Abstract
Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors that are implicated in the regulation of lipid and glucose homeostasis. PPAR agonists have been shown to control inflammatory processes, in part by inhibiting distinct proinflammatory genes (e.g. Il-1β and IFN-γ). IL-8 is a member of the proinflammatory chemokine family that is important for various functions, such as mediating the adhesion of eosinophilic granulocytes onto endothelial cells. The influence of PPARδ activators on the expression of IL-8 in noninduced quiescent endothelial cells is unclear. Therefore, we explored the influence of PPARδ activators on the expression of IL-8 in nonstimulated endothelial cells. PPARδ agonists induce IL-8 expression in human umbilical vein endothelial cells. This induction is demonstrated at the level of both protein and mRNA expression. Transcriptional activation studies using IL-8 reporter gene constructs and DNA binding assays revealed that PPARδ agonists mediated their effects via an NFκB binding site. It is well known that IL-8 is also regulated by mRNA stability. To provide further evidence for this concept, we performed mRNA stability assays and found that PPARδ agonists induce the mRNA stability of IL-8. In addition, we showed that PPARδ agonists induce the phosphorylation of ERK1/2 and p38, which are known to be involved in the increase of mRNA stability. The inhibition of these MAPK signaling pathways resulted in a significant suppression of the induced IL-8 expression and the reduced mRNA stability. Therefore, our data provide the first evidence that PPARδ induces IL-8 expression in nonstimulated endothelial cells via transcriptional as well as posttranscriptional mechanisms.
Keywords: Interleukin, mRNA, NFκB, PPAR, Promoters
Introduction
Peroxisome proliferator-activated receptors (PPARs)2 are members of the nuclear receptor/ligand-activated transcription factors superfamily and consist of three different subtypes: PPARα, PPARδ, and PPARγ. The role of these receptors was originally thought to be restricted to lipid and lipoprotein metabolism, glucose homeostasis, and cellular differentiation (1, 2). PPARs are activated by natural ligands, such as eicosanoids and fatty acids. In addition, synthetic antidiabetic thiazolidinediones and lipid-lowering fibrates have been shown to act as activators of PPARγ and PPARα, respectively (3, 4). To date, PPARδ, which is expressed in almost all tissues, is the subtype that still remains an interesting target for new pharmaceutical drugs.
New evidence suggests that PPARδ plays a crucial role in the regulation of differentiation, cell growth, and the metabolism of lipids and glucose (5, 6). With respect to the development of novel drugs, the effects and side effects of PPARδ activators are therefore of great importance.
In the last few years, knowledge concerning the impact of PPARδ in endothelial cell function has increased. During inflammation, proinflammatory stimuli such as LPS, TNFα, or IL-1β lead to a phenotypic change of the quiescent endothelium by induction of proinflammatory factors, such as IL-6, IL-8, MCP-1, or adhesion molecules, such as ICAM-1 or VCAM-1. Recently, Rival et al. demonstrated that the PPARδ activator L165041 suppresses TNFα-induced VCAM-1 and MCP-1 expression (7). Similar results were demonstrated by Fan et al., revealing that the PPARδ agonist GW501516 suppresses IL-1β-induced VCAM-1 and E-selectin expression in human umbilical vein endothelial cells (HUVECs) (8). Piqueras et al. demonstrated that PPARδ inhibits leukocyte recruitment and cell adhesion molecule expression (9). Recently, Liang et al. showed that the PPARδ agonist l-165041 suppresses C-reactive protein-induced IL-6 expression (10). The expression profile of both cytokines during treatment with various PPARδ agonist concentrations in the endothelial quiescent status has yet to be analyzed. Nonetheless, these studies showed that PPARδ agonists could suppress the inflammatory processes in activated endothelial cells. The impact of PPAR agonists on the normally quiescent endothelium, however, remains to be elucidated. This could be of significant importance due to the possible broad range of applications of PPARδ agonists in various diseases, such as chronic inflammation, glucose metabolism, dyslipidemia, obesity, and cancer therapy, to mention only a small number of possible medical applications.
During the process of inflammation, proinflammatory cytokines are produced by various cell types. IL-8 is one of these important cytokines. It is secreted at very low levels from noninduced cells, although the secretion is increased rapidly by a wide range of stimuli, such as TNF and IL-1 or bacterial products, such as LPS (11). There are two main methods of regulation: first, by transcriptional activation of the genes by NFκB, and second, by stabilization of the mRNA by the p38 MAPK pathway (12–16).
Besides the important proinflammatory action, IL-8 plays an important role in the tumor microenvironment. Secretion of IL-8 from cancer cells can aggravate the proliferation and survival of cancer cells, in part by autocrine signaling pathways (17). In addition, tumor-derived IL-8 can activate endothelial cells to promote angiogenesis. IL-8, however, is secreted not only by tumor cells, but also by endothelial cells, thereby enhancing endothelial cell survival, proliferation, and angiogenesis (18).
