Abstract
SalM is a short-chain dehydrogenase/reductase enzyme from the marine actinomycete Salinispora tropica that is involved in the biosynthesis of chloroethylmalonyl-CoA, a novel halogenated polyketide synthase extender unit of the proteasome inhibitor salinosporamide A. SalM was heterologously overexpressed in Escherichia coli and characterized in vitro for its substrate specificity, kinetics, and reaction profile. A sensitive real-time 13C NMR assay was developed to visualize the oxidation of 5-chloro-5-deoxy-d-ribose to 5-chloro-5-deoxy-d-ribono-γ-lactone in an NAD+-dependent reaction, followed by spontaneous lactone hydrolysis to 5-chloro-5-deoxy-d-ribonate. Although short-chain dehydrogenase/reductase enzymes are widely regarded as metal-independent, a strong divalent metal cation dependence for Mg2+, Ca2+, or Mn2+ was observed with SalM. Oxidative activity was also measured with the alternative substrates d-erythrose and d-ribose, making SalM the first reported stereospecific non-phosphorylative ribose 1-dehydrogenase.
Keywords: Bacteria, Carbohydrate Biosynthesis, Dehydrogenase, Enzyme Catalysis, NAD, NMR
Introduction
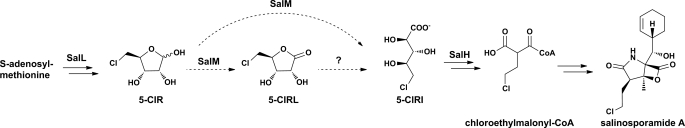
The marine actinomycete Salinispora tropica produces a suite of γ-lactam/β-lactone natural products identified as potent 20 S proteasome inhibitors (1). Exploration into the biosynthesis of the most bioactive family member, salinosporamide A, resulted in the characterization of a pathway for the biosynthesis of chloroethylmalonyl-CoA, a novel polyketide synthase substrate (Fig. 1) (2). Previous gene replacement of salM, which encodes a short-chain dehydrogenase/reductase (SDR)2 enzyme, dramatically and selectively reduced the production of salinosporamide A by ∼98% relative to the wild-type organism, whereas production of the non-chlorinated salinosporamide B remained unchanged. As salinosporamide B is alternatively produced from ethylmalonyl-CoA, the specific role of SalM in the biosynthesis of chloroethylmalonyl-CoA was established (2). Furthermore, the in vivo substrate of SalM was identified in this mutant strain as 5-chloro-5-deoxy-d-ribose (5-ClR) by detection of the accumulated fermentation product (2). Chemical complementation with 5-ClR to a separate upstream mutation in the chloroethylmalonyl-CoA pathway via the chlorinase SalL restored salinosporamide A production (2). Thus, on the basis of our understanding of the chloroethylmalonyl-CoA pathway, we predicted that SalM would oxidize 5-ClR at the anomeric carbon by acting as a pentose 1-dehydrogenase.
FIGURE 1.
Partial biosynthetic pathway to salinosporamide A in S. tropica and postulated enzymatic role(s) of the SDR SalM in the oxidation of 5-ClR. See Ref. 2 for the complete metabolic pathway.
It is intuitive to presume that SalM evolved from a primary metabolic ribose 1-dehydrogenase to oxidize a halogenated sugar derivative. However, despite the ubiquitous nature of ribose in biology, non-phosphorylative ribose 1-dehydrogenases (EC 1.1.1.115) have not been well characterized. Instead, pentose catabolism utilizes phosphorylated intermediates in the pentose phosphate pathway, nucleotide metabolism, and pentose-glucuronate conversion. Phosphorylated pentoses are also used in anabolic pathways such as the Calvin-Benson cycle and in the generation of nucleosides. The only previously reported “ribose 1-dehydrogenase” was isolated from pig liver and oxidized both d-ribose and d-xylose with approximately equal activity (3). Oxidative enzyme activity for ribose has been reported as an alternative substrate for other sugar oxidoreductase enzymes with broad substrate specificity (4–8); however, a non-phosphorylative pentose 1-dehydrogenase specific to the stereochemistry of ribose has yet to be reported.
Potentially related pentose 1-dehydrogenases such as l-arabinose 1-dehydrogenase and d-xylose 1-dehydrogenase have been shown to oxidize a cyclical hemiacetal substrate to the corresponding lactone (5, 6, 9, 10). Glucose 1-dehydrogenase has also been reported to possess “gluconolactonase” activity, catalyzing both the oxidation of glucose to gluconolactone and the subsequent hydrolysis to gluconate (6). The next anticipated enzyme in the chloroethylmalonyl-CoA biosynthetic pathway, SalH, is a dihydroxy-acid dehydratase and expected to accept 5-chloro-5-deoxyribonate as its substrate. Because the salinosporamide biosynthetic gene cluster (sal) does not encode a putative lactonase enzyme (2), we were compelled to determine whether a lactone intermediate exists and, if so, to decipher the fate of this pathway product. We thus set out to explore whether SalM produces a lactone or an acid or possesses bifunctional dehydrogenase/lactonase activity.
Traditional analysis of oxidoreductase enzymes such as SalM utilizes changes in optical absorption corresponding to the conversion of a cofactor such as NAD(P)(H) or FAD(H). Although this method provides a simple non-invasive way to monitor redox kinetics, it fails to identify the structure of the enzymatic product. Subsequent cofactor-independent reactions such as hydrolysis are thus not observed. Therefore, real-time visualization of product structures is imperative when transient intermediates are formed. A sensitive time-arrayed NMR approach was consequently developed to monitor the progress of the SalM reaction and to identify structures of intermediates and products. Here, we report a real-time 13C NMR-based characterization of SalM, a novel 5-chloro-5-deoxy-d-ribose 1-dehydrogenase.
