Abstract
We have previously shown that the hyperthermophilic archaeon, Sulfolobus solfataricus, catabolizes d-glucose and d-galactose to pyruvate and glyceraldehyde via a non-phosphorylative version of the Entner-Doudoroff pathway. At each step, one enzyme is active with both C6 epimers, leading to a metabolically promiscuous pathway. On further investigation, the catalytic promiscuity of the first enzyme in this pathway, glucose dehydrogenase, has been shown to extend to the C5 sugars, d-xylose and l-arabinose. In the current paper we establish that this promiscuity for C6 and C5 metabolites is also exhibited by the third enzyme in the pathway, 2-keto-3-deoxygluconate aldolase, but that the second step requires a specific C5-dehydratase, the gluconate dehydratase being active only with C6 metabolites. The products of this pathway for the catabolism of d-xylose and l-arabinose are pyruvate and glycolaldehyde, pyruvate entering the citric acid cycle after oxidative decarboxylation to acetyl-coenzyme A. We have identified and characterized the enzymes, both native and recombinant, that catalyze the conversion of glycolaldehyde to glycolate and then to glyoxylate, which can enter the citric acid cycle via the action of malate synthase. Evidence is also presented that similar enzymes for this pentose sugar pathway are present in Sulfolobus acidocaldarius, and metabolic tracer studies in this archaeon demonstrate its in vivo operation in parallel with a route involving no aldol cleavage of the 2-keto-3-deoxy-pentanoates but direct conversion to the citric acid cycle C5-metabolite, 2-oxoglutarate.
Keywords: Archaebacteria, Energy Metabolism, Enzyme Catalysis, Enzymes, Metabolism, Sulfolobus, Archaea, Extremophiles, Thermophiles
Introduction
Sulfolobus solfataricus and Sulfolobus acidocaldarius are hyperthermophilic archaea that grow optimally at 78–85 °C, pH 2–4, and are able to utilize a variety of carbon sources, including the four most-commonly occurring sugars in nature, d-glucose, d-galactose, d-xylose, and l-arabinose (1).
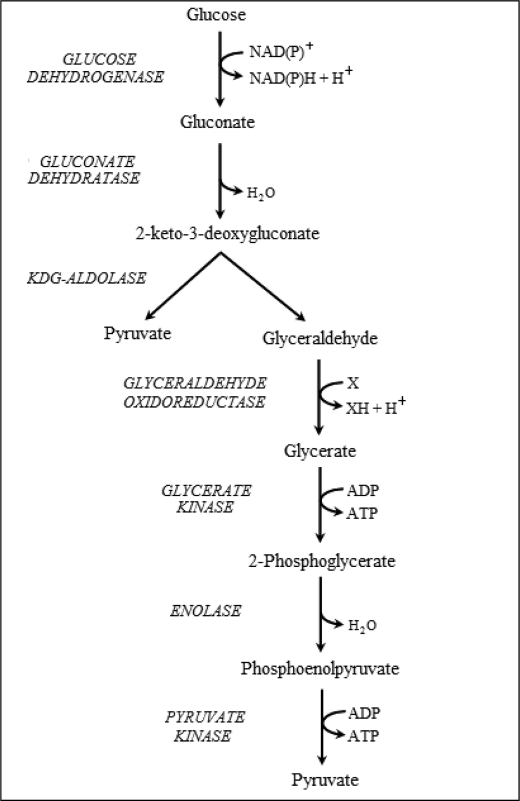
Metabolism of glucose in S. solfataricus and S. acidocaldarius proceeds via a non-phosphorylative variant of the Entner-Doudoroff pathway, which generates pyruvate with no net production of ATP (Fig. 1) (2–5). Glucose dehydrogenase catalyzes the conversion of glucose to gluconate, which is then dehydrated to 2-keto-3-deoxygluconate (KD-gluconate)4 by gluconate dehydratase. KD-gluconate in turn is cleaved to pyruvate and glyceraldehyde via 2-keto-3-deoxygluconate aldolase (KDG-aldolase), the glyceraldehyde generating a second molecule of pyruvate via the actions of glyceraldehyde oxidoreductase, glycerate kinase, enolase, and pyruvate kinase.
FIGURE 1.
The non-phosphorylative Entner-Doudoroff pathway for the catabolism of d-glucose and d-galactose in S. solfataricus.
In vitro kinetic analyses of glucose dehydrogenase, gluconate dehydratase, and KDG-aldolase from S. solfataricus showed these enzymes are also capable of catalyzing the catabolism of galactose, the C4 epimer of glucose, to pyruvate and glyceraldehyde, leading to the suggestion that the pathway exhibits a metabolic promiscuity toward these two hexose sugars (5–7). Because recombinantly produced glucose dehydrogenase has good activity with the pentose sugars d-xylose and l-arabinose (6) and KDG-aldolase catalyzes the condensation of the C-2 compound glycolaldehyde and pyruvate to form a 2-keto-3-deoxypentanoate,5 both enzymes are likely to have a role in the catabolism of the naturally occurring pentose sugars, d-xylose and l-arabinose, as well as of the hexose sugars d-glucose and d-galactose. However, the purified gluconate dehydratase was found to be specific for gluconate and galactonate (7).
A number of questions arise from these observations. Does Sulfolobus possess a dehydratase specific for d-xylonate and l-arabinonate, thereby establishing a pathway for C5-sugar catabolism that uses the same glucose dehydrogenase and KDG-aldolase involved in the utilization of C6 sugars? If so, what is the metabolic fate of glycolaldehyde? Finally, what is the relationship of this possible metabolic route to that proposed by Brouns et al. (8) for the catabolism of the non-natural isomer, d-arabinose, by S. solfataricus, whereby the 2-keto-3-deoxy-arabinonate is not aldol-cleaved but is converted to 2-oxoglutarate through the action of a KD-arabinonate dehydratase and 2,5-dioxopentanoate dehydrogenase?
In this paper we demonstrate that Sulfolobus possesses a C5-specific dehydratase that catalyzes the dehydration of d-xylonate and l-arabinonate to 2-keto-3-deoxy-d-xylonate (KD-xylonate) and 2-keto-3-deoxy-l-arabinonate (KD-arabinonate), respectively, and the enzymes glycolaldehyde oxidoreductase and glyoxylate reductase to catalyze the conversion of glycolaldehyde to glycolate and then to glyoxylate, which enters the citric acid cycle via the action of malate synthase. The genes for two of these three enzymes have been cloned and expressed, and the recombinant enzymes have been characterized. Furthermore, through in vivo metabolite tracer studies, we have shown that d-xylose is partitioned between this newly discovered pathway involving aldol cleavage and subsequent glyoxylate formation, and the pathway was identified for the metabolism of d-arabinose involving direct conversion to 2-oxoglutarate (8).