The present study investigated the influence of PPARδ activators on the production, secretion, and regulation of IL-8 in nonstimulated endothelial cells, revealing an induction of IL-8 expression conveyed by transcriptional, NFκB-dependent, and posttranscriptional mechanisms involving mRNA stability.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant human TNFα and IL-1β were purchased from R&D Systems (Minneapolis, MN). L165041, GW501546, PD98059, SB203580, LY294002, and actinomycin D were obtained from Sigma-Aldrich.
Cell Culture
HUVECs were purchased from PromoCell (Heidelberg, Germany) and were cultured until the fifth passage at 37 °C and 5% CO2 in endothelial cell growth medium (Cambrex, East Rutherford, NJ).
Enzyme-linked Immunosorbent Assay (ELISA)
The concentrations of IL-8 and IL-6 in cell culture supernatants were determined by ELISA. Commercially available ELISA kits (R&D Systems) for the quantification of IL-8 and IL-6 were used as described in the manufacturer's instructions.
Western Blot Analysis
Whole cell protein and cytoplasmic and nuclear protein extracts were prepared as described previously (19). Membranes were incubated with the indicated primary antibodies. Antibodies were as follows: anti-p-p65, anti-IκB-α, anti-phospho-IκB-α anti-p65, anti-ERK1/2, anti-phospho-ERK1/2, anti-phospho-p38, anti-p38, anti-phospho-Akt (Ser473) anti-phospho-Akt (Thr308), anti-Akt, and anti-SP1 from Santa Cruz Biotechnology (Heidelberg, Germany) and anti-tubulin from LabVision (Fremont, CA). Primary antibody application was followed by incubation with horseradish peroxidase-conjugated secondary antibodies (anti-mouse and anti-rabbit IgG, Amersham Biosciences; anti-goat, Dako, Glostrup, Denmark). Blots were developed using an enhanced chemiluminescence detection system (ECL) (Amersham Biosciences), according to the manufacturer's instructions.
Cytotoxicity Assay
The cytotoxic potential of L165041 was determined using the Cytotoxicity Detection kit (lactate dehydrogenase) from Roche, according to the manufacturer's instructions. Twenty-four hours after seeding, cells were exposed to L165041 for 24 h at the indicated concentrations.
RNA Extraction and RT-PCR
RT-PCR analyses were performed using total RNA (150 ng) extracted from subconfluent cell cultures. Total cellular mRNA was isolated by the RNeasy Mini Procedure (Qiagen, Hilden, Germany) after DNase digestion. RT-PCR analyses for IL-6, IL-8, and GAPDH were performed using the One-Step RT-PCR kit (Qiagen). PCR products were resolved by 1–2% agarose gel electrophoresis, and ethidium bromide-stained bands were visualized with an ultraviolet transilluminator. A densitometric analysis was used to quantify band intensities using the public domain Java image-processing program ImageJ (v1.29s). Optical densities (ODs) of the IL-8 bands were corrected for loading differences based on the corresponding GAPDH bands. The primer sets for IL-8, IL-6, and GAPDH have been published previously (20).
Transient Transfection and Analysis of Reporter Gene Expression
HUVECs (1.0 × 105 cells/well in 12-well plates) were transfected with 0.5 μg of the appropriate firefly luciferase construct and 0.1 μg of phRG-TK vector (Promega, Madison, WI) using the SuperFect transfection reagent (Qiagen). Human IL-8 reporter gene constructs were generously provided by Dr. Naofumi Mukaida (Division of Molecular Bioregulation, Cancer Research Institute, Kanazawa University, Japan). Twenty-four hours after transfection, cells were treated with vehicle (0.3% dimethyl sulfoxide) or with L165041 for 24 h. Luciferase activities were measured with the Dual-Luciferase Reporter Assay system (Promega).
Preparation of Nuclear Extracts and Gel Mobility Shift Analysis
HUVECs were treated with vehicle or incubated with L165041 for 60 min. Nuclear proteins were extracted as described previously (21). DNA binding reactions were performed with or without excess unlabeled NFκB consensus oligonucleotide (Promega) and p65 antibody (Santa Cruz Biotechnology). The electromobility shift analysis (EMSA) was performed as described previously (21).
