Abstract
Following the initial discovery of two legume-nodulating Burkholderia strains (L. Moulin, A. Munive, B. Dreyfus, and C. Boivin-Masson, Nature 411:948-950, 2001), we identified as nitrogen-fixing legume symbionts at least 50 different strains of Burkholderia caribensis and Ralstonia taiwanensis, all belonging to the β-subclass of proteobacteria, thus extending the phylogenetic diversity of the rhizobia. R. taiwanensis was found to represent 93% of the Mimosa isolates in Taiwan, indicating that β-proteobacteria can be the specific symbionts of a legume. The nod genes of rhizobial β-proteobacteria (β-rhizobia) are very similar to those of rhizobia from the α-subclass (α-rhizobia), strongly supporting the hypothesis of the unique origin of common nod genes. The β-rhizobial nod genes are located on a 0.5-Mb plasmid, together with the nifH gene, in R. taiwanensis and Burkholderia phymatum. Phylogenetic analysis of available nodA gene sequences clustered β-rhizobial sequences in two nodA lineages intertwined with α-rhizobial sequences. On the other hand, the β-rhizobia were grouped with free-living nitrogen-fixing β-proteobacteria on the basis of the nifH phylogenetic tree. These findings suggest that β-rhizobia evolved from diazotrophs through multiple lateral nod gene transfers.
Members of the Leguminosae, comprising about 18,000 species, play an important ecological role, with representatives in nearly every type of plant on Earth. Most species are able to form nitrogen-fixing symbioses with specific bacteria known as rhizobia. The recent identification of two β-proteobacterial strains of the genus Burkholderia able to nodulate legumes (10) changed the long-held dogma that only bacteria of the α subdivision are able to nodulate legumes (18, 23). These two strains were subsequently described as Burkholderia tuberum and Burkholderia phymatum (24). In addition, eight strains isolated from root nodules of Mimosa spp. were recently described as Ralstonia taiwanensis, also classified as β-proteobacteria (1), although their nodulation capacity was not confirmed. The terms α- and β-rhizobia were proposed to distinguish the rhizobial α- and β-proteobacteria, respectively (10). This unexpected discovery raised the question as to whether nodulation by β-proteobacteria is an extremely rare phenomenon or whether it had simply been overlooked until now. Moreover, the fact that the first two nodulating Burkholderia strains were isolated from Aspalathus and Machaerium spp., which are known to be associated with Bradyrhizobium (2, 12), may suggest that these β-proteobacteria are not the specific partners of the respective host legumes.
In this article, we confirm the widespread phylogenetic diversity of nitrogen-fixing legume symbionts by identifying as β-rhizobia an additional 2 Burkholderia strains from the species Burkholderia caribensis and a collection of at least 44 R. taiwanensis strains. These data increase to four the number of different β-rhizobial species identified so far, originating from three different continents. Moreover, we show that R. taiwanensis is the preferred partner of Mimosa pudica and Mimosa diplotricha in Taiwan. β-Rhizobia possess nod and nif genes which are very similar to those of α-rhizobia and which are located on a symbiotic plasmid. Phylogenetic analysis of available nodA and nifH genes from α- and β-proteobacteria suggests that β-rhizobia have evolved from diazotrophs through multiple lateral gene transfers.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
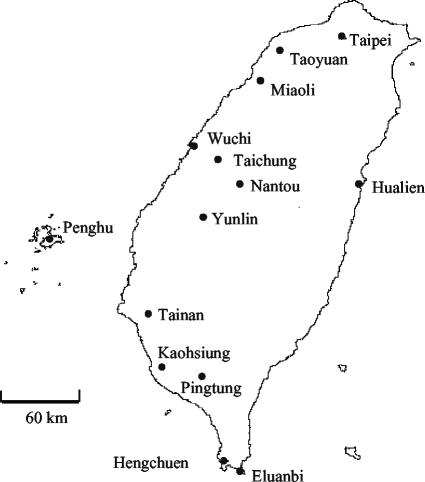
The Mimosa strains used in these studies are listed in Table 1. B. phymatum STM815, isolated from Machaerium lunatum in French Guiana, was previously described (10, 24). Mimosa strains were isolated from root nodules collected at 14 sites in Taiwan (Fig. 1) by using a previously described isolation procedure (1). Strains were maintained and grown on yeast extract-mannitol medium (18) at 28°C.
TABLE 1.