EXPERIMENTAL PROCEDURES
Growth of S. solfataricus and Preparation of Cell Extracts
Freeze-dried S. solfataricus P2 (DSM 1617) was obtained from DSMZ (Germany) and grown aerobically in Erlenmeyer flasks at 78 °C in basal salt medium (9) containing (per liter) 0.28 g of KH2PO4, 1.30 g of (NH4)2SO4, 0.25 g of MgSO4·7H20, 0.25 g of CaCl2·7H20 adjusted to pH 3.5. The medium also contained a 1% (v/v) trace element solution (per liter, 20.0 mg of FeCl3·6H20, 4.5 mg of Na2B4O7·10H20, 1.8 mg of MnCl2·4H20, 0.22 mg of ZnSO4·7H20, 0.05 mg of CuCl2·2H20, 0.03 mg of Na2MoO4·4H20, 0.03 mg of VOSO4·2H20, 0.01 mg of CoSO4·7H20), 0.05% (w/v) yeast extract, and 2% (w/v) d-glucose, d-xylose, or l-arabinose. Cells were harvested by centrifugation at 5000 × g for 10 min and resuspended at 0.2 g/ml in 50 mm Tris-HCl, pH 8.0, and incubated for 30 min. Cells were lysed by four 30-s bursts of sonication using a 150-watt Ultrasonic Disintegrator (MSE Scientific Instruments, Crawley, UK), and the soluble fraction of the cell extract was obtained by centrifugation at 12,500 × g for 30 min.
Growth of S. acidocaldarius and Preparation of Cell Extracts
S. acidocaldarius (DSM 639) was grown aerobically in Erlenmeyer flasks at 72 °C in the presence of 20 mm d-xylose. The medium contained (per liter) 0.25 g of MgSO4·7H2O, 1.7 g of K2SO4, 0.028 g of FeSO4·7H2O, 1 g of (NH4)2SO4, 0.28 g of KH2PO4, 0.07 g of CaCl2·2H2O and 1 ml of trace element solution (per liter: 15 g of MgSO4·7H2O, 1 g of MnSO4·H2O, 5 g of NaCl, 0.9 g of CoCl2·6 H2O, 0.05 g of H3BO3, 0.9 g of ZnSO4·7H2O, 0.05 g of Na2MoO4·2H2O, 0.13 g of NiCl2·6H2O, 0.0015 g of Na2SeO3·5H2O). The medium was adjusted to pH 2.5 with H2SO4. Cell extracts were prepared as described for S. solfataricus.
Enzyme Assays
Unless stated otherwise, the conditions stated below are those used for the assays of enzymes from S. solfataricus; minor variations were used for the assays at 50–60 °C of enzymes from S. acidocaldarius.
Xylose dehydrogenase activity was determined as previously described (6). Assays were performed at 70 °C in 1 ml of 100 mm HEPES, pH 8.0. The assay mixture contained 0.5 mm NADP+ and 5 mm d-xylose, and the reaction was started by the addition of enzyme. The production of NADPH was followed by the increase in absorbance at 340 nm (ϵ = 6220 m−1 cm−1). One unit of enzyme activity is defined as the amount of enzyme required to produce 1 μmol of NADPH per min.
Xylonate dehydratase activity was determined using the method of Lamble et al. (7). Assays were performed in 400 μl of 100 mm Bis-Tris, pH 7.0, containing 10 mm MgCl2 and 5 mm d-xylonate. The reaction was started by the addition of enzyme and was incubated for 10 min at 70 °C before being stopped by transferring 100 μl of the reaction mixture to 10 μl of 12% (w/v) trichloroacetic acid. After clarification by centrifugation at 16,000 × g for 5 min, the formation of KD-xylonate was quantified spectrophotometrically at 549 nm (ϵ = 67,800 m−1 cm−1) by the reaction with thiobarbituric acid (10).
2-Keto-3-deoxyxylonate aldolase activity was assayed in the direction of aldol synthesis using a thiobarbituric acid assay as described in Buchanan et al. (10). Assays were carried out at 70 °C in 50 mm sodium phosphate buffer, pH 6.0, with 50 mm pyruvate and 10 mm glycolaldehyde.
Two enzymatic activities were measured for the conversion of glycolaldehyde to glycolate. Aldehyde oxidoreductase was assayed at 70 °C in 1 ml of 100 mm Bis-Tris buffer, pH 6.8, 2 mm 2,6-dichlorophenolindophenol (DCPIP) and 5 mm glycolaldehyde; the assay was started by the addition of substrate, and the reduction of the DCPIP was followed at 595 nm (ϵ = 22,000 m−1 cm−1). Glycolaldehyde dehydrogenase activity was determined at 70 °C in 1 ml of 50 mm potassium phosphate buffer, pH 6.0. The assay mixture contained 1 mm NADP+ and 50 mm glycolaldehyde, and the reaction was started by the addition of enzyme. The reduction of NADP+ was followed by the increase in absorbance at 340 nm (ϵ = 6220 m−1 cm−1). One unit of enzyme activity is defined as the amount of enzyme required to produce 1 μmol of reduced DCPIP or NADPH per min.
Glycolate dehydrogenase (glyoxylate reductase) activity was determined as described in Yoshikawa et al. (11). Assays were performed at 70 °C in 1 ml of 100 mm HEPES, pH 7.5. The assay mixture contained 0.2 mm NADH and 10 mm glyoxylate, and the reaction was started by the addition of enzyme. The oxidation of NADH was followed by the decrease in absorbance at 340 nm. One unit of enzyme activity is defined as the amount of enzyme required to produce 1 μmol of NAD+ per min.
Malate synthase activity was determined as described in Uhrigshardt et al. (12). The assay was performed at 70 °C in 1 ml of 20 mm EPPS, pH 8.0, 100 mm KCl. The assay mixture contained 0.1 mm DTNB, 0.14 mm acetyl-coenzyme A (acetyl-CoA), and 10 mm glyoxylate, and the reaction was started by the addition of enzyme. The increase in absorbance at 412 nm (ϵ = 13,600 m−1 cm−1) was followed with time, one unit of enzyme activity being defined as the amount of enzyme required to produce 1 μmol of CoASH per min.
Isocitrate lyase activity was determined as described in Uhrigshardt et al. (12). The assay was performed at 70 °C in 1 ml of 100 mm Bis-Tris, pH 7.5, containing 100 mm NaCl and 20 mm MgCl2. The assay mixture contained 2.5 mm phenylhydrazine, 15 mm isocitrate, and the reaction was started by the addition of enzyme. The reaction was followed by the increase in absorbance at 324 nm (ϵ = 17,000 m−1 cm−1) corresponding to the production of 2-(phenylhydrazono)acetate. One unit of enzyme activity is defined as the amount of enzyme required to produce 1 μmol of glyoxylate per min.