Immunolocalization of p65 NFκB
For immunolocalization studies, HUVECs were plated onto 8-well chamber slides (Lab-Tek, Christchurch, New Zealand) at a density of 80%/well. The cells were grown in endothelial cell basal medium with 5% FCS for 24 h, fixed in 4% paraformaldehyde at room temperature for 10 min, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Permeabilized cells were rinsed three times with PBS and incubated in blocking solution (1% bovine serum albumin/PBS) for 30 min at room temperature to remove nonspecific binding of the antibody. All subsequent steps were carried out at room temperature, and cells were rinsed three times in 1% bovine serum albumin/PBS between each of the steps. RelA/p65 was detected using a rabbit polyclonal anti-RelA/p65 antibody (C-20; Santa Cruz Biotechnology) and an Alexa Fluor 594-conjugated goat anti-rabbit IgG secondary antibody. The slides were mounted in Vectashield immunofluorescence mouting medium (Vector Laboratories) and viewed with fluorescence microscopy. Cells were counterstained for nuclei with Hoechst staining.
Statistical Analyses
The data are expressed as means ± S.D. from at least three independent experiments. Statistical analyses were performed using Student's t test.
RESULTS
PPARδ Agonists Induce IL-8 Expression in Nonstimulated Endothelial Cells
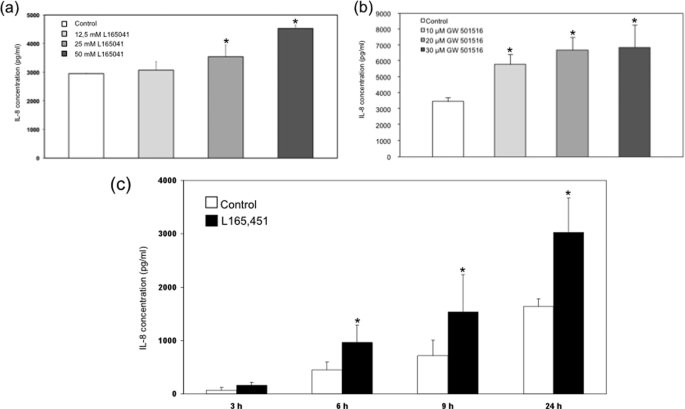
To evaluate the influence of the PPARδ agonists L165041 and GW501516 on quiescent nonstimulated endothelial cells, ELISA analyses were performed. Interestingly, these experiments demonstrated a time- and concentration-dependent induction of IL-8 expression in nonstimulated HUVECs treated with L165041 and GW501516, respectively (Fig. 1). The results were comparable for both PPARδ agonists, indicating that the effects are specific to PPARδ. To analyze whether the concentrations used have any cytotoxic effects, we performed cytotoxicity lactate dehydrogenase assays with our model compound, L165041, and found no relevant cytotoxic effects (supplemental Fig. 1). In further experiments we could demonstrate comparable results analyzing the expression of IL-6 in HUVEC treated with L165041 and GW501516, respectively (supplemental Fig. 2).
FIGURE 1.
Effects of GW501516 and L165041 on the IL-8 expression in supernatants of HUVEC. IL-8 protein content was assayed in culture supernatants by IL-8 ELISA (R&D Systems) according to the manufacturer's instructions. a and b, HUVECs were left untreated (solvent only, 0.1% dimethyl sulfoxide) or were treated with L165041 (a) and GW501516 (b) for 24 h at the indicated concentrations. c, HUVECs were left untreated (solvent only, 0.1% dimethyl sulfoxide) or were treated with L165041 for the indicated times. Mean values from triplicate experiments are depicted ± S.E. (error bars). *, p < 0.05 was considered significant.
PPARδ Agonists Up-regulate the Steady State of IL-8 mRNA
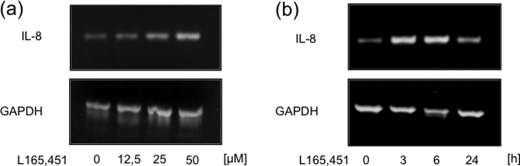
To determine whether activation of PPARδ plays a role in the steady state of IL-8 mRNA expression, we performed RT-PCR analysis after treatment with the PPARδ agonist L165041. Consistent with our protein expression data, application of the agonist induced IL-8 mRNA expression in a concentration- and time-dependent manner (Fig. 2). Comparable results could be demonstrated for IL-6 (supplemental Fig. 3). The expression of IL-8 can be regulated in a transcriptional and a posttranscriptional manner. We therefore first examined a possible transcriptional mechanism of control.
FIGURE 2.
IL-8 mRNA expression is induced by PPARδ agonists. RT-PCR analyses of total mRNA extracted from HUVECs that were treated with vehicle or L165041 for varying times (a) or varying concentrations (b) are shown. Results were confirmed in three independent sets of experiments.
PPARδ Agonists Up-regulate IL-8 Promoter Activity in an NFκB-dependent Manner
To analyze the underlying molecular mechanisms that mediate PPARδ-conveyed induction of IL-8 mRNA expression, luciferase reporter assays were employed. Luciferase reporter constructs containing the 5′-region of the IL-8 promoter and a series of deletion constructs were transiently transfected into vehicle- and PPARδ agonist-treated HUVECs.