Rhizobial strains and relevant characteristics
| Strain(s) (no. of isolates) | Host plant | Geographical origina | PCR-RFLP pattern
|
PFGE pattern | Reference or source | |
|---|---|---|---|---|---|---|
| 16S rDNA | nodA | |||||
| Ralstonia taiwanensis | ||||||
| LMG 19425 | Mimosa diplotricha | Pingtung | I | V | E | 1 |
| TJ1 to TJ4 (4) | Mimosa pudica | Pingtung | I | I | A | This study |
| TJ12 (1) | Mimosa pudica | Taoyuan | I | I | A | This study |
| TJ13 to TJ18 (6) | Mimosa pudica | Miaoli | I | V | A | This study |
| TJ19 to TJ28 (10) | Mimosa pudica | Taichung | I | I | A | This study |
| TJ29 (1) | Mimosa pudica | Nantou | I | II | B | This study |
| TJ30 to TJ35 (6) | Mimosa pudica | Nantou | I | I | A | This study |
| TJ36 to TJ39 (4) | Mimosa pudica | Tainan | I | V | A | This study |
| TJ41 to TJ48 (8) | Mimosa pudica | Kaohsiung | I | I | A | This study |
| TJ49 to TJ57 (9) | Mimosa pudica | Hengchuen | I | V | A | This study |
| TJ60 and TJ61 (2) | Mimosa pudica | Eluanbi | I | V | I | This study |
| TJ62 to TJ64 (3) | Mimosa pudica | Hualien | I | I | A | This study |
| TJ65 to TJ77 (13) | Mimosa pudica | Penghu | I | I | A | This study |
| TJ78 to TJ86 (9) | Mimosa diplotricha | Pingtung | I | I | A | This study |
| TJ87 (1) | Mimosa diplotricha | Pingtung | I | V | E | This study |
| TJ89 to TJ97 (9) | Mimosa diplotricha | Taipei | I | V | A | This study |
| TJ99 to TJ107 (9) | Mimosa diplotricha | Taoyuan | I | V | A | This study |
| TJ108 to TJ110 (3) | Mimosa diplotricha | Taichung | I | III | O | This study |
| TJ112 to TJ119 (8) | Mimosa diplotricha | Taichung | I | IV | P | This study |
| TJ120 to TJ122 (3) | Mimosa diplotricha | Taichung | I | V | N | This study |
| TJ123 and TJ124 (2) | Mimosa diplotricha | Wuchi | I | V | A | This study |
| TJ125 (1) | Mimosa diplotricha | Wuchi | I | V | D | This study |
| TJ128 to TJ130 (3) | Mimosa diplotricha | Nantou | I | IV | D | This study |
| TJ131 and TJ132 (2) | Mimosa diplotricha | Nantou | I | V | E | This study |
| TJ137 and TJ138 (2) | Mimosa diplotricha | Nantou | I | V | A | This study |
| TJ141 (1) | Mimosa diplotricha | Yunlin | I | V | D | This study |
| TJ142 to TJ144 (3) | Mimosa diplotricha | Tainan | I | V | B | This study |
| TJ145 to TJ153 (9) | Mimosa diplotricha | Kaohsiung | I | I | A | This study |
| TJ154 (1) | Mimosa diplotricha | Kaohsiung | I | I | J | This study |
| TJ160 (1) | Mimosa diplotricha | Hengchuen | I | V | L | This study |
| TJ161 and TJ162 (2) | Mimosa diplotricha | Hengchuen | I | V | A | This study |
| TJ163 and TJ164 (2) | Mimosa diplotricha | Hengchuen | I | IV | M | This study |
| TJ190 to TJ199 (10) | Mimosa diplotricha | Penghu | I | I | A | This study |
| LMG 19426 | Mimosa pudica | Pingtung | II | IV | K | 1 |
| TJ40 (1) | Mimosa pudica | Kaohsiung | II | II | K | This study |
| TJ58 and TJ59 (2) | Mimosa pudica | Eluanbi | II | II | K | This study |
| TJ111 (1) | Mimosa diplotricha | Taichung | II | IV | G | This study |
| TJ126 and TJ127 (2) | Mimosa diplotricha | Nantou | II | IV | C | This study |
| TJ133 and TJ134 (2) | Mimosa diplotricha | Nantou | II | IV | F | This study |
| TJ135 and TJ136 (2) | Mimosa diplotricha | Nantou | II | IV | B | This study |
| TJ139 and TJ140 (2) | Mimosa diplotricha | Yunlin | II | IV | G | This study |
| TJ155 to TJ159 (5) | Mimosa diplotricha | Hengchuen | II | IV | K | This study |
| LMG 19424 | Mimosa pudica | Pingtung | III | V | H | 1 |
| TJ5 to TJ10 (6) | Mimosa pudica | Pingtung | III | V | H | This study |
| TJ98 (1) | Mimosa diplotricha | Taipei | III | V | H | This study |
| LMG 19430 | Mimosa diplotricha | Pingtung | IV | V | R | 1 |
| TJ11 (1) | Mimosa pudica | Pingtung | IV | V | Q | This study |
| TJ88 (1) | Mimosa diplotricha | Pingtung | IV | V | R | This study |
| Rhizobium sp. | ||||||
| TJ167b to TJ169 (3) | Mimosa diplotricha | Taoyuan | V | ND | T | This study |
| TJ173b to TJ176 (4) | Mimosa diplotricha | Nantou | V | ND | X | This study |
| TJ171b (1) | Mimosa diplotricha | Hualien | VI | ND | V | This study |
| TJ172b (1) | Mimosa diplotricha | Hualien | VII | ND | W | This study |
| TJ189 (1) | Mimosa diplotricha | Pingtung | VII | ND | W | This study |
| Burkholderia caribensis | ||||||
| TJ182b (1) | Mimosa diplotricha | Pingtung | VIII | ND | Y | This study |
| TJ183 (1) | Mimosa pudica | Pingtung | VIII | ND | Y | This study |
| Sinorhizobium sp. strain TJ170b (1) | Mimosa pudica | Penghu | IX | ND | U | This study |
Sites were located in Taiwan (Fig. 1).