2,5-Dioxopentanoate dehydrogenase (2-oxoglutarate semialdehyde dehydrogenase) assays were performed at 70 °C in 1 ml of 100 mm HEPES, pH 7.5, as described in Brouns et al. (8). The assay mixture contained 1 mm NADP+ and 10 mm pentanedial, and the reaction was started by the addition of enzyme. The reduction of NADP+ was followed by the increase in absorbance at 340 nm. One unit of enzyme activity is defined as the amount of enzyme required to produce 1μmol of NADPH per min.
Gene Cloning and Expression of Recombinant Enzymes
All genes in this study were PCR-amplified from S. solfataricus genomic DNA. PCR-amplification was carried out using Phusion followed by A-tailing with Taq polymerase. Amplified DNA fragments were purified by electrophoresis in a 1% (w/v) agarose gel, extracted using the Qiaex II Gel Extraction kit (Qiagen Ltd, Crawley, UK) and, unless otherwise stated, ligated into the pGEMT-Easy intermediate cloning vector. After sequence determination to ensure PCR fidelity, fragments were ligated into pET vectors as detailed below; unless otherwise stated, restriction sites were used that removed the His tag from the vector.
d-Xylonate Dehydratase
A putative second dehydratase gene (SS02665) was identified through a BLAST search of the S. solfataricus genome using the gluconate dehydratase gene sequence (SSO3198). The gene was cloned into pET3a and pET19b vectors (Novagen, Nottingham, UK) using the Nde1 and BamH1 sites to produce both untagged and His-tagged constructs, respectively. The plasmids were transformed into a series of host Escherichia coli strains including BL21(DE3), KRX, Rosetta, C41, and C43. Strains containing either vector were grown in LB or Overnight Express media (Novagen) containing appropriate antibiotics. LB cultures were grown with and without isopropyl 1-thio-β-d-galactopyranoside induction.
2,5-Dioxopentanoate (2-Oxoglutarate Semialdehyde) Dehydrogenase
The 2,5-dioxopentanoate dehydrogenase gene (SSO3117) annotated in the genome of S. solfataricus was cloned into pET28a (Novagen) using the Nde1 and NcoI sites. The gene was expressed in E. coli Rosetta cells grown at 37 °C in Overnight Express medium. A cell extract was prepared as described previously and then incubated at 70 °C for 10 min to precipitate E. coli proteins.
The gene was also cloned from S. acidocaldarius into pET17b using NdeI and BamHI sites. The gene was expressed in E. coli Rosetta-pLysS cells grown at 37 °C for 4 h. After preparation of a cell extract, the recombinant protein was purified by heat precipitation at 72 °C for 30 min followed by gel filtration chromatography on a Superdex 200 column (1 × 60 cm) in 100 mm Tris, pH 7.4, containing 150 mm NaCl.
Glyoxylate Reductase
A putative glyoxylate reductase gene (SSO3187) was identified in the genome of S. solfataricus using a BLAST search with the glyoxylate reductase sequence from Pyrococcus horikoshii (11). The gene was cloned into pET3a using the NcoI and XhoI sites. Expression of the recombinant enzyme was carried out in E. coli Rosetta cells grown at 37 °C overnight in Overnight Express medium. Cells were harvested by centrifugation at 5000 × g for 10 min and resuspended at 0.2 g/ml in 50 mm Tris-HCl, pH 8.0. After 30 min at room temperature, the cells were lysed by three 30-s bursts of sonication, and cell debris was removed by centrifugation at 12,500 × g for 30 min. The resulting cell extract was then incubated at 80 °C for 15 min to remove the majority of E. coli proteins before being subjected to gel filtration chromatography on a GE Healthcare Superdex 200 column (1 × 30 cm) using 50 mm Tris-HCl buffer, pH 8.5, containing 100 mm NaCl.
Malate Synthase
The putative malate synthase gene (SSO1334) annotated in the genome of S. solfataricus was cloned into pET19b using the NcoI and XhoI sites. The gene was expressed in E. coli Rosetta cells grown at 37 °C in Overnight Express medium to produce ∼50% soluble recombinant enzyme. Cells were harvested, and a cell extract was prepared and heat-treated at 80 °C for 10 min as described above. The recombinant malate synthase was then purified by anion exchange chromatography on a HiTrap Q-Sepharose column (GE Healthcare) using 50 mm Tris-HCl, pH 8.5, and an elution gradient of 0–1 m NaCl followed by gel filtration on a Superdex 200 column (1 × 30 cm) in 50 mm Tris-HCl, pH 8.5.
Isocitrate Lyase
The putative isocitrate lyase gene (SSO1333) annotated in the genome of S. solfataricus was cloned into pET19b using the NcoI and XhoI sites. The gene was expressed in RosettaTM grown at 37 °C overnight in Overnight Express medium both singularly and co-expressed with the malate synthase gene. A cell extract was prepared as described previously and was then incubated at 70 °C for 10 min.
Protein Concentration and Xylose Concentration
Protein concentrations were determined by the method of Bradford (13) using a calibration curve constructed with bovine serum albumin. Xylose concentrations in growth supernatants were determined according to Johnsen and Schönheit (14) using a calibration curve constructed with d-xylose.
13C-Labeling Experiments
S. acidocaldarius was grown aerobically at 72 °C with shaking at 150 rpm in 100-ml Erlenmeyer flasks containing 20 ml of medium, pH 2.5, containing 25 mm d-[1-13C]-xylose or d-[2-13C]-xylose. Cell aliquots were harvested during midexponential growth by centrifuging 2 ml of culture broth at 8000 × g for 10 min at 10 °C. The dried biomass pellet was hydrolyzed in 1.5 ml of 6 m HCl for 24 h at 110 °C in sealed 2-ml Eppendorf tubes and desiccated overnight in a heating block at 85 °C under a constant air stream. The hydrolysate was dissolved in 50 μl of 99.8% (v/v) dimethyl formamide and transferred to a new Eppendorf cup within a few seconds. For derivatization, 30 μl of N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide were added, which readily silylates hydroxyl groups, thiols, primary amines, amides, and carboxyl groups (15), and the mixture was shaken at 550 rpm and 85 °C for 60 min. 1 μl of the derivatized sample was then injected into a 6890N Network GC system, combined with a 5975 Inert XL Mass Selective Detector (Agilent Technologies, Böblingen, Germany), and analyzed as described in Fischer and Sauer (16) and Zamboni et al. (17). The GC temperature profile was 160 °C for 1 min, increased to 320 °C at 20 °C per min, and held at 320 °C for 1 min. The injector temperature was set to 230 °C, the split ratio to 1:10, and the flow rate to 1.5 ml/min; the carrier gas was helium in an HP-5MS column (30 m × 0.25 mm, 0.25 μm coated) (Agilent Technologies).
Mass spectra of the derivatized amino acids alanine, aspartate, glutamate, proline, serine, and threonine were corrected for the natural abundance of all stable isotopes and for unlabeled biomass from the inoculum. Glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, tyrosine, and valine were not used in this study, and arginine, asparagine, cysteine, glutamine, and tryptophan were not detectable.