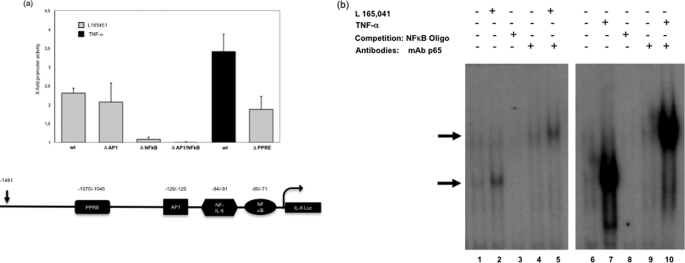
Analysis of the expression of luciferase in vehicle and PPARδ agonist-treated cells revealed an approximate 2.3-fold induction of basal luciferase activity with the wild type IL-8 construct (Fig. 3a). Deletion of the AP1 site did not significantly influence IL-8 promoter activity, whereas the deletion of NFκB led to a complete loss (Fig. 3a). Recently, the IL-8 promoter was shown to contain a PPRE site. To exclude the possibility that this site is also an important point of action of PPARδ agonists, we analyzed an IL-8 promoter construct with a deletion of the PPRE site. Interestingly, the deletion did not significantly influence luciferase promoter activity during treatment with PPARδ agonists, demonstrating that the inducing effect of IL-8 is conveyed only by the NFκB site (Fig. 3a).
FIGURE 3.
PPARδ agonists induce IL-8 transcription in an NFκB-dependent manner. a, analyses of deletional IL-8 luciferase (Luc) reporter constructs in HUVECs. The Luc activities are expressed as x-fold activity compared with untreated controls (mean ± S.D. (error bars) of five independent triplicate assays). b, representative EMSAs using nuclear extracts of vehicle- and L165041-treated (50 μmol/liter, 1 h) HUVECs. Competition with unlabeled excess double-stranded NFκB consensus oligonucleotides is shown in lanes 3 and 8 (0.35 μmol/liter). A representative autoradiography from three independent experiments is presented. Supershift analyses were performed by addition of a p65-specific antibody (lanes 4 and 5 and lanes 9 and 10; 100 ng/μl). Arrows on the left indicate NFκB-containing transcription factor-binding complexes. Comparable results were obtained from three independent experiments.
NFκB-dependent Binding to the IL-8 Promoter Is Induced by PPARδ Agonist Treatment
To determine the nuclear factors that bind to the NFκB binding site, we performed EMSAs using nuclear extracts from HUVECs and a 32P-labeled NFκB oligonucleotide probe. In untreated HUVECs, a distinct complex was observed to bind and shift the migration pattern of the oligonucleotide (Fig. 3b, lane 1). A significant increase in DNA binding activity, however, was observed in the lysates of cells treated with L165041 (Fig. 3b, lane 2) and in cells treated with TNFα as a control (Fig. 3b, lane 6). A supershift was observed upon addition of the p65 antibody (Fig. 3b, lanes 4 and 5 and lanes 9 and 10), confirming that p65 binds to the IL-8 promoter. Competition assays using excess unlabeled oligonucleotide supported the assumption that nuclear proteins bind to the IL-8 promoter sequence in an NFκB-specific manner (Fig. 3b, lanes 3 and 8).
PPARδ Agonists Lead to the Phosphorylation and Nuclear Translocation of NFκB
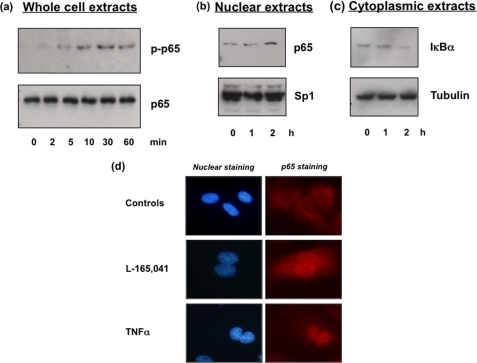
After obtaining data suggesting that PPARδ agonists convey the IL-8 induction in an NFκB-dependent manner, we analyzed the phosphorylation status of p65 in our PPARδ agonist-treated HUVECs. Interestingly, an increase in p65 phosphorylation and p65 nuclear translocation was detected by Western blot analysis (Fig. 4, a and b). Consistent with these results, we could demonstrate an increase in IκBα degradation (Fig. 4c). In addition, we found a significant nuclear translocation of p65 by immunofluorescence staining of NFκB p65 in HUVECs treated with PPARδ agonists (Fig. 4d). The increase in p65 staining was comparable with the influence of TNFα.
FIGURE 4.