16S rDNA of the indicated strain was sequenced as part of this study. ND, not determined.
FIG. 1.
Sampling sites for M. pudica and M. diplotricha in Taiwan.
DNA manipulation.
For pulsed-field gel electrophoresis (PFGE) genotyping, agarose plugs containing intact bacterial genomic DNA were digested with XbaI (Boehringer Mannheim) and subjected to electrophoresis on 1.2% SeaKem GTG agarose (FMC) gels in 0.5× Tris-borate-EDTA buffer for 24 h at 14°C with a pulse ramp of 5 to 35 s at 200 V (LKB 2015 system; Pharmacia). For PFGE genome organization analysis, intact genomic DNA in agarose plugs was electrophoresed on an 0.8% agarose gel in Tris-agarose-EDTA for 41 h with a pulse time of 500 s at 100 V (CHEF-Mapper XA system; Bio-Rad). PFGE agarose gels were blotted on nylon membranes (Hybond), hybridized with 32P-labeled nodA, nodC, and nifH PCR products for 17 h at 65°C, and washed at 55°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate.
Nearly full-length 16S ribosomal DNA (rDNA) was amplified and sequenced as previously described (1). 16S rDNA PCR-restriction fragment length polymorphism (RFLP) analysis was performed as described previously (1), except that AluI, CfoI, HinfI, and MspI were used. nodA amplification and sequencing were performed with pairs of primers with the sequences 5′-TGGARVBTNYSYTGGGAAA-3′ and5′-TCAYARYTCNGRNCCRTTYC-3′ (strains LMG 19424, LMG 19425, TJ171, and TJ182), 5′-TGGARVBTNYSYTGGGAAA-3′ and 5′-GGRTKNGGNCCRTCRTCRAANGT-3′ (strains TJ167, TJ172, and TJ173), and 5′-TGCRGTGGAARNTRBVYTGGGAAA-3′ and 5′-TCACARCTCKGGCCCGTTCCG-3′ (strain STM815). nodA PCR-RFLP analysis was performed with a 531-bp PCR product obtained with a primer pair with the sequences 5′-ATCTTGAACTCTCCGACC-3′ and 5′-GTTCGATTGTTTCGCCG-3′ and digested with AluI, CfoI, HinfI, MspI, and NdeII.
A 520-bp fragment containing part of the nodH and nodA genes of strain LMG 19424 was amplified and sequenced with primers with the sequences 5′-GCCATCCACATCATCGATG-3′ and 5′-CGGCTTCGCATTGAAAGGC-3′. A 2.1-kb fragment containing the nodB gene and part of the nodC gene of strain LMG 19424 was amplified with primers with the sequences 5′-CAGATCNAGDCCBTTGAARCGCA-3′ and 5′-CTNCGNGCCCARCGNAGTTG-3′. A 1.2-kb overlapping fragment containing part of the nodC and nodI genes was amplified with primers with the sequences 5′-GTATGTTCCTAACGCTATCGCGGC-3′ and 5′-TCTTCCATVAWRTGVGTNGTCA-3′. These fragments were further sequenced with pairs of degenerate primers based on available nodB, nodC, and nodI alignments.
A 640-bp fragment containing part of the nifH gene was amplified and sequenced with primers with the sequences 5′-CGCIWTYTACGGIAARGGIGG-3′ and 5′-GGIKCRTAYTSGATIACIGTCAT-3′.
PCR products of 440 to 636 bp, used as probes for PFGE hybridization, were amplified from LMG 19424 and STM815 with primer pairs with the sequences 5′-AARGGNGGNATYGGHAARTC-3′ and 5′-GCRTAVAKNGCCATCATYTC-3′ (for nifH), 5′-GGTTCCACGTAAGCTTCCCTCWCCGAYCAYWTSGARTTGGC-3′ and 5′-GCGATTACCCTGTACACCCACAGSTYKGGYCCCCGTTCCG-3′ (for nodA), and 5′-GGTTCCACGTAAGCTTCCCGACATGGAGTACTGGCTCGC-3′) and 5′-GCGATTACCCTGTACACCCGACAGCCAATCGCTATTTCCG-3′ (for nodC).
Phylogenetic analysis.
Multiple alignments were performed with CLUSTAL X (19) and manually corrected by using GeneDoc (11). Phylogenetic analysis was carried out with a maximum-likelihood (ML) approach by using PAUP version 4.0b10 (17). Two types of substitution (the substitution matrix being estimated by ML), three classes of site rate variation based on the codon structure of the DNA sequence, and base frequencies were estimated from the data by the ML approach. The same model was applied for both nifH and nodA phylogenies. Node robustness was esimated by bootstrap analysis by combining the Seqboot and DnaML programs from the PHYLIP package (5).
Plant tests.
Seeds were surface sterilized with concentrated sulfuric acid for 10 min and then with 3% sodium hyperchlorite for 10 min. Plant cultivation and nodulation tests were carried out as described previously (8). Nitrogen fixation was estimated by visual observation of the vigor and foliage color of 60-day-old plants. Fresh nodules were observed under an Olympus SHZ 10 stereomicroscope. Sections 80 μm thick were prepared by using a Leica VT1000S Vibratome. Microscopic preparations were cleared with sodium hypochlorite and stained with methylene blue as described by Truchet et al. (21).