RESULTS AND DISCUSSION
A Proposed Catabolic Pathway for d-Xylose and l-Arabinose
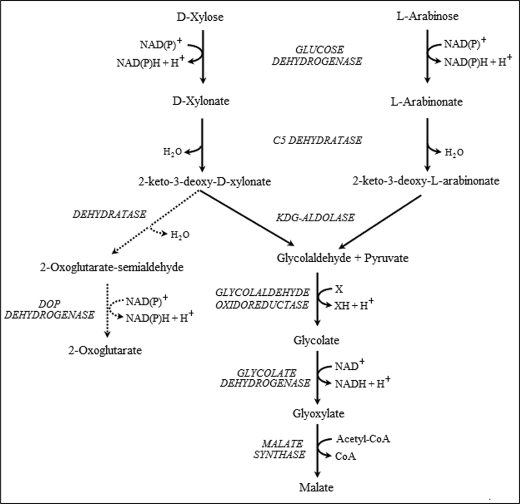
Our previous findings that S. solfataricus glucose dehydrogenase can catalyze the oxidation of both d-xylose and l-arabinose (6) and that the KDG-aldolase can catalyze the aldol cleavage of 2-keto-3-deoxypentanoate (until now, assayed only in the condensation direction) led us to consider what might be the catabolic fate of these two C5 sugars. Based on known enzymatic activities in other organisms, a proposed scheme is shown in Fig. 2. This scheme predicts that Sulfolobus will possess a C5-specific dehydratase, the enzymes glycolaldehyde oxidoreductase or glycolaldehyde dehydrogenase, glycolate dehydrogenase (glyoxylate reductase), and malate synthase, and that the aldolase will catalyze the cleavage of both KD-xylonate and KD-arabinonate. The malate formed would then enter the citric acid cycle for complete oxidation to CO2 and reducing equivalents. Also shown in Fig. 2 is a possible parallel pathway whereby KD-xylonate and KD-arabinonate are converted to 2-oxoglutarate. This pathway is identical to that proposed by Brouns et al. (8) for the catabolism of d-arabinose in S. solfataricus, the 2-keto-3-deoxy-d-arabinonate of that pathway being identical to 2-keto-3-deoxy-d-xylonate produced in the above scheme.
FIGURE 2.
The proposed non-phosphorylative Entner-Doudoroff pathway for the catabolism of d-xylose and l-arabinose in S. solfataricus. The scheme proposed in this paper is shown in solid arrows. The xylose dehydrogenase is the same protein as that labeled glucose dehydrogenase in Fig. 1, the one enzyme being catalytically active with d-glucose, d-galactose, d-xylose, and l-arabinose. The KDG-aldolase is proposed to catalyze the aldol cleavage of KD-gluconate and KD-galactonate (see Fig. 1) and of KD-xylonate and KD-arabinonate. The direct conversion of KD-xylonate to 2-oxoglutarate, as proposed by Brouns et al. (8), is shown with dotted arrows. DOP dehydrogenase, 2,5-dioxopentanoate dehydrogenase.
As detailed below, we present several lines of evidence for the pathways shown in Fig. 2. First, with respect to the newly proposed pathway for the metabolism of C5-sugars via glycolaldehyde, the enzyme activities have been assayed and found in cell extracts of S. solfataricus and S. acidocaldarius grown on either C6 or C5 sugars as a sole carbon source, and nearly all the respective S. solfataricus genes have been cloned and expressed and the recombinant enzymes characterized. Second, with respect to the in vivo partitioning of metabolites between the two pathways, we report [13C]metabolic labeling studies.
Enzyme Assays of Cell Extracts
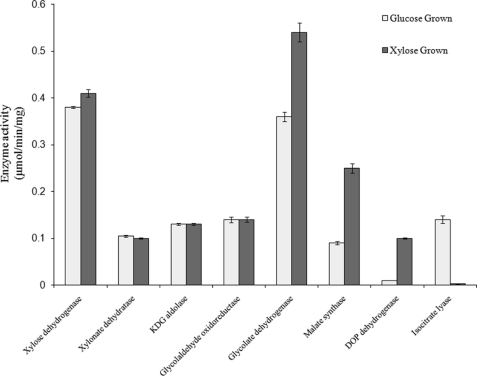
The first critical test for the proposed pathway of C5-sugar catabolism in S. solfataricus is to assay for the native enzyme activities in cell extracts of this organism. S. solfataricus was grown on either d-glucose or d-xylose as the sole carbon source, and cell extracts were prepared as described under “Experimental Procedures.” Enzyme activities were measured in these unfractionated extracts and are shown in Fig. 3. It should be noted that these activities are those measured in the forward direction as defined by the scheme shown in Fig. 2, except for KDG-aldolase and glycolate dehydrogenase. In the case of glycolate dehydrogenase (also known as glyoxylate reductase), the equilibrium is far toward glycolate, and therefore the enzyme was assayed using glyoxylate and NAD+ as substrates.
FIGURE 3.
Enzyme activities in cell extracts of S. solfataricus. Cell extracts were prepared from S. solfataricus grown on d-glucose and on d-xylose. The catalytic activities in these two extracts of the enzymes in Fig. 2 were assayed as described under “Experimental Procedures.” Glycolaldehyde oxidoreductase was assayed with DCPIP as electron acceptor. DOP dehydrogenase is 2,5-dioxopentanoate dehydrogenase and was assayed with pentanedial as substrate.
Activities of xylose dehydrogenase and KDG-aldolase did not differ between glucose- and xylose-grown cells. This is not unexpected in that glucose dehydrogenase (encoded by gene SSO3003) has comparable activity with d-glucose, d-galactose, d-xylose, and l-arabinose (6). Furthermore, activities of glucose dehydrogenase and xylose dehydrogenase were found not to be additive in cell extracts of either S. solfataricus or S. acidocaldarius (data not shown), which provides in vivo evidence that indeed both sugars are oxidized by the same dehydrogenase, i.e. glucose/xylose dehydrogenase. The KDG-aldolase (SSO3197) is also active with the 2-keto-3-deoxy-derivatives of these four sugars (see below). Glycolaldehyde oxidation to glycolate was found with DCPIP or NADP+ as electron acceptors. The activity with DCPIP is likely to be the glyceraldehyde oxidoreductase reported by Kardinahl et al. (18) in S. acidocaldarius, whereas that with NADP+ was shown to be an activity of the 2,5-dioxopentanoate dehydrogenase identified by Brouns et al. (8) as part of the d-arabinose degradation pathway (see “Recombinant Enzyme Characterization” below). The oxidoreductase activity in both S. solfataricus and S. acidocaldarius is 7–10-fold higher than that of the dehydrogenase, and so would be the more significant enzyme candidate for the oxidation of glycolaldehyde in the proposed pathway.