PPARδ agonists induce p65 phosphorylation and nuclear translocation. Representative Western blot analysis of HUVECs that were treated with vehicle or L165041 (50 μm) for the indicated times is shown. Protein was separated by 8% SDS-PAGE, and proteins were detected by enhanced chemiluminescence. All comparable results were obtained from three independent experiments. a, total cellular protein showing the amount of phospho-p65 and p65. b, nuclear protein showing the amount of p65. Sp1 protein served as a control. c, cytoplasmic protein showing the amount of IκBα. Tubulin protein served as a control. d, representative immunofluorescent analysis of p65 in HUVECs that were treated with vehicle, L165041 (50 μm), or TNFα (20 ng/ml). Comparable results were obtained from three independent experiments.
PPARδ Agonists Induce IL-8 mRNA Stability
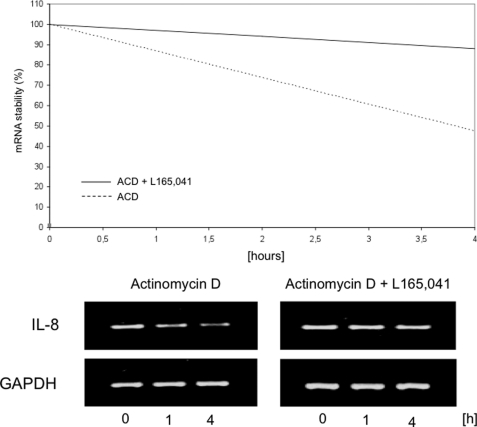
IL-8 expression is not only controlled on the transcriptional level, but can be also influenced by the stability of IL-8 mRNA. Therefore, we next analyzed IL-8 mRNA stability in solvent- and PPARδ agonist-treated endothelial cells. Surprisingly, a significant increase in IL-8 mRNA stability was found (Fig. 5), providing an additional mechanism for the induction of IL-8 expression.
FIGURE 5.
IL-8 mRNA half-life is induced by PPARδ agonists. HUVEC were incubated with vehicle or L165041 (50 m) for 1 h, followed by incubation with fresh medium containing actinomycin D (10 μg/ml) for 0, 1, and 4 h. RT-PCR analyses for IL-8/GAPDH of total RNA extracted from subconfluent cell cultures were performed. The PCR products were separated by 2% agarose gel electrophoresis, and ethidium bromide-stained bands were visualized using an ultraviolet transilluminator. IL-8 bands were quantified by densitometric scanning, the results of which were normalized to amounts of GAPDH mRNA. Comparable results were obtained from three independent experiments.
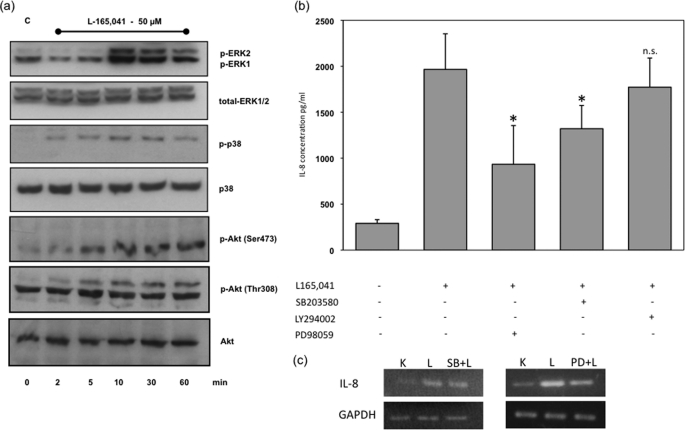
PPARδ Agonists Induce ERK1/2, p38, and Akt Phosphorylation
It is known that the p38 and ERK1/2 MAPK signaling pathways are involved in the regulation of the mRNA stability of IL-8. We therefore analyzed the phosphorylation status of MAPK ERK1/2 and p38 as well as Akt by Western blot analysis. Interestingly, the MAPK ERK1/2 and p38 were quickly phosphorylated after treatment with PPARδ agonists, comparable results could be demonstrated for Akt (Fig. 6a). To analyze whether these signaling pathways influence PPARδ-induced IL-8 protein expression, we blocked the ERK1/2, p38, and Akt signaling pathways using the specific inhibitors PD98059, SB203580, and LY294002, respectively. Except for Akt signaling, these treatments led to partial and significant neutralization of PPARδ agonist-induced IL-8 expression, demonstrating the importance of the p38 and ERK1/2 MAPK signaling pathways in PPARδ-mediated IL-8 expression (Fig. 6b). The phosphorylation of Akt seems to have no significant influence on IL-8 expression.
FIGURE 6.