Nucleotide sequence accession numbers.
EMBL accession numbers for the 16S rRNA genes are as follows: AJ505296 (TJ167), AJ505297 (TJ170), AJ505298 (TJ171), AJ505299 (TJ172), AJ505300 (TJ173), and AJ505301(TJ182). Accession numbers for the nifH genes are as follows: AJ505312 (TJ173), AJ505313 (TJ172), AJ505314 (TJ171), AJ505315 (TJ170), AJ505316 (TJ167), AJ505317 (TJ182), AJ505319 (STM815), AJ505320 (LMG 19424), and AJ505321 (LMG 19425). Accession numbers for the nodA genes are as follows: AJ505304 (LMG 19425), AJ505305 (TJ167), AJ505306 (TJ171), AJ505307 (TJ172), AJ505308 (TJ173), AJ505309 (TJ182), AJ55310 (STM1441), AJ505311 (LMG 19424), and AJ505318 (STM815). The accession number for nodBCI of LMG 19424 is AJ505303.
RESULTS
Most rhizobia isolated from M. pudica and M. diplotricha in Taiwan belong to the genus Ralstonia.
To further examine the taxonomic diversity of Mimosa nodule isolates, 190 new isolates were recovered from root nodules of M. pudica and M. diplotricha plants growing in 14 different areas in Taiwan (Table 1).
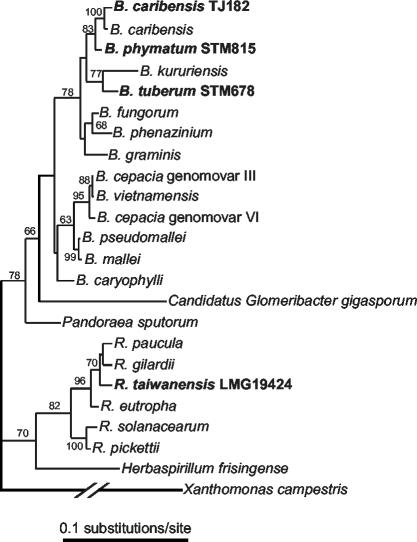
16S rDNA PCR-RFLP analysis grouped 177 isolates together with R. taiwanensis reference strains (Table 1). Additional PFGE and nodA PCR-RFLP (see below) analyses showed that these R. taiwanensis isolates represented at least 44 different strains (Table 1). The remaining 13 isolates fell into six PFGE pattern groups. 16S rDNA sequencing of representative strains of these groups showed that they belonged to the genus Burkholderia (strains TJ182 and TJ183) (Fig. 2), the genus Rhizobium (TJ167 to TJ169, TJ171 to TJ176, and TJ189) (data not shown), or the genus Sinorhizobium (TJ170) (data not shown) (Table 1). Further taxonomical analysis identified TJ182 and TJ183 as B. caribensis (24) (Fig. 2).
FIG. 2.
16S rDNA tree showing phylogenetic positions of legume-nodulating Ralstonia and Burkholderia species within the β-proteobacteria. The ML tree (base frequencies estimated, mutation rates drawn from an γ + INVdistribution, four classes of mutations) was reconstructed by using PAUP. Xanthomonas campestris was used as an outgroup. Legume symbionts are shown in bold type. Nodulating Burkholderia strains are named according to Vandamme et al. (24). GenBank/EMBL accession numbers for the 16S rDNA sequences were AF175314 (B. cepacia genomovar VI), AF148556 (B. cepacia genomovar III), U96928 (B. vietnamensis), U91839 (B. pseudomallei), AF110188 (B. mallei), AB021423 (B. carophylli), AJ302312 (B. phymatum), AJ505301 (B. caribensis), Y17009 (B. caribensis), AF215705 (B. fungorum), AB021394 (B. phenazinium), U96939 (B. graminis), AB024310 (B. kuruiensis), AJ302311 (B. tuberum), AF139176 (P. sputorum), AF139176 (G. gigasporum), AJ238359 (H. frisingense), AL646072 (R. solanacearum), AB004790 (R. pickettii),AF085226 (R. paucula), AF300324 (R. taiwanensis), AF076645 (R. gilardii), M32021 (R. eutropha), and AF188831 (X. campestris).
Representatives of the Mimosa isolate collection were double-checked for their ability to nodulate M. pudica under axenic laboratory conditions. The four R. taiwanensis strains tested, LMG 19424, LMG 19425, LMG 19426, and LMG 19430, formed nitrogen-fixing nodules (Fig. 3a and b) from which the original inoculated bacteria could be reisolated. The nodules displayed a genuine nodule structure, with central infected tissue containing cells with intracellular bacteria and peripheral tissue with vascular bundles (Fig. 3c). B. caribensis TJ182 and TJ183, as well as Rhizobium sp. strains TJ167 and TJ173, were able to effectively nodulate M. pudica (data not shown). On the other hand, Sinorhizobium sp. strain TJ170 was unable to nodulate either M. pudica or M. diplotricha (data not shown).
FIG. 3.