It was not surprising to find that the activities of glyoxylate reductase and malate synthase are higher in cells grown on xylose than on glucose, as these enzymes are proposed to be involved in C5 sugar catabolism, but not C6. On the other hand, the levels of glycolaldehyde oxidoreductase would not be expected to change between growth on glucose and xylose if this enzyme activity is indeed from the glyceraldehyde oxidoreductase that functions in C6 sugar metabolism. However, unexpectedly, the levels of xylonate dehydratase were similar in glucose- and xylose-grown S. solfataricus even though this enzyme has no detectable activity with gluconate or galactonate.
Malate synthase and isocitrate lyase are usually coincidentally produced in aerobic microorganisms growing on acetate as the sole carbon source, the glyoxylate cycle being required for the biosynthesis of C4 compounds from the C2 nutrient (19). Significantly, in Sulfolobus growing on xylose, malate synthase is produced in the absence of an isocitrate lyase, consistent with the metabolic scheme proposed (Fig. 2), although the latter enzyme was induced in cells grown on glucose. As pointed out by Uhrigshardt et al. (12), significant levels of acetate have been found in S. acidocaldarius growing on glucose, and this may explain the induction of both glyoxylate cycle enzymes in the presence of this C6 substrate. All the enzyme activities for the pathway are also found in cell extracts of S. acidocaldarius grown on xylose (Table 1), and the same potential catabolic routes of C5-sugars are, therefore, likely for this species.
TABLE 1.
Catalytic activities and kinetic parameters of enzymes involved in C5 sugar metabolism
Enzyme activities in cell extracts are those measured under Vmax conditions. S.E on all kinetic parameters are <10% of the mean values. ND, no data.
| Enzyme | Substrate |
S. solfataricus |
S. acidocaldarius native enzyme (cell extract from xylose-grown cells) activity | |||
|---|---|---|---|---|---|---|
| Native enzyme (cell extract from xylose-grown cells) |
Recombinant enzyme |
|||||
| Km | Enzyme activity | Km | Vmax | |||
| mm | units/mg protein | mm | units/mg protein | units/mg protein | ||
| Xylose dehydrogenase | d-Xylose | 0.18 | 0.41 | 0.18 | 71 | 0.25 |
| l-Arabinose | 0.47 | 0.16 | 0.50 | 62 | ||
| Xylonate dehydratase | d-Xylonate | 0.28a | 0.08 | ND | ND | 0.024 |
| l-Arabinonate | 0.17a | 0.05 | ND | ND | ||
| KDG-aldolase | Pyruvate | 2.7a | 0.13 | 2.8a | 31a | 0.07 |
| Glycolaldehyde | 2.0a | 2.9a | ||||
| Aldehyde oxidoreductase (DCPIP) | Glycolaldehyde | 0.37 | 0.14 | ND | ND | 0.034 |
| Glyoxylate reductase | Glyoxylate | 5.0 | 0.54 | 5.0 | 150 | 0.12 |
| NADH+ | 0.1 | 0.1 | ||||
| Malate synthase | Glyoxylate | 0.05 | 0.25 | 0.06 | 14 | 0.072 |
| Acetyl-CoA | 0.002 | 0.002 | ||||
| Isocitrate lyase | Isocitrate | b | b | 0.96 | 8 | ND |
| 2,5-Dioxopentanoate dehydrogenase (NADP+) | Pentanedial | ND | 0.1 | 3.3 | 35 | 0.15c |
a These are apparent Km and Vmax values due to enzyme inhibition in the presence of high substrate concentration.
b Enzyme absent in cell extracts of xylose-grown cells.
c The recombinant enzyme of S. acidocaldarius was characterized as a homotetramer with subunit Mr values = 52,000. When assayed at 70 °C and pH 7.0, Vmax = 50 units/mg and Km = 0.14 mm (pentanedial) and 0.04 mm (NADP+). The specific activity with 2,5, dioxopentanoate (12 mm) was 1.5 units/mg. Enzyme activity was 15-fold higher in cells grown on xylose as compared to glucose.
Characterization of Xylonate-arabinonate Dehydratase
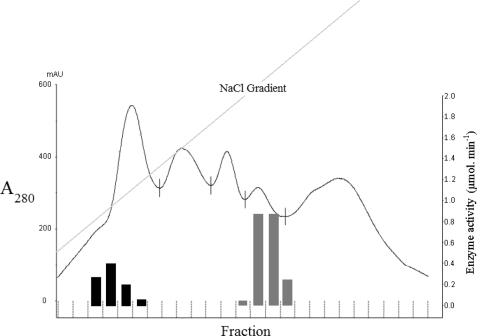
Given the promiscuous nature of glucose dehydrogenase and KDG-aldolase for both C6- and C5-metabolites, it was important to determine whether there was also a single dehydratase or two separate enzymes catalyzing the dehydration of gluconate/galactonate and xylonate/arabinonate. When cell extracts of S. solfataricus were subjected to either gel filtration or ion-exchange chromatography, the xylonate dehydratase was clearly separated from the gluconate dehydratase (Fig. 4). The former enzyme was also active with l-arabinonate and the latter with d-galactonate (7), suggesting that the two enzymes are promiscuous for C5 and C6 sugar acids, respectively. As shown in Fig. 4, the two enzymes are produced in approximately equal catalytic amounts.
FIGURE 4.
Anion exchange chromatography of a cell extract from S. solfataricus. A cell extract was prepared from S. solfataricus grown on d-xylose and subjected to anion exchange chromatography on a HiTrap Q-Sepharose column using 50 mm Tris-HCl, pH 8, and an elution gradient of 0–0.7 M NaCl. The elution profile (A280 nm) between 0.13 and 0.35 m NaCl is shown. All fractions were assayed for gluconate dehydratase (gray columns) and xylonate dehydratase (black columns), and the total activity (μmol product per min) in each fraction is displayed. mAU, milliabsorbance units.
Using the semipurified xylonate dehydratase after ion-exchange chromatography, the dependence of measured velocity on substrate concentration showed that the enzyme exhibited what appeared to be pronounced substrate inhibition with both d-xylonate and l-arabinonate. However, the data do not give a good fit to the classical substrate inhibition equation, indicating that other factors may influence the dependence of enzyme velocity on substrate concentration. Interestingly, the purified d-arabinonate dehydratase identified in S. solfataricus is also subject to what appears to be substrate inhibition (8).
Gel filtration chromatography showed that the gluconate dehydratase appeared to be active as a monomer of Mr ≈ 48,000. On the other hand, xylonate dehydratase activity eluted with an Mr ≈ 165,000, indicating that the active enzyme is a homotetramer.
Recombinant Enzyme Characterization
To confirm the activities and substrate specificities of the enzymes in the proposed pathway for C5 sugar catabolism (Fig. 2) and to determine their kinetic parameters, the genes encoding the enzymes have been cloned from S. solfataricus and expressed in E. coli. In each case, heat treatment of the cell extract gave recombinant enzyme that was of >90% purity, although in some cases a chromatography step was then carried out to achieve a homogeneous preparation. The data are recorded in Table 1 and are compared with those found with enzyme assays from cell extracts.