EXPERIMENTAL PROCEDURES
Chemicals
All purchased chemicals were reagent-grade from Sigma-Aldrich unless noted otherwise. Isopropyl β-d-1-thiogalactopyranoside was obtained from Denville Scientific, d-erythrose from Alfa Aesar as a 70% (w/v) syrup, [U-13C]ribose (98% 13C) from Cambridge Isotope Laboratories, and nickel-nitrilotriacetic acid (Ni-NTA) from Qiagen. The putative SalM substrate and products 5-ClR (11), 5-chloro-5-deoxy-d-ribono-γ-lactone (5-ClRL) (12), and 5-chloro-5-deoxy-d-ribonate (5-ClRI) (2) were all synthesized according to literature procedures.
Expression and Purification of Recombinant SalM
Genomic DNA was obtained from cultures of S. tropica CNB-440 as described previously and used as a template for PCR (13). The 768-bp salM gene (Stro_1027) was PCR-amplified from genomic DNA using Pfu polymerase (Stratagene, La Jolla, CA) with forward primer 5′-CGTGGTTCCCATGGCATGACGAACGGTGGGCGCC-3′ and reverse primer 5′-GCTCGAATTCAAGCTTTCACTGCGCGAGGTAACCTC-3′. The PCR product was digested with NcoI and HindIII (the introduced restriction sites are underlined) and ligated into NcoI/HindIII-digested pHIS8 (14), and its sequence was verified (SeqXcel, San Diego, CA). Plasmid preparation and isolation were performed in Escherichia coli DH5α as described previously (13). N-terminally His8-tagged SalM was overexpressed in E. coli BL21(DE3). A 10-ml starter culture was grown overnight from a single colony in Terrific broth with 50 μg/ml kanamycin sulfate at 37 °C with shaking and then used to inoculate 1 liter of Terrific broth medium at 28 °C with 50 μg/ml kanamycin sulfate. Growth was monitored to an absorbance of 0.47, and then 0.2 mm isopropyl β-d-1-thiogalactopyranoside was added to induce protein expression. The culture was grown overnight at 28 °C with shaking.
All protein purification steps took place at 4 °C. Protein purification buffers contained 300 mm NaCl, 50 mm sodium phosphate adjusted to pH 8.0, and increasing concentrations of imidazole. Buffers A (lysis), B (wash), and C (elution) contained 10, 20, and 250 mm imidazole, respectively. Cells were pelleted at 6300 × g for 45 min, resuspended in buffer A, and lysed with six 30-s bursts of probe sonication with resting periods of 30 s. The lysate was centrifuged for 30 min at 10,000 × g. Soluble protein was collected and purified on an Ni-NTA column by washing with several volumes of buffer B and eluting with 2.5 ml of buffer C. Eluant was desalted using a PD-10 desalting column (GE Healthcare) and resuspended in 50 mm sodium phosphate buffer adjusted to pH 8.0. Desalted protein was concentrated on a Vivaspin 6 10-kDa membrane centrifuge concentrator (Sartorius Stedim Biotech S.A., Aubagne Cedex, France) and then subjected to size exclusion chromatography on a Superdex 200 column (GE Healthcare) with 100 mm Tris-HCl adjusted to pH 8.0, 500 mm NaCl, and 2 mm dithiothreitol.
Construction of C-terminal Mutants
SalM C-terminal mutants were PCR-amplified from genomic DNA with forward primer 5′-GCATACCATAGAATTCATGACGAACGGTGGGCGCCTAT-3′ and the following reverse primer for the specified mutant: Q255E, 5′-GCTCGAATTCAAGCTTTCACTCCGCGAGGTAACCTC-3′; Q255N, 5′-ATTGAGAGCTGCGGCCGCTCAGTTCGCGAGGTAACCTCCGTCGA-3′; Q255S, 5′-ATTGAGAGCTGCGGCCGCTCAGCTCGCGAGGTAACCTCCGTCGA-3′; Q255V, 5′-ATTGAGAGCTGCGGCCGCTCACACCGCGAGGTAACCTCCGTCGA-3′; extension 256N, 5′-ATTGAGAGCTGCGGCCGCTCAGTTCTGCGCGAGGTAACCTCCGTCGA-3′; Q255V/extension 256Q, 5′-ATTGAGAGCTGCGGCCGCTCACTGCACCGCGAGGTAACCTCCGTCGA-3′; A254-Q255 deletion, 5′-ATTGAGAGCTGCGGCCGCTCAGAGGTAACCTCCGTCGA-3′; and Q255 deletion, 5′-ATTGAGAGCTGCGGCCGCTCACGCGAGGTAACCTCCGTCGA-3′.
With the exception of the Q255E mutant, PCR products were digested with EcoRI and NotI (the introduced restriction sites are underlined) and ligated into EcoRI/NotI-digested pHIS8, and the sequence was verified. The Q255E mutant was constructed as described above with HindIII in place of NotI. Proteins were expressed via the autoinduction expression system Overnight Express I (EMD Chemicals, Gibbstown, NJ) in 1 liter of Luria broth in 50 μg/ml kanamycin sulfate at 28 °C for 24 h. A wild-type SalM control was concurrently expressed under the same conditions to verify expression and solubility. Protein purification was carried out in a manner analogous to that described for wild-type SalM.
Enzyme Assays
In vitro enzyme assays were performed in a Greiner 96-well half-area microtiter plate. Conversion of NAD+ to NADH was monitored at a wavelength of 340 nm using a SpectraMax M2 spectrometer (Molecular Devices, Sunnyvale, CA). All microplate assays were performed at 30 °C in a 50-μl volume with 100 mm Tris-HCl (pH 7.5) unless noted otherwise.
Divalent Cation Analysis
To identify suitable metal cofactors, SalM was assayed for activity with 0.5 mm 5-ClR, 0.5 mm NAD+, and 2.4 μg (0.048 mg/ml) SalM. 2 mm FeSO4, NiSO4, ZnSO4, CuCl2, CaCl2 MnCl2, MgSO4, or MgCl2 or no divalent cation was added.