Nodules of M. pudica 4 weeks after inoculation with R. taiwanensis LMG 19424. (a) Nodulated roots. (b) Root segment with pink nodules. (c) Longitudinal section showing the structure of a nodule. Plant tissue was cleared with sodium hypochlorite and stained with methylene blue as described by Truchet et al. (21). ic, infected cells; vb, vascular bundles.
Ralstonia and Burkholderia nod and nif genes are very similar to those of α-rhizobia.
In α-rhizobia, the ability to nodulate requires the nodABC genes, responsible for the synthesis of the Nod factor core structure (13, 14). These three genes are all present in the first β-rhizobium identified, B. tuberum STM678. To investigate whether the other β-rhizobial species identified so far also possess essential nod genes, we searched for the presence of nodA in a collection of β-rhizobial strains. So far, sequences homologous to nodA have not been identified in nonrhizobial bacteria, and this gene therefore constitutes—together with the nodBC genes—a molecular signature for rhizobia. We found that an internal nodA sequence (from bp 44 to 574) could be amplified from 181 Ralstonia isolates (177 new isolates and 4 R. taiwanensis reference strains) (data not shown). The nodA genes of R. taiwanensis LMG 19424 and LMG 19425 were sequenced, as were those of B. caribensis TJ182 and B. phymatum STM815. Partial nodA sequences of four α-rhizobial isolates were also determined. Sequence similarities between α- and β-rhizobial NodA proteins ranged from 60% (B. caribensis and Azorhizobium caulinodans) to 72% (B. phymatum and Rhizobium sp. strain TJ172) (Table 2), confirming that the nodA genes of β-proteobacteria are very similar to those of α-rhizobia. Interestingly, α- and β-rhizobial symbionts of M. diplotricha possess unrelated NodA sequences (Table 2).
TABLE 2.
Sequence identities among NodA amino acid sequencesa
| Sequence | % Identity to the following sequence:
|
||||||
|---|---|---|---|---|---|---|---|
| Bca | Bp | Ac | Sme | WM9 | TJ167 | TJ172 | |
| Rt | 99.4 | 79.8 | 60.1 | 63.0 | 66.5 | 69.4 | 68.2 |
| Bca | 80.2 | 60.0 | 63.2 | 66.8 | 70.0 | 67.9 | |
| Bp | 64.3 | 68.7 | 69.2 | 69.8 | 72.0 | ||
| Ac | 55.6 | 56.1 | 57.4 | 58.4 | |||
| Sme | 61.7 | 64.5 | 61.6 | ||||
| WM9 | 69.5 | 70.0 | |||||
| TJ167 | 81.6 | ||||||
Rt, R. taiwanensis LMG 19425; Bca, B. caribensis TJ182; Bp, B. phymatum STM815; Ac, A. caulinodans ORS571; Sme, Sinorhizobium meliloti 2011; WM9, Bradyrhizobium sp. strain WM9; TJ167, Rhizobium sp. strain TJ167; TJ172, Rhizobium sp. strain TJ172. NodA sequence lengths ranged from 170 amino acids (partial sequences) to 197 amino acids (complete sequences). Percent identities were calculated from 170-amino-acid sequences.
We also searched for the presence of other nodulation genes, besides nodA, in R. taiwanensis LMG 19424 by PCR amplification (see Materials and Methods). Analysis of the amplified DNA sequences revealed the presence of the common nodBC genes preceded by a NodD-dependent regulatory sequence (nod box), as well as part of the nodH gene, involved in Nod factor sulfation, and the nodI gene, presumably involved in Nod factor transport. Sequencing indicated that nodA is separated from nodBC. Such genetic nonlinkage of nodABC was found previously for B. tuberum (10) as well as for several rhizobia (25). Sequence similarity with rhizobial Nod proteins available in databases ranged from 32% (A. caulinodans) to 58% (Mesorhizobium sp. strain N33) for NodB and from 49% (Rhizobium gallicum) to 74% (Rhizobium etli) for NodC.
Part of the nifH gene, encoding dinitrogenase reductase, a key enzyme in nitrogen fixation, was also amplified and sequenced in representative strains of R. taiwanensis, B. caribensis, B. phymatum, and Rhizobium spp. (data not shown).
nod and nif genes are located on a plasmid in R. taiwanensis and B. phymatum.
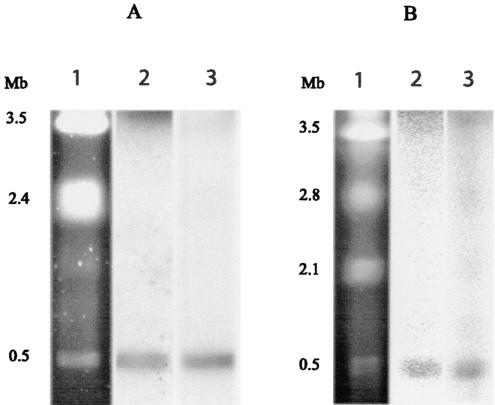
Genes required for nodulation and symbiotic nitrogen fixation are often clustered and located on large plasmids (9) or mobile symbiotic islands (15). To determine the locations of symbiotic genes in the genomes of the new β-rhizobia, we first examined the genome organization of Ralstonia and Burkholderia representatives by using PFGE (Fig. 4). Two high-molecular-weight replicons with apparent sizes of 3.5 and 2.4 Mb and a smaller replicon of about 0.5 Mb were identified for R. taiwanensis LMG 19424, while B. phymatum STM815 possesses replicons of approximately 3.5, 2.8, 2.1, and 0.5 Mb. No readable PFGE profile could be obtained with B. tuberum. To determine which replicons carry the symbiotic genes, Southern blots of PFGE agarose gels were hybridized with nifH and either nodA or nodC probes. Both nod and nif probes hybridized with the smallest 0.5-Mb replicons of R. taiwanensis and B. phymatum (Fig. 4). These symbiotic replicons did not hybridize with parental strain 16S rRNA, suggesting that they are genuine plasmids (data not shown).