PPARδ agonists activate MAPK signaling pathways. a, representative Western blot analysis of HUVECs that were treated with vehicle or L165041 (50 μm) for the indicated times up to 60 min. Total cellular protein was separated by 8% SDS-PAGE. ERK1/2, phospho-ERK1/2, p38, phospho-p38, Akt and phospho-Akt (Thr308 and Ser473) protein were detected by enhanced chemiluminescence. Comparable results were obtained from three independent experiments. b, effects of MAPK signaling pathway inhibition on L165041-induced IL-8 expression in supernatants of HUVEC. HUVECs were pretreated for 30 min with 30 μm PD98059, 1 μm SB203580, and 10 μm LY294002 and then exposed to 50 μm L165041 for 6 h. The amount of IL-8 released by HUVECs into the culture medium was assessed by ELISA. Values are expressed as mean ± S.E. (error bars) (n = 4). *, p < 0.05 was considered significant. c, effects of MAPK signaling pathway inhibition on L165041-induced IL-8 mRNA expression in HUVECs. HUVECs were pretreated 30 min with 30 μm PD98059 and 1 μm SB203580 and then exposed to 50 μm L165,041 for 6 h. The amount of IL-8 mRNA expression by HUVEC was assessed by RT-PCR. Results were confirmed in three independent sets of experiments.
These results could also be bolstered by mRNA analysis. To analyze whether p38 and ERK1/2 influence PPARδ-induced IL-8 mRNA expression, we blocked the ERK1/2 and p38 signaling pathways using the specific inhibitors PD98059 and SB203580, respectively. These treatments led to partial neutralization of PPARδ agonist-induced IL-8 mRNA expression (Fig. 6c).
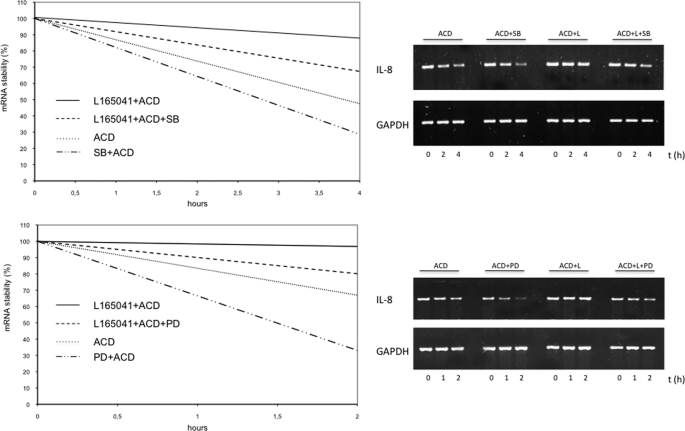
Inhibition of MAPK Signaling Pathways p38 and ERK1/2 Results in Decreased IL-8 mRNA Stability
To provide evidence that the MAPK signaling pathways influence PPARδ-induced IL-8 mRNA expression and stability, we performed mRNA stability assays while inhibiting the MAPK signaling pathways. We demonstrated that blockade of the ERK1/2 and p38 signaling pathways using PD98059 and SB203580, respectively, resulted in a reduced mRNA stability compared with the PPARδ agonist only (Fig. 7).
FIGURE 7.
Inhibition of MAPK signaling pathways reduces IL-8 mRNA stability. Top, HUVECs were incubated with vehicle, L165041 (50 μm), SB203580 (30 mm), and L165041 + SB203580 for 30 min, followed by incubation with fresh medium containing actinomycin D (10 μg/ml) for 0, 1, and 4 h. RT-PCR analyses for IL-8/GAPDH of total RNA extracted from subconfluent cell cultures were performed. The PCR products were separated by 2% agarose gel electrophoresis, and ethidium bromide-stained bands were visualized using an ultraviolet transilluminator. IL-8 bands were quantified by densitometric scanning, the results of which were normalized to amounts of GAPDH mRNA. Comparable results were obtained from three independent experiments. Bottom, HUVECs were incubated with vehicle, L165041 (50 μm), PD98059 (30 mm), and L165041 + PD98059 for 30 min, followed by incubation with fresh medium containing actinomycin D (10 μg/ml) for 0, 1, and 2 h. RT-PCR analyses for IL-8/GAPDH of total RNA extracted from subconfluent cell cultures were performed. The PCR products were separated by 2% agarose gel electrophoresis, and ethidium bromide-stained bands were visualized using an ultraviolet transilluminator. IL-8 bands were quantified by densitometric scanning, the results of which were normalized to amounts of GAPDH mRNA. Comparable results were obtained from three independent experiments.
DISCUSSION
In the last decade, knowledge concerning PPAR activators has increased rapidly. The research community has primarily focused on PPARα and PPARγ agonists, which are already in clinical use. In the last 2 years, however, PPARδ agonists have raised significant interest, even though the research concerning this nuclear receptor and its ligands has been limited. Emerging evidence has demonstrated that PPARδ plays an important role in the regulation of differentiation, cell growth, and lipid and glucose metabolism (5, 22). Recently, the impact of PPARδ agonists in endothelial cell function and angiogenesis has been addressed (24). These results highlight the growing importance and the possible expanding area of employing PPARδ agonists for clinical use.