Xylose Dehydrogenase
The recombinant glucose dehydrogenase from S. solfataricus has been previously reported from our group and shown to have comparable activity with d-glucose, d-galactose, d-xylose, and l-arabinose (6). In the context of the pathway for C5 sugar catabolism, the kinetic parameters with d-xylose and l-arabinose were determined (Table 1).
Xylonate Dehydratase
As described under “Experimental Procedures,” a putative second dehydratase gene (SS02665) was identified in the genome sequence, with its translated protein product having 62% amino acid sequence identity to the gluconate dehydratase. The gene was cloned, but attempts to generate a soluble and active recombinant enzyme using a variety of E. coli, baculovirus, and fungal expression systems have been unsuccessful.
KDG-aldolase
Recombinant KDG-aldolase from S. solfataricus has been previously characterized in our laboratory with respect to its aldol-cleavage activity with KD-gluconate and KD-galactonate (6) and with its ability to catalyze the aldol condensation of pyruvate and glyceraldehyde. KD-xylonate and KD-arabinonate are not commercially available, and therefore the kinetics of only the aldol condensation activity of this enzyme with pyruvate and the C2 compound glycolaldehyde have been investigated (Table 1). The Vmax of 31 μmol min−1 mg of protein−1 is 2-fold greater than that with glyceraldehyde. NMR analysis of the product generated confirmed it to be a 2-keto-3-deoxy-pentanoate; however, it clearly could not reveal the relative amounts of the two possible enantiomers.
To demonstrate the aldol cleavage activity of KDG-aldolase with both KD-d-xylonate and KD-l-arabinonate, coupled assays were carried out. d-Xylonate and l-arabinonate were incubated separately with the semipurified xylonate dehydratase described above, and the production of KD-xylonate and KD-arabinonate was measured spectrophotometrically by the reaction with thiobarbituric acid (10). Incubation of the products from the two dehydratase reactions with pure recombinant KDG-aldolase verified that the latter enzyme catalyzed the aldol cleavage of both enantiomers to pyruvate and glycolaldehyde. The data are, therefore, consistent with the one aldolase enzyme being involved in the catabolism of both the C6 and C5 sugars (Fig. 2).
Glycolaldehyde Oxidoreductase and Glycolaldehyde Dehydrogenase
Glycolaldehyde DCPIP oxidoreductase most likely reflects a side activity of the glyceraldehyde DCPIP oxidoreductase reported by Kardinahl et al. (18). Because the enzyme is extremely oxygen-sensitive and found to contain a molybdenum cofactor, no attempts were made to clone and express heterologously the genes (SS02636, SS02637, and SS02639) encoding the three enzyme components.
Brouns et al. (8) reported that S. solfataricus gene SSO3117 encodes an aldehyde dehydrogenase that, when recombinantly expressed, possessed catalytic activity with 2,5-dioxopentanoate, glyceraldehyde, and glycolaldehyde. We have confirmed this observation, found that the enzyme is specific for NADP+, and report the kinetic parameters for the substrates pentanedial (as an analog of 2,5-dioxopentanoate) and glycolaldehyde (Table 1).
Glycolate Dehydrogenase (Glyoxylate Reductase)
The gene annotated as glyoxylate reductase (SS03187) in the S. solfataricus genome was cloned and expressed in E. coli to give a soluble, active recombinant protein that was purified by anion exchange chromatography and gel filtration. The enzyme was found to be NAD-dependent, and the kinetic parameters were determined (Table 1).
Malate Synthase and Isocitrate Lyase
Purified malate synthase and semipurified isocitrate lyase have been characterized previously in S. acidocaldarius, and evidence for a functional glyoxylate cycle in this organism has been presented (12). In the current paper the S. solfataricus genes annotated as malate synthase (SS01334) and isocitrate lyase (SS01333) were cloned and expressed in E. coli, and the proteins were purified as described under “Experimental Procedures.” The kinetic parameters (Table 1) were similar to those reported for the corresponding S. acidocaldarius enzymes as were the dependence of isocitrate lyase activity on the presence of Mg2+ and the oligomeric nature of the two enzymes as judged by SDS-PAGE and gel filtration (malate synthase: homodimer of Mr ≈ 189,000; isocitrate lyase: homotetramer of Mr ≈ 190,000).
In both S. acidocaldarius and S. solfataricus, the malate synthase and isocitrate lyase genes are found 484 and 461 base pairs apart, respectively, and TATA boxes (20) are found upstream of both genes, indicating that they do not exist in an operon. Furthermore, no Shine-Dalgarno sequence could be identified upstream of either gene; such sequences are not found upstream of isolated genes in S. solfataricus (21), with most transcripts completely lacking 5′-untranslated regions (22). As stated above, isocitrate lyase activity is found in cells grown on glucose (12), which may be linked with the finding in S. acidocaldarius of acetate production under these growth conditions.
At this point, the data presented from assays in cell extracts and with the purified recombinant enzymes are consistent with the metabolism of d-xylose and l-arabinose via the two possible pathways outlined in Fig. 2. It was, therefore, important to investigate their operation in vivo and the extent to which metabolites are partitioned between them. This was performed using metabolic labeling studies with 13C sugars.
Metabolic Labeling Studies
Fig. 2 shows the two proposed pathways for the catabolism of d-xylose and l-arabinose. It is important to note that Brouns et al. (8) studied the catabolism of d-arabinose in S. solfataricus and found that the resulting 2-keto-3-deoxy-d-arabinonate is converted to 2-oxoglutarate through the action of KD-arabinonate dehydratase and 2,5-dioxopentanoate dehydrogenase. However, 2-keto-3-deoxy-d-arabinonate is identical to 2-keto-3-deoxy-d-xylonate and, therefore, can also undergo aldol cleavage to pyruvate and glycolaldehyde as we have proposed for the catabolism of d-xylose and l-arabinose. Of the two Sulfolobus species considered here, the ability of S. acidocaldarius to grow on minimal media in the absence of yeast extract (whereas S. solfataricus cannot) makes it ideal for metabolic labeling studies; that is, S. acidocaldarius will grow on 13C-labeled sugars as a sole carbon source, thereby permitting the flow of C5 sugars via these two possible pathways to be determined.
To determine the relative in vivo activities of xylose degradation pathways in S. acidocaldarius, we performed labeling experiments using d-[1-13C]- and d-[2-13C]xylose. Label in the proteinogenic amino acids was determined by GC-MS, which gives direct information about the labeling patterns of the precursors such as 3-phosphoglycerate, pyruvate, oxaloacetate or 2-oxoglutarate. The absence of the pentose phosphate pathway was confirmed by the absence of labeling in serine (Table 2), as catabolism of labeled xylose via xylulose-5P and 3-phosphoglycerate would lead to label in this amino acid.
TABLE 2.