FIG. 4.
Locations of nodA and nifH genes on replicons of R. taiwanensis LMG 19424 (A) and B. phymatum STM815 (B). Lane 1, PFGE of undigested genomic DNA stained with ethidium bromide; lanes 2 and 3, autoradiographs of blotted PFGE gels hybridized with nodA and nifH probes, respectively. Sizes of replicons are indicated on the left.
Hence, the clustering of nodulation and nitrogen fixation genes is a common feature of α- and β-rhizobia.
Phylogenetic analysis of nodA and nifH genes of α- and β-rhizobia.
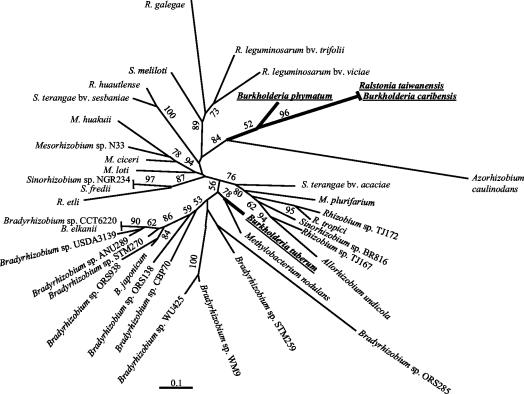
Phylogenetic analysis of 42 nodA sequences—including most available α-rhizobial and four β-rhizobial sequences—resulted in the ML tree shown in Fig. 5. The four β-proteobacteria fell into two strongly supported clades. B. phymatum, B. caribensis, and R. taiwanensis strains clustered in the same clade. The nodA sequence closest to this clade comes from the highly divergent and atypical A. caulinodans, although this finding may have resulted from a long branch attraction artifact (4). B. tuberum and Methylobacterium nodulans fell into a separate and strongly supported cluster. Interestingly, the β-rhizobia R. taiwanensis and B. caribensis and the α-rhizobia Rhizobium sp. strains TJ167 and TJ172 isolated from M. diplotricha clustered separately in the nodA tree, suggesting that their nodulation genes have different origins.
FIG. 5.
Unrooted nodA phylogenetic tree of rhizobia. β-Proteobacterial strains are in shown in bold type and underlined. The tree was reconstructed by using an ML approach based on a 597-bp alignment (excluding the additional segment at the N-terminal part). Sequence lengths included ranged from 558 bp (B. phymatum STM815; partial sequence) to 597 bp (most strains). Values along branches indicate bootstrap percentages higher than 50%, based on 100 replicates. nodA sequences for published bacteria are available from GenBank. EMBL accession numbers and nodA sequences for unpublished bacteria were AJ505318 (B. phymatum), AJ505311 (R. taiwanensis), AJ505309 (B. caribensis), AJ300229 (S. terangae bv. acaciae), AJ300249 (M. plurifarium), AJ505307 (TJ172), AJ300234 (BR816), AJ505305 (TJ167), AJ300242 (A. undicola), AJ302321 (B. tuberum), AJ303088 (STM259), AJ430707 (WU425), AJ430730 (CBP70), AJ430715 (ORS938), AJ430712 (USDA3139), AJ430728 (CCT6220), AJ300260 (STM270), AJ300247 (M. ciceri), AJ300228 (S. terangae bv. sesbaniae), and J300235 (R. huautlense).
The clustering of the β-rhizobial sequences in different nodA lineages intertwined with α-rhizobial sequences suggested that multiple nod gene transfers have occurred between the two subclasses of proteobacteria. Indeed, a single transfer of nodulation genes between α- and β-proteobacteria would have led to a single branch of β-proteobacteria within the rest of the tree, which is composed of α-proteobacteria. Constraining the four β-rhizobial symbionts to the same clade led to a tree that was only marginally less likely than the ML tree (P value, 0.083, as determined by the Shimodeira-Hasegawa test implemented in PAUP) and thus did not clearly support or infer the hypothesis of multiple nod gene transfers. On the other hand, the different lengths of the NodA proteins from B. tuberum and R. taiwanensis that clustered in two clades support the hypothesis of different origins for the corresponding genes. The NodA sequence from B. tuberum possesses at the N terminus an additional 13-amino-acid segment that is characteristic of bradyrhizobial NodA sequences (9a), while R. taiwanensis and the genera Azorhizobium, Sinorhizobium, Mesorhizobium, and Rhizobium all lack this NodA N-terminal extension. Moreover, the similarity between B. caribensis TJ182 and R. taiwanensis LMG 19424 nodA sequences (97.4% identity) indicates that nod gene transfer may have occurred between β-proteobacteria, as already suggested for α-proteobacteria (16).