In the current study, we demonstrated that PPARδ agonists effectively induced IL-8 expression in nonstimulated quiescent endothelial cells. We identified two responsible mechanisms underlying this effect: a transcriptional mechanism conveyed by NFκB and a posttranscriptional mechanism mediated by a p38- and ERK1/2-dependent increase in mRNA stability. The influence of the major transcription factor NFκB on IL-8 expression is an established method of regulation (11, 25). Important proinflammatory cytokines, such as TNFα or IL-1β, as well as LPS are known to induce IL-8 expression via NFκB-containing transcription factor complexes targeting the IL-8 promoter (25–27). To our knowledge, this is the first study to demonstrate that PPARδ agonists lead to a concentration-dependent phosphorylation and nuclear translocation of p65 in nonstimulated endothelial cells, thereby increasing NFκB DNA binding. In addition, we found that the PPRE element at the IL-8 promoter did not significantly influence IL-8 promoter activity. These data are contrary to those obtained for stimulated endothelial cells. Liang et al. convincingly demonstrated that C-reactive protein in stimulated endothelial cells suppresses NFκB phosphorylation when the cells are treated with the PPAR agonist GW501516, thereby resulting in reduced IL-6 and IL-8 expression (10). A similar result was demonstrated by Rodriguez-Calvo et al. for LPS-stimulated adipocytes, in which reduced IL-6 expression and NFκB DNA binding activity were detected after the previously LPS-stimulated adipocytes were treated with the PPARδ activator GW501516 (28). The same group also showed an increase of IL-6 mRNA and protein in the control group treated only with GW501516 at very low PPARδ concentrations (28). These results were bolstered by our analysis concerning the expression of IL-6 in endothelial cells. In addition, Rival et al. demonstrated that the PPARδ activator L165041 inhibits TNFα-induced VCAM-1 and MCP1 expression at concentrations up to 100 μm (7). The important difference between those studies and the current work is that nonstimulated quiescent endothelial cells were used in the present study, revealing the possibility of a dual function of the PPARδ agonist in endothelial cells that is dependent on the activation status of the cell. This novel information may be important, because this would implicate a differentiated use of these compounds regarding their application in the treatment of various diseases. It is possible that this dual action also explains the different results concerning the influence of PPARδ activators in the treatment and development of various tumor entities. Recently, Bility et al. demonstrated that PPARδ agonists inhibit chemically induced skin carcinogenesis, whereas Kim et al. demonstrated that chemically induced skin carcinogenesis is not influenced by PPARδ (29, 30). Han et al. demonstrated that PPARδ agonists induce human lung cancer cell proliferation, whereas He et al. found that they do not influence lung cancer cell proliferation (31, 32). It is quite possible that the activation status of the cancer cells or the cancer microenvironment, including macrophages, fibroblasts, endothelial cells, and others, influences the way PPARδ exerts its effects.
It was recently demonstrated that PPARδ activation induces endothelial cell proliferation, VEGF production, and angiogenesis (33). These results are consistent with those of our study; however, we employed quiescent cells instead of cytokine-induced endothelial cells. We showed that PPARδ activation led to a proangiogenic and therefore a proinflammatory effect and that PPARδ activation led to the induction of VEGF expression. The induction of the proinflammatory and proangiogenic cytokine IL-8 by PPARδ agonists supports these results, as Li et al. demonstrated that IL-8 enhances endothelial cell survival and proliferation, regulates the production of matrix metalloproteinases, and can induce angiogenesis (18).
Aside from the mechanism involving the conveying of NFκB, we demonstrated an influence of PPARδ on the mRNA stability of IL-8. The regulation of IL-8 mRNA stability is a well accepted and important mechanism of IL-8 mRNA expression in various cell types and conditions (11, 34, 35). Many authors have demonstrated that the p38 MAPK signaling pathway, and to a minor extent ERK1/2, control IL-8 mRNA stability (13, 35–37). Here, we also demonstrated that the induction of p38 and ERK1/2 by PPARδ agonists resulted in an increase in IL-8 mRNA stability and therefore IL-8 production that could be partially abolished by inhibiting the MAPK signaling pathway. A comparable mechanism has already been described for a PPARα agonist. Ren et al. demonstrated that the activation of PPARα led to reduced mRNA stability and therefore a decrease of nephrin expression in kidney epithelial cells (23). Interestingly, PPARδ agonists activated Akt but failed to influence IL-8 expression by this signaling pathway.
Taken together, we demonstrated that in nonstimulated quiescent HUVECs, treatment with PPARδ agonists resulted in an NFκB-dependent transcriptional and posttranscriptional MAPK signaling pathway-dependent induction of IL-8 expression. These novel findings reveal that the effects of PPARδ agonists may depend on the activation status of endothelial cells and that this status is responsible for a more pro- or antiinflammatory action. These findings may influence the development of PPARδ agonists for clinical use.