As MgCl2, CaCl2, and MnCl2 were identified as accelerating the SalM-catalyzed reaction, an activity versus concentration assay was performed with the cation concentration varying between 1 and 50 mm. 1 mm 5-ClR, 1 mm NAD+, and 2.4 μg (0.048 mg/ml) SalM were used. These assays were performed in triplicate and averaged. The maximum velocity under steady-state conditions for each concentration was fitted with a linear line using SigmaPlot 11.0 (Systat Software, Inc., Chicago, IL).
Comparative Substrate Assay
10 sugars were assayed for activity with SalM: d-ribose, 2-deoxy-d-ribose, d-ribose 5-phosphate, 5-ClR, d-erythrose, d-allose, d-glucose, d-xylose, d-arabinose, and l-arabinose. A final concentration of 2 mm carbohydrate was used for all substrates with excess NAD+ cofactor at 2.5 mm. SalM (1.6 μg, 0.032 mg/ml) was added to each 50-μl reaction buffered with 100 mm Tris-HCl (pH 8.0) containing 2 mm MgCl2. After enzyme addition, absorbance measurements were recorded every minute for 3 h.
Kinetics Assays
Kinetic data were determined for d-ribose, d-erythrose, and 5-ClR. All reactions contained 2.4 μg SalM (0.048 mg/ml), presoaked in 10 mm MgCl2, and 4 mm NAD+ cofactor. Substrate concentrations versus initial velocities were plotted on SigmaPlot 11.0 and fit with a nonlinear Michaelis-Menten curve. Concentrations of d-ribose ranged from 0.5 to 200 mm, representing a range of 0.3–10.8 Km, whereas concentrations of d-erythrose ranged from 0.10 to 40 mm, representing a range of 0.4–16 Km. However, the Km for 5-ClR was at the lower limit of detection for NADH absorbance. Therefore, kinetic assays with 5-ClR were repeated on a 100-μl scale to increase the absorbance path length. The concentration of SalM was reduced to 1.2 μg/reaction (0.012 mg/ml). Concentrations tested for 5-ClR ranged from 10 μm to 10 mm, representing a range of 0.6–600 Km.
Lactone Opening Assay
A colorimetric assay for the detection of functionalized carboxylic acids was used to monitor the hydrolysis of 5-ClRL to 5-ClRI. 10 mm synthetically prepared 5-ClRL was dissolved in 100 mm Tris-HCl (pH 7.5), and 3 ml was aliquoted into two identical tubes. Active or denatured (boiled for 10 min) SalM (0.008 mg/ml), both presoaked with 10 mm MgCl2, was added to the 5-ClRL solution. Two 200-μl aliquots were removed from each tube at regular intervals and subjected to derivatization and colorimetric analysis as described previously (15). The experiment was repeated with 0.5 mm NAD+ and 0.5 mm NADH present in the buffer. Absorbance measurements were converted to 5-ClRL concentration by reference to a standard curve generated at the time of the assay. To determine the hydrolysis rate constants, a linear line was fit to the plot of the natural logarithm of lactone concentration versus time using SigmaPlot 11.0.
NMR-based Assays
Carbon-detected NMR experiments were measured on a Varian VX500 spectrometer equipped with an XSensTM Cold Probe. All assays were performed with the sample chamber set at a constant temperature of 30 °C. Carbon-free 60 mm sodium phosphate buffer at pH 7.5 was used instead of Tris-HCl. The final reaction volume was 250 μl in a 3-mm diameter NMR tube.
Uniformly 13C-Labeled Ribose
A 3 mm solution of [U-13C]ribose, 3.5 mm NAD+, and 2 mm MgCl2 in 200 μl of 62.5 mm sodium phosphate buffer (pH 7.5) with ∼30% deuterium oxide was placed in the NMR tube. To the reaction was added 35 μg of SalM (in a 50-μl volume of 50 mm sodium phosphate buffer at pH 7.5) for a final enzyme concentration of 0.140 mg/ml. One-dimensional 13C NMR spectra were measured using 256 scans with a 1-s T1 relaxation time. A spectrum was taken before enzyme addition, and then following enzyme addition, spectra were recorded every 10–15 min for 4 h. Additional spectra were taken 21 and 72 h after enzyme addition during which the sample was exposed to ambient temperature.
Unlabeled 5-ClR Distortionless Enhancement of Polarization Transfer (DEPT) NMR Assay
4 mm unlabeled 5-ClR, 3.5 mm NAD+, and 2 mm MgCl2 were dissolved into 200 μl of 62.5 mm sodium phosphate buffer (pH 7.5). To the reaction was added 24 μg of SalM (in a 50-μl volume of 50 mm sodium phosphate buffer at pH 7.5) for a final enzyme concentration of 0.120 mg/ml. The final deuterium oxide concentration was ∼50%. A 2048-scan DEPT135 spectrum with a T1 of 1 s was recorded before enzyme addition and then repeatedly following enzyme addition for the first four spectra. Each acquisition required ∼68 min. A final spectrum was started 8 h after enzyme addition.
RESULTS
Bioinformatics Analysis
Amino acid sequence similarity to SalM was used to identify potential enzymatic homologs. BLAST analysis of the 255-amino acid sequence of SalM indicated a classical SDR enzyme (16). On the basis of previously reported phylogenetic analyses of the SDR superfamily, it was expected that SalM should perform a simple ketone/alcohol redox reaction and would not participate in any additional chemistry such as epimerization, decarboxylation, or dehydration (16, 17). The highest scoring sequence was an uncharacterized 67% identical protein (accession number YP_638874) from several terrestrial Mycobacterium species (strains KMS, MCS, and JCS). SalM shows 40% sequence similarity to annotated glucose 1-dehydrogenases from Listeria grayi DSM 20601 and Brevibacillus brevis NBRC 100599 (accession numbers ZP_04443055 and YP_002770578, respectively).