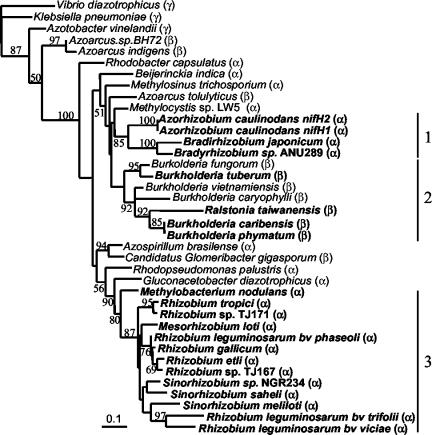
Interestingly, the phylogeny of the nitrogen fixation gene nifH provides a representation of the rhizobia different from that of the phylogeny of nodA (Fig. 6). Indeed, some of the groupings within the nifH tree corresponded to the phylogeny of the organisms as deduced from comparative 16S rDNA analysis, although α- and β-proteobacteria did not form distinct and monophyletic clades. Moreover, the nifH tree grouped together free-living and symbiotic nitrogen-fixing Burkholderia and Ralstonia strains. An example of a representative organism is B. tuberum, which grouped with M. nodulans in the nodA phylogeny but grouped with other β-proteobacteria in the nifH phylogeny. Constraining either α-and β-rhizobia or M. nodulans and B. tuberum to the same clade led to a tree that was statistically less probable than the ML tree (both with P values of <10−4). These results suggest that nod and nif genes of β-rhizobia have different origins.
FIG. 6.
nifH phylogenetic tree. The tree was reconstructed by using an ML approach based on an 800-bp alignment matrix (partial and full sequence lengths ranged from 336 to 797 bp). Values along branches indicate bootstrap percentages higher than 50%. The tree was rooted by using sequences from V. diazotrophicus, K. pneumoniae, and A. vinelandii. Rhizobia are shown in bold type, and the α-, β-, or γ-proteobacterial classification is indicated in parentheses. Clusters 1 and 3 contain α-rhizobia only, while cluster 2 includes both symbiotic and nonsymbiotic diazotrophic β-proteobacteria. nifH sequences for published bacteria are available from GenBank EMBL. EMBL accession numbers and nifH sequences for unpublished bacteria were AJ302315 (B. tuberum), AJ505320 (R. taiwanensis), AJ512206 (B. vietnamensis), AJ512207 (B. caryophylli), AJ505317 (B. caribensis), AJ505319 (B. phymatum), and AJ512205 (M. nodulans). nifH sequences from B. fungorum, R. palustris, and R. leguminosarum were from partially completed genome Web sites.
DISCUSSION
In this study, we have confirmed and extended the phylogenetic diversity of rhizobia initially presented in articles by Moulin et al. (10) and Chen et al. (1). We have identified as rhizobia two additional Burkholderia strains as well as at least 48 different R. taiwanensis strains isolated from M. pudica and M. diplotricha. Representative Burkholderia and Ralstonia strains fix nitrogen in symbiosis with their respective host plants, demonstrating that the root nodule β-proteobacteria are indeed true rhizobia. Detailed studies have shown that R. taiwanensis-induced nodule ontogeny and development are similar to those described for other, mimosa-like legumes (1a). Moreover, R. taiwanensis is the favored partner of M. pudica and M. diplotricha in Taiwan, indicating that nodulation by β-proteobacteria is not a rare phenomenon exhibited by certain opportunistic strains. In this respect, it should be noted that R. taiwanensis strains also have been isolated from M. pudica in India (20).
The widespread character of nodulation by β-proteobacteria is also attested to by the phylogenetic diversity of the β-rhizobia identified so far, including one Ralstonia species (1) andthree Burkholderia species (B. caribensis, B. tuberum, and B. phymatum) (24), as well as the fact that they have been isolated from Asia, Africa, and South America. Since many legumes and environments remain to be explored, it is highly likely that further characterization of rhizobia will reveal an even greater diversity. For many decades, standard isolation procedures have been used for rhizobia, and identification as legume symbionts through nodulation tests has required the availability of host plant seeds. These traditional approaches, coupled with the difficulty of obtaining seeds for many tropical legumes, have probably contributed to masking of the natural diversity of rhizobia.