Mass isotopomer distribution in alanine, serine, glutamate, proline, aspartate, and threonine in S. acidocaldarius
The data are from 100% [1-13C] and [2-13C]xylose experiments.
| Fractional label | m0a | m1 | m2 | m3 | m4 | m5 |
|---|---|---|---|---|---|---|
| 100% [1-13C]Xylose | ||||||
| Serine (M-57)+b | 0.962c | 0.032 | 0.001 | 0.002 | ||
| Serine (M-85)+ | 0.996 | 0.006 | 0.000 | |||
| Alanine (M-57)+ | 0.475 | 0.512 | 0.011 | 0.000 | ||
| Alanine (M-85)+ | 0.984 | 0.017 | 0.000 | |||
| Glutamate (M-57)+ | 0.457 | 0.512 | 0.023 | 0.002 | 0.000 | 0.000 |
| Glutamate (M-85)+ | 0.966 | 0.034 | 0.000 | 0.000 | 0.000 | |
| Proline (M-57)+ | 0.425 | 0.550 | 0.020 | 0.000 | 0.000 | 0.000 |
| Proline (M-85)+ | 0.961 | 0.035 | 0.000 | 0.000 | 0.000 | |
| Aspartate (M-57)+ | 0.961 | 0.032 | 0.002 | 0.001 | 0.000 | |
| Aspartate (M-85)+ | 0.988 | 0.011 | 0.000 | 0.002 | ||
| Threonine (M-57)+ | 0.959 | 0.039 | 0.000 | 0.001 | 0.000 | |
| Threonine (M-85)+ | 0.986 | 0.017 | 0.000 | 0.001 | ||
| 100% [2-13C]Xylose | ||||||
| Alanine (M-57)+ | 0.309 | 0.659 | 0.026 | 0.004 | ||
| Alanine (M-85)+ | 0.456 | 0.536 | 0.006 | |||
| Glutamate (M-57)+ | 0.140 | 0.711 | 0.140 | 0.004 | 0.000 | 0.000 |
| Glutamate (M-85)+ | 0.209 | 0.776 | 0.012 | 0.000 | 0.000 | |
| Proline (M-57)+ | 0.120 | 0.755 | 0.118 | 0.002 | 0.000 | 0.001 |
| Proline (M-85)+ | 0.216 | 0.754 | 0.027 | 0.000 | 0.000 | |
| Aspartate (M-57)+ | 0.227 | 0.735 | 0.032 | 0.002 | 0.000 | |
| Aspartate (M-85)+ | 0.609 | 0.383 | 0.004 | 0.001 | ||
| Threonine (M-57)+ | 0.223 | 0.756 | 0.015 | 0.001 | 0.001 | |
| Threonine (M-85)+ | 0.608 | 0.385 | 0.004 | 0.000 | ||
am0 is the fractional abundance of the fragments with the lowest mass, and mi>0 is the abundance of molecules with higher masses.
b Cracking of the derivatized amino acids leads to the following fragments: (M-57)+ with loss of a tert-butyl group; (M-85)+, with loss of the CO of the amino acid (C-1 position) and a tert-butyl group. Thus, (M-57)+ and (M-85)+ correspond to the 1–3, 1–4 or 1–5, and the 2–3, 2–4 or 2–5 fragments, respectively.
c Abundances are from triplicate experiments and generally with <5% deviation.
The first of the two alternative pathways postulated for xylose catabolism (via KDG-aldolase) would lead to incorporation of C1 to C3 of xylose into pyruvate, the direct precursor of alanine. The second pathway via 2,5-dioxopentanoate leaves the carbon backbone unchanged. Our data from tracer experiments using [1-13C]xylose show that the first pathway contributes to about half of the pyruvate formation, yielding 50% unlabeled alanine and 50% alanine labeled at position one (Table 2). We also observed labeled glutamate and proline, but this cannot be explained by anaplerotic reactions from pyruvate to oxaloacetate and then to glutamate because the amino acids threonine and aspartate, derived from oxaloacetate, were completely unlabeled (Table 2). The activity of pyruvate ferredoxin oxidoreductase can be excluded because the labeled C1 of pyruvate would be lost as CO2. Therefore, we conclude that the second pathway degrading xylose via 2,5-dioxopentanoate directly to glutamate takes place simultaneously with the first pathway via KDG-aldolase and accounts for at least 50% of the glutamate. Because of the loss of label in both pyruvate and 2-oxoglutarate ferredoxin oxidoreductase reactions, we cannot exclude reformation of 2-oxoglutarate through the citric acid cycle or the so-called pyruvate shunt, and therefore the 50% labels are a lower limit.
The glyoxylate formed in the degradation of xylose to pyruvate via the KDG-aldolase route is proposed to end up in malate. The C1 label of pyruvate is lost as CO2 in the formation of acetyl-CoA required to condense with glyoxylate, and therefore, the malate formed through this pathway is unlabeled and may account for the unlabeled alanine fraction obtained by gluconeogenic reactions from malate/oxaloacetate to pyruvate (Table 2). In addition, unlabeled malate can also originate from glutamate since the C1 label of glutamate is lost as CO2 in the 2-oxoglutarate oxidoreductase reaction.
In addition to the main conclusion from the [1-13C]xylose tracer studies, that both pathways account for about half of the formation of pyruvate or 2-oxoglutarate respectively, we tried to differentiate between formation of unlabeled malate from glyoxylate or from glutamate. To obtain additional positional labeling information within the citric acid cycle intermediates, we conducted tracer experiments using [2-13C]xylose. Glyoxylate condenses with acetyl-CoA to form malate, which now carries the label at position 2. If malate is formed from glutamate labeled at position 2, the label ends up in position 1 or 4 of malate and leads to alanine labeled at position 1, as observed to be about 10% (Table 2). The threonine and aspartate are labeled at both positions 1 and 4; however, within the resolution of this analysis, we can only estimate the possible label at position 2, which would give a 15–30% range for the formation of malate from glyoxylate (Table 2).
Conclusion
The in vivo labeling studies with S. acidocaldarius indicate that d-xylose (and probably, therefore, l- arabinose) is degraded simultaneously via two routes, each at about 50%. One route involves aldol cleavage to pyruvate and glycolaldehyde, which is further converted to glyoxylate, glycolate, and finally malate. In the second route, d-xylose is converted to 2-oxoglutarate, the KD-xylonate being dehydrated to 2-oxoglutarate semialdehyde, which is subsequently oxidized to 2-oxoglutarate. The enzymes of the first route, which constitutes a novel pathway of pentose degradation, have been described and characterized from S. solfataricus and S. acidocaldarius in the current paper.
It has also been proposed that the Entner-Doudoroff pathway in S. solfataricus is branched (3) whereby KD-gluconate is phosphorylated to 2-keto-3-deoxy-6-phosphogluconate via a KDG-kinase; KDG-aldolase then catalyzes the cleavage to pyruvate and glyceraldehyde-3P, generating a pathway that operates in parallel with the non-phosphorylative route. Through kinetic and structural studies of KDG-kinase and KDG-aldolase, we have shown that this part-phosphorylative pathway in S. solfataricus is also promiscuous for the metabolism of both glucose and galactose (23, 24); however, we currently have no evidence for a similar semi-phosphorylative pathway being involved in the catabolism of the C5 sugars.