Enzyme Purification and Cofactor Identification
Recombinant SalM was expressed in E. coli BL21(DE3) for in vitro characterization. The N-terminal His8-tagged enzyme was purified by Ni-NTA affinity chromatography and afforded ∼20 mg/liter recombinant protein at >90% purity. All enzyme activity assays utilized the tagged protein without further purification because SalM was prone to aggregation and eluted over a very broad range of sizes via size exclusion chromatography.
To assay SalM, we first identified the appropriate redox cofactor. The SDR family of enzymes contain a Rossmann fold for the binding of dinucleotide cofactors (17), with many SDR enzymes having a preference for either the phosphorylated or non-phosphorylated cofactor (16). Although no activity was observed with NADP+, the addition of NAD+ as cofactor resulted in its conversion to NADH as monitored spectrophotometrically. The preference for NAD+ is further supported by the bioinformatics analysis of the primary sequence of the cofactor-binding region (16). Alignment of SalM with 3α,20β-hydroxysteroid dehydrogenase (Protein Data Bank code 2hsd) revealed a conserved aspartic acid residue at position 40 equivalent to Asp36 of 2hsd. This would place SalM into the cD1d subfamily of SDRs, in which NAD+ is the expected enzyme cofactor. Asp40 of SalM likely forms hydrogen bonds with the 2′- and 3′-hydroxyls of the adenine ribose moiety (18).
Our initial attempts to assay SalM with 5-ClR and NAD+ resulted in minimum activity. While optimizing assay conditions, we observed that the addition of the divalent metal cation magnesium, manganese, or calcium increased its activity severalfold at low millimolar concentrations.
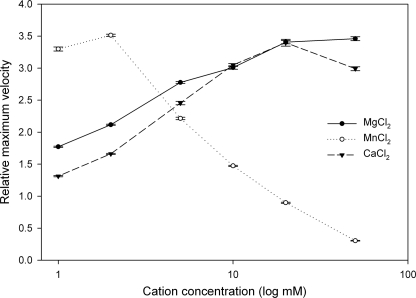
Increasing concentrations of Mg2+ and Ca2+ were shown to have a positive relationship with activity (Fig. 2). Maximum activity was reached by 20 mm. Additional cation failed to increase activity or was inhibitory. At all concentrations, enzyme activity was slow to reach the linear kinetic phase. However, presoaking concentrated enzyme stock solutions with 10 mm MgCl2 for several days at −20 °C and then adding enzyme to a Mg2+-free buffer at the time of the assay resulted in equivalent activity. Additionally, this method allowed steady-state kinetics to be reached much sooner compared with adding divalent cation at the start of the assay, suggesting that the metal ion is a slow binding structural component that reaches saturation.
FIGURE 2.
Metal dependence of SalM activity. The SalM enzyme was assayed for activity with 1 mm NAD+ and 1 mm 5-ClR in the presence of various concentrations of MgCl2, MnCl2, or CaCl2 between 1 and 50 mm. The maximum velocity of the reaction at steady state was normalized to the maximum velocity of the SalM reaction without the addition of metal (activity = 1).
When using the cation-presoaked enzyme, subsequent addition of metal ions to the assay buffer was found to inhibit activity, indicating that the cation reached saturation and then became inhibitory. As expected, the addition of EDTA to the assay mixture significantly inhibited enzyme activity.
Mn2+ was shown to stimulate SalM activity strongly in the 1–2 mm range but was inhibitory at higher concentrations (Fig. 2). At 2 mm MnCl2, activity was equal to that of 20 mm MgCl2. However, at concentrations above 20 mm, activity was less compared with SalM devoid of divalent cation.
Fe2+, Cu2+, Zn2+, and Ni2+ were also tested but were found to be inactive or inhibitory (data not shown). The addition of MgCl2 or MgSO4 resulted in equivalent activity, indicating that the counterion was not responsible for changes in enzyme activity.
C-terminal Mutations
Metal dependence within the SDR family is rare; however, there is precedence. Two distinct isolated cases of structural metal dependence in SDR enzymes have been previously characterized structurally. In the first case, dTDP-6-deoxy-l-lyxo-4-hexulose reductase (RmlD) from Salmonella enterica (Protein Data Bank code 1kbz) was shown to require Mg2+ for dimerization (19). Its high resolution crystal structure showed that the magnesium ion was bound by two glutamate residues per monomer to stabilize the dimer (20). In the second case, R-specific alcohol dehydrogenase from Lactobacillus brevis (Protein Data Bank code 1nxq) was shown to be a homotetramer stabilized by two structured magnesium ions per tetramer (21). The carboxylate of the C-terminal glutamine residue coordinates water molecules that bind magnesium. As with SalM, R-specific alcohol dehydrogenase had a slow binding rate with Mg2+, and Mn2+ was also shown to be a suitable cofactor (activity versus concentration was not reported).
Although we were unable to discern the monomeric state of SalM due to extensive aggregation, SalM does contain a C-terminal glutamine residue, as with R-specific alcohol dehydrogenase. To probe the importance of Gln255 in SalM, eight mutants, including truncations (Q255 deletion and A254-Q255 deletion), substitutions (Q255E, Q255S, Q255N, and Q255V), and extensions (256V and Q255N/extension 256Q), were generated. In all cases, highly expressed yet entirely insoluble protein was produced, suggesting an important structural role of Gln255.
Substrate Specificity and Kinetics
To identify enzyme substrate specificity, 10 different sugars were assayed. Only 5-ClR, d-ribose, and d-erythrose showed activity, with 5-ClR being the preferred substrate. Sugars tested and found inactive (<2% activity relative to 5-ClR) included 2-deoxy-d-ribose, d-ribose 5-phosphate, d-xylose, d-arabinose, l-arabinose, d-allose, and d-glucose. The Km differed significantly among the three substrates, with 5-ClR binding to SalM being 2 orders of magnitude greater than d-erythrose and 3 orders of magnitude greater than d-ribose (Table 1). Vmax values were comparable for all three substrates, indicating that the Km is the driver of differential activity among the three preferred substrates. kcat values were not calculated due to enzyme aggregation, which led to an unknown fraction of the total SalM enzyme being inactive.