Nitrogen fixation, which is widespread in eubacteria and archaea, is thought to be an ancestral function now lost by many bacteria (3). Conversely, nodulation is thought to have appeared recently in evolution, at the same time as the appearance of legumes on Earth, about 70 to 130 millions years ago. At that period of history, the α- and β-proteobacteria and the different rhizobial lineages already had diverged (22). The genes required for legume nodulation are thought to have been acquired subsequently by lateral transfer from undefined sources, thus converting soil saprophytes into symbionts (7). This hypothesis has been confirmed by recent data (6, 15). The presence in α- and β-rhizobia of very similar and phylogenetically related nodABC genes strongly supports the hypothesis of a unique origin for the common nod genes. However, it is not clear whether a single transfer event was responsible for the spread of nodulation genes from one subclass to the other or whether recurrent transfers occurred between the two subclasses. Our phylogenetic and NodA length analyses together suggest the occurrence of at least two lateral transfers between these two unrelated subclasses of proteobacteria, although statistical analysis did not allow this hypothesis to be ascertained. Further identification of other β-rhizobia may be useful for confirming such a hypothesis. Moreover, a comparative analysis of the nodA, nifH, and 16S rDNA trees suggests that β-rhizobia emerged through the transfer of nod genes to diazotrophic β-proteobacteria. Since the nod and nif genes are located on the same plasmid in the β-rhizobia investigated, it is possible that exogenous nod and nif genes were cotransferred prior to the replacement of the exogenous nifH gene by the indigenous gene. This level of complexity is in line with the highly complex evolutionary history of the legume-rhizobium symbiosis.
Acknowledgments
We thank Y. Prin for help in microscopy studies and M. Dukhan for help with photographs. We also thank D. Barker and J. Batut for comments and suggestions.
W.-M. Chen was supported by a grant from the National Science Council, Taipei, Taiwan, Republic of China (NSC 91-2320-B-022-001).
REFERENCES
- 1.Chen, W. M., S. Laevens, T. M. Lee, T. Coenye, P. de Vos, M. Mergeay, and P. Vandamme. 2001. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51:1729-1735. [DOI] [PubMed] [Google Scholar]
- 1a.W.-M. Chen, E. K. James, A. R. Prescott, M. Kierans, and J. I. Sprent. Nodulation of Mimosa spp. by the β-proteobacterium Ralstonia taiwanensis. Mol. Plant-Microbe Interact., in press. [DOI] [PubMed]
- 2.Deschodt, C. C., and B. W. Strijdom. 1976. Effective nodulation of Aspalathus linearis by rhizobia from other Aspalathus species. Phytophylactica 8:103-104. [Google Scholar]
- 3.Fani, R., R. Gallo, and P. Lio. 2000. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J. Mol. Evol. 51:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27:401-410. [Google Scholar]
- 5.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. University of Washington, Seattle.
- 6.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lalaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch, A. M., M. R. Lum, and J. A. Downie. 2001. What makes the rhizobia-legume symbiosis so special? Plant Physiol. 127:1484-1492. [PMC free article] [PubMed] [Google Scholar]
- 8.Lortet, G., N. Méar, J. Lorquin, B. Dreyfus, P. de Lajudie, C. Rosenberg, and C. Boivin. 1996. Nod factor thin-layer chromatography profiling as a tool to characterize symbiotic specificity of rhizobial strains: application to Sinorhizobium saheli, S. teranga and Rhizobium sp. strains isolated from Acacia and Sesbania. Mol. Plant-Microbe Interact. 9:736-747. [Google Scholar]
- 9.Martinez-Romero, E., and J. Caballero-Mellado. 1996. Rhizobium phylogenies and bacterial genetic diversity. Crit. Rev. Plant Sci. 15:113-140. [Google Scholar]
- 9a.L. Moulin, G. Béna, C. Boivin-Masson, and T. Stpkowski. Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 10.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 11.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 12.Parker, M. A., and A. Lunk. 2000. Relationships of bradyrhizobia from Platypodium and Machaerium (Papilionoideae: tribe Dalbergieae) on Barro Colorado Island, Panama. Int. J. Syst. Evol. Microbiol. 50:1179-1186. [DOI] [PubMed] [Google Scholar]
- 13.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultze, M., and A. Kondorosi. 1998. Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32:33-57. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suominen, L., C. Roos, G. Lortet, L. Paulin, and K. Lindstrom. 2001. Identification and structure of the Rhizobium galegae common nodulation genes: evidence for horizontal gene transfer. Mol. Biol. Evol. 18:907-916. [DOI] [PubMed] [Google Scholar]
- 17.Swofford, D. 1998. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 18.Sy, A., E. Giraud, P. Jourand, N. Garcia, A. Willems, P. de Lajudie, Y. Prin, M. Neyra, M. Gillis, C. Boivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi, A. K. 2002. Rhizobia of the β-subclass of Proteobacteria: a tale of losing the race. Curr. Sci. 82:8-9. [Google Scholar]
- 21.Truchet, G., S. Camut, F. de Billy, R. Odorico, and J. Vasse. 1989. The Rhizobium-legume symbiosis. Protoplasma 149:82-88. [Google Scholar]
- 22.Turner, S. L., and J. P. W. Young. 2000. The glutamine synthetases of rhizobia: phylogenetics and evolutionary implications. Mol. Biol. Evol. 17:309-319. [DOI] [PubMed] [Google Scholar]
- 23.van Berkum, P., and B. D. Eardly. 1998. Molecular evolutionary systematics of the Rhizobiaceae, p. 1-24. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Molecular biology of plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 24.Vandamme, P., J. Goris, W. M. Chen, P. de Vos, and A. Willems. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X. X., S. L. Turner, X. W. Guo, H. J. Yang, F. Debellé, G. P. Yang, J. Dénarié, J. P. W. Young, and F. D. Li. 2000. The common nodulation genes of Astragalus sinicus rhizobia are conserved despite chromosomal diversity. Appl. Environ. Microbiol. 66:2988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]