pathol. 207, 32–42 [DOI] [PubMed] [Google Scholar]
- 14.Bühling F., Peitz U., Krüger S., Küster D., Vieth M., Gebert I., Roessner A., Weber E., Malfertheiner P., Wex T. (2004) Biol. Chem. 385, 439–445 [DOI] [PubMed] [Google Scholar]
- 15.Obermajer N., Magister S., Kopitar A. N., Tepes B., Ihan A., Kos J. (2009) Eur. J. Cell Biol. 88, 461–471 [DOI] [PubMed] [Google Scholar]
- 16.Nascimento F. D., Rizzi C. C., Nantes I. L., Stefe I., Turk B., Carmona A. K., Nader H. B., Juliano L., Tersariol I. L. (2005) Arch. Biochem. Biophys. 436, 323–332 [DOI] [PubMed] [Google Scholar]
- 17.Lechner A. M., Assfalg-Machleidt I., Zahler S., Stoeckelhuber M., Machleidt W., Jochum M., Nägler D. K. (2006) J. Biol. Chem. 281, 39588–39597 [DOI] [PubMed] [Google Scholar]
- 18.Sivaraman J., Nägler D. K., Zhang R., Ménard R., Cygler M. (2000) J. Mol. Biol. 295, 939–951 [DOI] [PubMed] [Google Scholar]
- 19.Vasiljeva O., Papazoglou A., Krüger A., Brodoefel H., Korovin M., Deussing J., Augustin N., Nielsen B. S., Almholt K., Bogyo M., Peters C., Reinheckel T. (2006) Cancer Res. 66, 5242–5250 [DOI] [PubMed] [Google Scholar]
- 20.Krueger S., Hundertmark T., Kuester D., Kalinski T., Peitz U., Roessner A. (2007) Pathol Res. Pract. 203, 433–444 [DOI] [PubMed] [Google Scholar]
- 21.Marchetti M., Aricò B., Burroni D., Figura N., Rappuoli R., Ghiara P. (1995) Science 267, 1655–1658 [DOI] [PubMed] [Google Scholar]
- 22.Sevenich L., Schurigt U., Sachse K., Gajda M., Werner F., Müller S., Vasiljeva O., Schwinde A., Klemm N., Deussing J., Peters C., Reinheckel T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halangk W., Lerch M. M., Brandt-Nedelev B., Roth W., Ruthenbuerger M., Reinheckel T., Domschke W., Lippert H., Peters C., Deussing J. (2000) J. Clin. Invest. 106, 773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naumann M., Crabtree J. E. (2004) Trends Microbiol. 12, 29–36 [DOI] [PubMed] [Google Scholar]
- 25.Gerber A., Wille A., Welte T., Ansorge S., Bühling F. (2001) J. Interferon Cytokine Res. 21, 11–19 [DOI] [PubMed] [Google Scholar]
- 26.Goll R., Husebekk A., Isaksen V., Kauric G., Hansen T., Florholmen J. (2005) Scand. J. Immunol. 61, 92–97 [DOI] [PubMed] [Google Scholar]
- 27.Junnila S., Kokkola A., Mizuguchi T., Hirata K., Karjalainen-Lindsberg M. L., Puolakkainen P., Monni O. (2010) (2009)Genes Chromosomes Cancer 49, 28–39 [DOI] [PubMed] [Google Scholar]
- 28.El-Tanani M. K. (2008) Front Biosci. 13, 4276–4284 [DOI] [PubMed] [Google Scholar]
- 29.Wang K. X., Denhardt D. T. (2008) Cytokine Growth Factor Rev. 19, 333–345 [DOI] [PubMed] [Google Scholar]
- 30.Levy S., Todd S. C., Maecker H. T. (1998) Annu. Rev. Immunol. 16, 89–109 [DOI] [PubMed] [Google Scholar]
- 31.Obermajer N., Repnik U., Jevnikar Z., Turk B., Kreft M., Kos J. (2008) Immunology 124, 76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong B. L., Zhu S. L., Huang X. R., Ma J., Xia H. H., Bucala R., Wong B. C., Lan H. Y. (2009) Am. J. Pathol. 174, 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]