TABLE 1.
Kinetic values for accepted substrates of SalM
| Km | Vmax | |
|---|---|---|
| mm | mmol min−1 mg−1 | |
| d-Ribose | 19 ± 3 | 4.8 ± 0.2 |
| d-Erythrose | 2.5 ± 0.5 | 5.4 ± 0.4 |
| 5-ClR | 0.02 ± 0.01 | 6.8 ± 0.1 |
Carbon NMR Assays of SalM
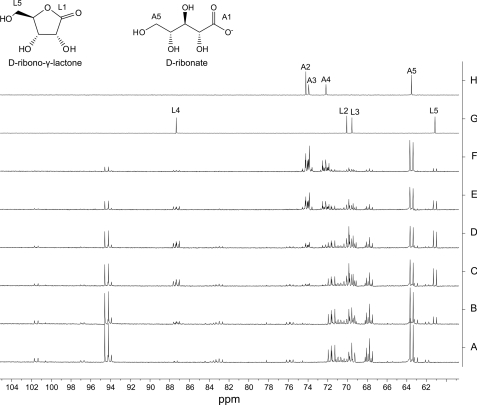
To explore the product structure(s) of SalM, we first assayed activity with [U-13C]ribose in an arrayed NMR experiment (Fig. 3). A standard 1H-decoupled 13C NMR spectrum of 256 scans was recorded of the reaction mixture immediately prior to the addition of SalM. After enzyme addition, equivalent scans were repeated at selected time points extending up to 72 h. The short scan time of ∼9 min allowed only the labeled ribose carbon signals to be readily detected in the assay mixture that also contained NAD+ and its two ribose residues. However, as ribose adopts four cyclical anomeric forms in solution, its NMR spectrum is rather complex for a five-carbon molecule. The six-membered α- and β-pyranoses account for ∼21.5 and 58.5%, respectively, of the total sugar at a temperature of 30 °C, whereas the five-membered α- and β-furanoses account for the remaining 6.5 and 13.5%, respectively (22). On the other hand, the open-chain aldehyde is only a transient intermediate and thus not observed by NMR analysis. Upon oxidation of the anomeric C1, we anticipated that the spectrum would significantly simplify as the reaction progressed to give a single product.
FIGURE 3.
Partial 125-MHz 13C NMR spectra of [U-13C]ribose and NAD+ assayed with SalM. Spectra acquired over 9 min were taken prior to the addition of enzyme (trace A) and then after enzyme addition at 45 min (trace B), 115 min (trace C), 210 min (trace D), 21 h (trace E), and 72 h (trace F). Standards of unlabeled ribono-γ-lactone (trace G) and ribonate (trace H) are provided for reference, and C2–C5 are labeled as L2–L5 and A2–A5, respectively. Lactone formation is clearly apparent with the emergence of L4 and L5 at 87.5 and 61.0 ppm, respectively, beginning with trace B. As the lactone peaks fade over time, several new peaks emerge at 70–72 ppm that correspond to A2–A4 of ribonate. Resonances for C1 of each of the pentose molecules are not shown.
Upon the addition of SalM, the consumption of ribose was observed as the four 13C doublets between 93 and 102 ppm representing the anomeric C1 positions of the ribose congeners decreased in intensity over time. With the oxidation of C1, a new doublet of weak intensity likewise emerged at 178.9 ppm. Unfortunately, because the chemical shifts of the C1 carbonyls of ribono-γ-lactone and ribonate standards are nearly identical, we turned our attention to other more diagnostic signals for analysis. Significantly, two clear signals emerged characteristic of ribono-γ-lactone: a doublet of doublets centered at 87.4 ppm and a doublet centered at 61.1 ppm corresponding to C4 and C5, respectively. No peaks corresponding to ribono-δ-lactone were observed, suggesting that the less abundant furanose is the preferred enzyme substrate. As the reaction progressed further, these characteristic lactone peaks decreased in intensity with the concomitant emergence of a new cluster of signals at 72–74 ppm corresponding to C2–C4 of ribonate. It is evident that the initial product of SalM was a five-membered lactone, which was then hydrolyzed to an acid. However, the role of SalM in lactone hydrolysis remained unclear.
We next explored the putative natural substrate 5-ClR using a complementary NMR spectroscopic strategy. Because we instead used unlabeled material, we utilized the coherence transfer spectroscopic technique DEPT, which resulted in enhanced 4-fold sensitivity (23). The DEPT135 experiment allowed the visualization of all protonated carbons with differential phasing of methylene versus methyl and methine carbons.
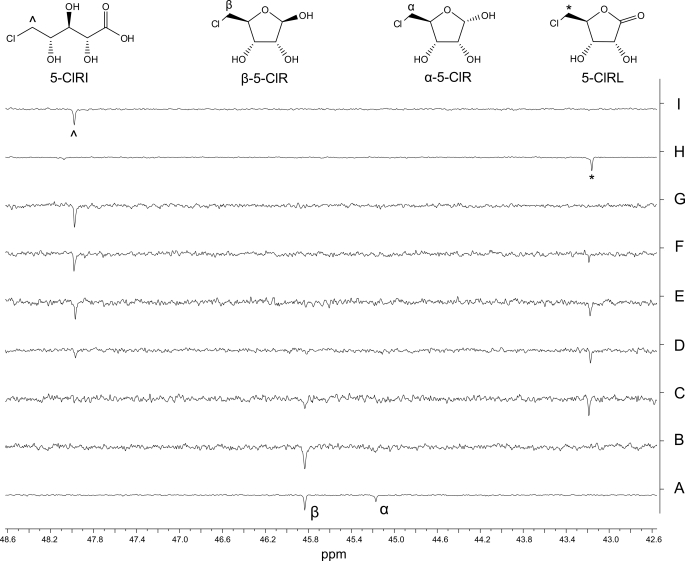
Although the use of 5-ClR simplified the NMR spectrum by eliminating the carbon signals pertaining to the two pyranose anomers, the increased scan time of this assay from 9 to 68 min complicated the analysis by allowing the two ribose moieties per NAD(H) cofactor molecule to be equally visible. This scenario posed a challenge to differentiate the product profile from that of the cofactor. To simplify this dilemma, we identified a diagnostic set of signals to monitor throughout the enzymatic reaction pertaining to the C5 ribose methylene carbons. C5 of the chlorinated sugar substrate was significantly upfield-shifted in relation to the phosphate-attached cofactor riboses. This was true as well in the potential products 5-ClRL and 5-ClRI standards (Fig. 4). C5 of the β-anomer of 5-ClR was clearly visible at 45.8 ppm in the first time point before the addition of SalM, with the less prevalent α-anomer at 45.2 ppm being less visible under these conditions. After enzyme addition, a new peak appeared at 43.2 ppm correlating to C5 of 5-ClRL, which eventually gave way to a second product peak at 48.0 ppm relating to C5 of 5-ClRI, confirming the result of the labeled ribose experiment.