The existence of a promiscuous central metabolic pathway in S. solfataricus and the constitutive production of the component enzymes may indicate a primitive evolutionary feature in this hyperthermophilic archaeon, or it may be an adaptation to survival in its extreme environment, allowing it to scavenge efficiently for energy substrates comprising both hexose and pentose sugars. However, when both nutrients are present, it may be that hexoses are preferentially used, as glucose in the growth medium of S. solfataricus has been found to repress the expression of the arabinose transport genes (25).
The catabolism of C5 sugars in Sulfolobus contrasts with sugar metabolism in other Archaea. In Haloferax volcanii, for example, d-xylose is metabolized via the pathway leading directly to 2-oxoglutarate, not utilizing the branch involving aldol cleavage of 2-keto-3-deoxy-d-xylonate and subsequent conversion to malate (26). Furthermore, the xylose dehydrogenase of H. volcanii is highly specific; d-glucose was used at a 130-fold lower catalytic efficiency than d-xylose, and no significant activity was measured with l-arabinose. Although we have previously provided structural explanations for the catalytic promiscuity exhibited by the glucose dehydrogenase (27) and KDG-aldolase (28) from S. solfataricus, the metabolic significance of partitioning between the two possible catabolic routes for C5 sugars and its regulation remain to be explored.
H. J. Lamble, D. W. Hough, S. D. Bull, and M. J. Danson, unpublished data.
- KD-
- 2-keto-3-deoxy-
- KDG-aldolase
- 2-keto-3-deoxygluconate aldolase
- DCPIP
- 2,6-dichlorophenolindophenol
- EPPS
- 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Grogan D. W. (1989) J. Bacteriol. 171, 6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Rosa M., Gambacorta A., Nicolaus B., Giardina P., Poerio E., Buonocore V. (1984) Biochem. J. 224, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed H., Ettema T. J., Tjaden B., Geerling A. C., van der Oost J., Siebers B. (2005) Biochem. J. 390, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siebers B., Schönheit P. (2005) Curr. Opin. Microbiol. 8, 695–705 [DOI] [PubMed] [Google Scholar]
- 5.Danson M. J., Lamble H. J., Hough D. W. (2007) Archaea: Molecular and Cellular Biology (Cavicchioli R. ed) pp. 260–287, American Society for Microbiology, Washington, DC [Google Scholar]
- 6.Lamble H. J., Heyer N. I., Bull S. D., Hough D. W., Danson M. J. (2003) J. Biol. Chem. 278, 34066–34072 [DOI] [PubMed] [Google Scholar]
- 7.Lamble H. J., Milburn C. C., Taylor G. L., Hough D. W., Danson M. J. (2004) FEBS Lett. 576, 133–136 [DOI] [PubMed] [Google Scholar]
- 8.Brouns S. J., Walther J., Snijders A. P., van de Werken H. J., Willemen H. L., Worm P., de Vos M. G., Andersson A., Lundgren M., Mazon H. F., van den Heuvel R. H., Nilsson P., Salmon L., de Vos W. M., Wright P. C., Bernander R., van der Oost J. (2006) J. Biol. Chem. 281, 27378–27388 [DOI] [PubMed] [Google Scholar]
- 9.Brock T. D., Brock K. M., Belly R. T., Weiss R. L. (1972) Arch. Microbiol. 84, 54–68 [DOI] [PubMed] [Google Scholar]
- 10.Buchanan C. L., Connaris H., Danson M. J., Reeve C. D., Hough D. W. (1999) Biochem. J. 343, 563–570 [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshikawa S., Arai R., Kinoshita Y., Uchikubo-Kamo T., Wakamatsu T., Akasaka R., Masui R., Terada T., Kuramitsu S., Shirouzu M., Yokoyama S. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 357–365 [DOI] [PubMed] [Google Scholar]
- 12.Uhrigshardt H., Walden M., John H., Petersen A., Anemüller S. (2002) FEBS Lett. 513, 223–229 [DOI] [PubMed] [Google Scholar]
- 13.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 14.Johnsen U., Schönheit P. (2004) J. Bacteriol. 186, 6198–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dauner M., Sauer U. (2000) Biotechnol. Prog. 16, 642–649 [DOI] [PubMed] [Google Scholar]
- 16.Fischer E., Sauer U. (2003) Eur. J. Biochem. 270, 880–891 [DOI] [PubMed] [Google Scholar]
- 17.Zamboni N., Fendt S. M., Rühl M., Sauer U. (2009) Nat. Protoc. 4, 878–892 [DOI] [PubMed] [Google Scholar]
- 18.Kardinahl S., Schmidt C. L., Hansen T., Anemüller S., Petersen A., Schäfer G. (1999) Eur. J. Biochem. 260, 540–548 [DOI] [PubMed] [Google Scholar]
- 19.Cozzone A. J. (1998) Annu. Rev. Microbiol. 52, 127–164 [DOI] [PubMed] [Google Scholar]
- 20.Reiter W. D., Hüdepohl U., Zillig W. (1990) Proc. Natl. Acad. Sci. 87, 9509–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tolstrup N., Sensen C. W., Garrett R. A., Clausen I. G. (2000) Extremophiles 4, 175–179 [DOI] [PubMed] [Google Scholar]
- 22.Wurtzel O., Sapra R., Chen F., Zhu Y., Simmons B. A., Sorek R. (2010) Genome Res. 20, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamble H. J., Theodossis A., Milburn C. C., Taylor G. L., Bull S. D., Hough D. W., Danson M. J. (2005) FEBS Lett. 579, 6865–6869 [DOI] [PubMed] [Google Scholar]
- 24.Potter J. A., Kerou M., Lamble H. J., Bull S. D., Hough D. W., Danson M. J., Taylor G. L. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1283–1287 [DOI] [PubMed] [Google Scholar]
- 25.Lubelska J. M., Jonuscheit M., Schleper C., Albers S. V., Driessen A. J. (2006) Extremophiles 10, 383–391 [DOI] [PubMed] [Google Scholar]
- 26.Johnsen U., Dambeck M., Zaiss H., Fuhrer T., Soppa J., Sauer U., Schönheit P. (2009) J. Biol. Chem. 284, 27290–27303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milburn C. C., Lamble H. J., Theodossis A., Bull S. D., Hough D. W., Danson M. J., Taylor G. L. (2006) J. Biol. Chem. 281, 14796–14804 [DOI] [PubMed] [Google Scholar]
- 28.Theodossis A., Walden H., Westwick E. J., Connaris H., Lamble H. J., Hough D. W., Danson M. J., Taylor G. L. (2004) J. Biol. Chem. 279, 43886–43892 [DOI] [PubMed] [Google Scholar]