FIGURE 4.
Partial 125-MHz DEPT NMR spectra of the SalM assay with unlabeled 5-ClR. A DEPT135 NMR assay was used to monitor the oxidation of 4 mm unlabeled 5-ClR by SalM. C5 resonances are shown. Spectra acquired over 1 h were taken prior to the addition of SalM (trace B) and then sequentially after enzyme addition at 0–1 h (trace C), 1–2 h (trace D), 2–3 h (trace E), 3–4 h (trace F), and 8–9 h (trace G). Standards of 5-ClR (trace A), 5-ClRL (trace H), and 5-ClRI (trace I) are shown for reference. C5 of the substrate 5-ClR is populated between two resonances at 45.2 and 45.8 ppm and relates to the α- and β-anomers, respectively (trace A). 5-ClRL appears within the 1st h after the addition of SalM as noted with the characteristic emergence of C5 at 43.2 ppm (*). C5 of 5-ClRI (∧) subsequently appears in the 2nd h and increases in intensity to become the sole product after 9 h.
Lactone Opening Assay
The NMR assays established that the SalM reaction involves the enzymatic oxidation of a furanose hemiacetal to a lactone. To identify the role of SalM in the subsequent hydrolysis of the lactone to the corresponding carboxylic acid, a colorimetric assay was employed. Active and boiled SalM enzymes were separately added to solutions of 10 mm 5-ClRL in 100 mm Tris (pH 7.5) and analyzed colorimetrically for lactone concentration at periodic time points. At all time points, the concentration of lactone was approximately equal regardless of whether the active or boiled control enzyme was added (supplemental Tables S1 and S2), indicating that SalM does not actively participate in the hydrolysis of the lactone.
The hydrolysis rate of lactones in aqueous solution is known to follow second-order kinetics, dependent on both lactone concentration and hydroxide concentration (pH) (24). Lactone hydrolysis may liberate a proton in basic solutions, which alters the pH. However, if a sufficiently strong buffer is used, this effect is minimum, converting the hydrolysis rate to a pseudo first-order equation. The hydrolysis rates of 5-ClRL in the presence of active SalM and in the absence of SalM were −0.0035 ± 0.0001/min and −0.0036 ± 0.0002/min, respectively (supplemental Fig. S1).
DISCUSSION
Substrate Specificity and Kinetic Analysis
In this study, we have shown that SalM accepts 5-ClR, d-ribose, and d-erythrose as substrates with varying activity (Fig. 5). In addition to 5-ClR, the 5-fluoro and 5-bromo analogs are presumed as substrates based on previous in vivo experiments with the upstream SalL mutant to produce fluoro- and bromosalinosporamide (13, 25). However, examination of non-accepted substrates can be equally informative in structure-activity relationship analysis. Carbohydrates provide a unique opportunity to individually probe minor alterations in substrate structure and stereochemistry. Inversion of stereochemistry at either C2 or C3 (d-arabinose and d-xylose, respectively) led to abolishment of activity, indicating that SalM is specific to the stereochemistry of ribose. However, it should be noted that d-arabinose and d-xylose exist primarily in pyranose conformations in aqueous solution (22). The biologically relevant 2-deoxy-d-ribose was also found to be an inactive substrate, indicating that the C2 hydroxyl of ribose is required for activity and possibly forms key binding interactions with SalM at this position.
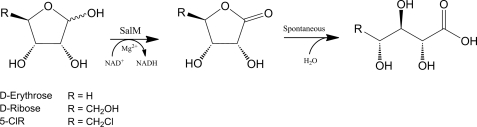
FIGURE 5.
SalM-mediated transformation of select furanoses. SalM oxidizes C1 of furanose carbohydrates with stereochemistry of d-ribose at C2 and C3 to the corresponding γ-lactone. The four-carbon d-erythrose was accepted, whereas the six-carbon d-allose was not, thereby indicating a limit to the size of the C4 furanose substituent. Lactone hydrolysis was found not to be mediated by SalM.
Carbon chain length and ring size also influence SalM activity. Both 5-ClR and d-erythrose are capable of forming only five-membered rings, establishing furanoses as valid substrates. The observation of d-ribose being converted solely to the γ-lactone also supports the exclusive acceptance of five-membered rings. SalM did not accept d-allose, the hexose with identical stereochemistry to d-ribose at C2, C3, and C4. As d-allose adopts a furanose form of 8–10% at the assay temperature, this observation suggests that the enzyme cannot accommodate more than one carbon extending from C4 of the furanose ring (22).
To further analyze the structure-activity relationship of SalM, we compared the kinetic parameters of the three accepted substrates. The minor differences in Vmax indicate that enzyme-substrate binding accounts for the majority of change in activity. The significantly lower Km of 5-ClR over ribose likely has two sources. First, the replacement of the C5 hydroxyl with a chloro group in 5-ClR prevents the formation of a pyranose ring. As the true enzyme substrate appears to be one of the furanose anomers, which comprise only 20% of total ribose in solution at 30 °C, the effective substrate concentration of ribose is actually 5-fold lower compared with 5-ClR. Second, the switch from chloro to the more polar hydroxyl group likely creates unfavorable binding interactions with the enzyme. Like 5-ClR, erythrose adopts only a furanose ring structure yet has a 100-fold increase in Km. The lack of a fifth carbon and attached chloride extending from C4 of the furanose ring eliminates the possibility of any favorable binding interactions that 5-ClR may generate with SalM at this site.
Metal Dependence and Lactonase Activity
Convergent evolution has produced multiple strategies for catalyzing the oxidation of hydroxyls to carbonyls. Two of the most prominent families of such enzymes are the SDRs and the medium-chain dehydrogenases/reductases. Although the reactions catalyzed may be similar, their mechanisms are distinct. Metal dependence is synonymous within the medium-chain dehydrogenase/reductase family, with zinc acting as a catalytic component to activate a coordinated water molecule for abstraction of the hydroxyl proton of the substrate (26). Glucose 1-dehydrogenase from the medium-chain dehydrogenase/reductase family has been reported to oxidize glucose to gluconolactone, followed by “lactonase” activity to hydrolyze the lactone (7, 27, 28). However, no mechanism has been reported for catalysis of this additional functionality.
Unlike the medium-chain dehydrogenase/reductase family, the metal-independent SDRs are typically catalyzed by a lysine-activated tyrosine (17). Because the mechanism of classical SDRs is well established to be metal-independent and because SalM possesses the highly conserved YX3K catalytic group, it is likely that the metal ion is not contributing to substrate oxidation (17). Our initial speculation as to the atypical metal dependence of SalM included the possibility of additional lactonase activity. Lactonase enzymes such as Drp35 from Staphylococcus aureus bind a catalytic zinc cation to activate water for hydrolysis of lactones (29). This enzyme was also shown to exhibit lactonase activity when bound to Mg2+ or Mn2+. However, when SalM was assayed without Mg2+, 5-ClR was not oxidized to 5-ClRL, indicating that the metal ion is required for the first step of the reaction and not the latter. As SalM does possess a C-terminal glutamine, as in the case of R-specific alcohol dehydrogenase from L. brevis, we anticipate the divalent metal cation to play a similar structural role. This hypothesis is supported by the total loss of solubility for all C-terminal mutants of SalM.
Having established that SalM does not participate in lactone hydrolysis, we explored the possibility of a missing chloroethylmalonyl-CoA biosynthetic enzyme. In metabolic pathways that require lactone hydrolysis, a lactonase is often employed to facilitate the reaction (5, 10, 30, 31). Although the salinosporamide gene cluster does not contain a lactonase, a search of the total genome sequence of S. tropica CNB-440 (32) resulted in one annotated gluconolactonase. This gene (Stro_0658) is located ∼400 open reading frames from the sal locus. Although it is not known if this enzyme participates in the lactone opening of 5-ClRL, it seems unlikely to be specialized for this reaction because Salinispora arenicola CNS-205, the closest sequenced relative of S. tropica, contains a 92% similar gluconolactonase yet does not contain the salinosporamide gene cluster (33). It is therefore possible that the biosynthesis of chloroethylmalonyl-CoA depends on the spontaneous hydrolysis of 5-ClRL. This may result in a specific bottleneck in salinosporamide A production, suggesting that fermentation yields of this prospective drug candidate may be increased by engineering a lactonase into S. tropica.
Evolution of SalM and the Chloroethylmalonyl-CoA Pathway
Previously characterized pentose dehydrogenases for d-arabinose (EC 1.1.1.117), l-arabinose (EC 1.1.1.46), and d-xylose (EC 1.1.1.179) have been linked to non-phosphorylative pentose catabolism (5). In such pathways, the pentose is oxidized to a sugar lactone, followed by lactonase-mediated hydrolysis to the pentonic acid. A pentonic-acid dehydratase then creates a 2-keto-3-deoxypentonic acid, which may be subsequently oxidized to α-ketoglutarate or pyruvate (31). The transformation of 5-ClR in salinosporamide biosynthesis follows a strikingly similar route. In the initial step, 5-ClR is oxidized to 5-ClRL by SalM, followed by hydrolysis to 5-ClRI. The acid dehydratase SalH then putatively dehydrates 5-ClRI to 5-chloro-4-hydroxy-2-oxopentanoate (2).
It is tempting to envision this portion of chloroethylmalonyl-CoA biosynthesis as being recruited from non-phosphorylative pentose oxidation. SalM has been shown here to act as a furanose 1-dehydrogenase with activity for both d-ribose and d-erythrose. Neither substrate has a previously characterized stereospecific 1-dehydrogenase. The lack of activity for SalM with the pentoses l-arabinose and d-xylose implies that SalM was not likely recruited from previously identified pathways. If SalM did evolve from a pentose oxidation pathway, it would likely be specific to d-ribose. As the enzymes of such a putative pathway have yet to be elucidated, it creates the potential to use secondary metabolic enzymes, SalM and SalH, as probes for primary metabolic non-phosphorylative ribose oxidation pathways.
Supplementary Material
Acknowledgments
We thank W. Fenical and P. R. Jensen (Scripps Institution of Oceanography) for S. tropica strains and A. Mrse (University of California at San Diego) for NMR spectroscopy assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant CA127622 (to B. S. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
- SDR
- short-chain dehydrogenase/reductase
- 5-ClR
- 5-chloro-5-deoxy-d-ribose
- Ni-NTA
- nickel-nitrilotriacetic acid
- 5-ClRL
- 5-chloro-5-deoxy-d-ribono-γ-lactone
- 5-ClRI
- 5-chloro-5-deoxy-d-ribonate
- DEPT
- distortionless enhancement of polarization transfer.
REFERENCES
- 1.Gulder T. A., Moore B. S. (2010) Angew. Chem. Int. Ed., in press [Google Scholar]
- 2.Eustáquio A. S., McGlinchey R. P., Liu Y., Hazzard C., Beer L. L., Florova G., Alhamadsheh M. M., Lechner A., Kale A. J., Kobayashi Y., Reynolds K. A., Moore B. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12295–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiwara H. W., Domschke W., Domagk G. F. (1968) Hoppe-Seyler's Z. Physiol. Chem. 349, 1575–1581 [PubMed] [Google Scholar]
- 4.Johnsen U., Schönheit P. (2004) J. Bacteriol. 186, 6198–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouns S. J.., Walther J., Snijders A. P., van de Werken H. J., Willemen H. L., Worm P., de Vos M. G., Andersson A., Lundgren M., Mazon H. F., van den Heuvel R. H., Nilsson P., Salmon L., de Vos W. M., Wright P. C., Bernander R., van der Oost J. (2006) J. Biol. Chem. 281, 27378–27388 [DOI] [PubMed] [Google Scholar]
- 6.Watanabe S., Kodaki T., Kodak T., Makino K. (2006) J. Biol. Chem. 281, 2612–2623 [DOI] [PubMed] [Google Scholar]
- 7.Bonete M. J., Pire C., LLorca F. I., Camacho M. L. (1996) FEBS Lett. 383, 227–229 [DOI] [PubMed] [Google Scholar]
- 8.Scher B. M., Horecker B. L. (1966) Arch. Biochem. Biophys. 116, 117–128 [DOI] [PubMed] [Google Scholar]
- 9.Milburn C. C., Lamble H. J., Theodossis A., Bull S. D., Hough D. W., Danson M. J., Taylor G. L. (2006) J. Biol. Chem. 281, 14796–14804 [DOI] [PubMed] [Google Scholar]
- 10.Stephens C., Christen B., Fuchs T., Sundaram V., Watanabe K., Jenal U. (2007) J. Bacteriol. 189, 2181–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki Y. (1974) Bull. Chem. Soc. Jpn. 47, 2077–2078 [Google Scholar]
- 12.Bouchez V., Stasik I., Beaupere D., Uzan R. (1997) Carbohydr. Res. 300, 139–142 [DOI] [PubMed] [Google Scholar]
- 13.Eustáquio A. S., Pojer F., Noel J. P., Moore B. S. (2008) Nat. Chem. Biol. 4, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jez J. M., Ferrer J. L., Bowman M. E., Dixon R. A., Noel J. P. (2000) Biochemistry 39, 890–902 [DOI] [PubMed] [Google Scholar]
- 15.Hestrin S. (1949) J. Biol. Chem. 180, 249–261 [PubMed] [Google Scholar]
- 16.Kallberg Y., Oppermann U., Jörnvall H., Persson B. (2002) Eur. J. Biochem. 269, 4409–4417 [DOI] [PubMed] [Google Scholar]
- 17.Kavanagh K. L., Jörnvall H., Persson B., Oppermann U. (2008) Cell. Mol. Life Sci. 65, 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wierenga R. K., Terpstra P., Hol W. G. (1986) J. Mol. Biol. 187, 101–107 [DOI] [PubMed] [Google Scholar]
- 19.Graninger M., Nidetzky B., Heinrichs D. E., Whitfield C., Messner P. (1999) J. Biol. Chem. 274, 25069–25077 [DOI] [PubMed] [Google Scholar]
- 20.Blankenfeldt W., Kerr I. D., Giraud M. F., McMiken H. J., Leonard G., Whitfield C., Messner P., Graninger M., Naismith J. H. (2002) Structure 10, 773–786 [DOI] [PubMed] [Google Scholar]
- 21.Niefind K., Müller J., Riebel B., Hummel W., Schomburg D. (2003) J. Mol. Biol. 327, 317–328 [DOI] [PubMed] [Google Scholar]
- 22.Angyal S. J., Pickles V. A. (1972) Aust. J. Chem. 25, 1695–1710 [Google Scholar]
- 23.Jacobsen N. A. (2007) NMR Spectroscopy Explained, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 24.Jermyn M. A. (1960) Biochim. Biophys. Acta 37, 78–92 [DOI] [PubMed] [Google Scholar]
- 25.Eustáquio A. S., Moore B. S. (2008) Angew. Chem. Int. Ed. 47, 3936–3938 [DOI] [PubMed] [Google Scholar]
- 26.Baker P. J., Britton K. L., Fisher M., Esclapez J., Pire C., Bonete M. J., Ferrer J., Rice D. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelov A., Fütterer O., Valerius O., Braus G. H., Liebl W. (2005) FEBS J. 272, 1054–1062 [DOI] [PubMed] [Google Scholar]
- 28.Lamble H. J., Heyer N. I., Bull S. D., Hough D. W., Danson M. J. (2003) J. Biol. Chem. 278, 34066–34072 [DOI] [PubMed] [Google Scholar]
- 29.Tanaka Y., Morikawa K., Ohki Y., Yao M., Tsumoto K., Watanabe N., Ohta T., Tanaka I. (2007) J. Biol. Chem. 282, 5770–5780 [DOI] [PubMed] [Google Scholar]
- 30.Mathias A. L., Rigo L. U., Funayama S., Pedrosa F. O. (1989) J. Bacteriol. 171, 5206–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe S., Shimada N., Tajima K., Kodaki T., Makino K. (2006) J. Biol. Chem. 281, 33521–33536 [DOI] [PubMed] [Google Scholar]
- 32.Udwary D. W., Zeigler L., Asolkar R. N., Singan V., Lapidus A., Fenical W., Jensen P. R., Moore B. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penn K., Jenkins C., Nett M., Udwary D. W., Gontang E. A., McGlinchey R. P., Foster B., Lapidus A., Podell S., Allen E. E., Moore B. S., Jensen P. R. (2009) ISME J. 3, 1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.