Supplementary Material
This work was supported by the Wilhelm Sander Stiftung (M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- PPAR
- peroxisome proliferator-activated receptor
- HUVEC
- human umbilical vein endothelial cell
- ICAM
- intercellular adhesion molecule
- MCP
- monocyte chemoattractant protein
- VCAM
- vascular cellular adhesion molecule
- PPRE
- PPAR response element.
REFERENCES
- 1.Hansen M. K., Connolly T. M. (2008) Curr. Opin. Investig. Drugs 9, 247–255 [PubMed] [Google Scholar]
- 2.Torra I. P., Chinetti G., Duval C., Fruchart J. C., Staels B. (2001) Curr. Opin. Lipidol. 12, 245–254 [DOI] [PubMed] [Google Scholar]
- 3.Duan S. Z., Usher M. G., Mortensen R. M. (2009) Curr. Opin. Nephrol. Hypertens. 18, 128–133 [DOI] [PubMed] [Google Scholar]
- 4.Gross B., Staels B. (2007) Best Pract. Res. Clin. Endocrinol. Metab. 21, 687–710 [DOI] [PubMed] [Google Scholar]
- 5.Reilly S. M., Lee C. H. (2008) FEBS Lett. 582, 26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seedorf U., Aberle J. (2007) Biochim. Biophys. Acta 1771, 1125–1131 [DOI] [PubMed] [Google Scholar]
- 7.Rival Y., Benéteau N., Taillandier T., Pezet M., Dupont-Passelaigue E., Patoiseau J. F., Junquéro D., Colpaert F. C., Delhon A. (2002) Eur. J. Pharmacol. 435, 143–151 [DOI] [PubMed] [Google Scholar]
- 8.Fan Y., Wang Y., Tang Z., Zhang H., Qin X., Zhu Y., Guan Y., Wang X., Staels B., Chien S., Wang N. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 315–321 [DOI] [PubMed] [Google Scholar]
- 9.Piqueras L., Sanz M. J., Perretti M., Morcillo E., Norling L., Mitchell J. A., Li Y., Bishop-Bailey D. (2009) J. Leukocyte Biol. 86, 115–122 [DOI] [PubMed] [Google Scholar]
- 10.Liang Y.J., Liu Y. C., Chen C. Y., Lai L. P., Shyu K. G., Juang S. J., Wang B. W., Leu J. G. (2010) Int. J. Cardiol. 143, 361–367 [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. (2002) J. Leukocyte Biol. 72, 847–855 [PubMed] [Google Scholar]
- 12.Henness S., van Thoor E., Ge Q., Armour C. L., Hughes J. M., Ammit A. J. (2006) Am. J. Physiol. Lung Cell. Mol. Physiol. 290, L1283–L1290 [DOI] [PubMed] [Google Scholar]
- 13.Subramaniam D., Ramalingam S., May R., Dieckgraefe B. K., Berg D. E., Pothoulakis C., Houchen C. W., Wang T. C., Anant S. (2008) Gastroenterology 134, 1070–1082 [DOI] [PubMed] [Google Scholar]
- 14.Clifton D. R., Rydkina E., Huyck H., Pryhuber G., Freeman R. S., Silverman D. J., Sahni S. K. (2005) Int. J. Med. Microbiol. 295, 267–278 [DOI] [PubMed] [Google Scholar]
- 15.Miyazawa K., Mori A., Miyata H., Akahane M., Ajisawa Y., Okudaira H. (1998) J. Biol. Chem. 273, 24832–24838 [DOI] [PubMed] [Google Scholar]
- 16.Yang H. T., Cohen P., Rousseau S. (2008) Cell. Signal. 20, 375–380 [DOI] [PubMed] [Google Scholar]
- 17.Waugh D. J., Wilson C. (2008) Clin. Cancer Res. 14, 6735–6741 [DOI] [PubMed] [Google Scholar]
- 18.Li A., Varney M. L., Valasek J., Godfrey M., Dave B. J., Singh R. K. (2005) Angiogenesis 8, 63–71 [DOI] [PubMed] [Google Scholar]
- 19.Meissner M., Stein M., Urbich C., Reisinger K., Suske G., Staels B., Kaufmann R., Gille J. (2004) Circ. Res. 94, 324–332 [DOI] [PubMed] [Google Scholar]
- 20.Meissner M., Pinter A., Michailidou D., Hrgovic I., Kaprolat N., Stein M., Holtmeier W., Kaufmann R., Gille J. (2008) J. Invest. Dermatol. 128, 2084–2091 [DOI] [PubMed] [Google Scholar]
- 21.Meissner M., Reichenbach G., Stein M., Hrgovic I., Kaufmann R., Gille J. (2009) Cancer Res. 69, 1976–1984 [DOI] [PubMed] [Google Scholar]
- 22.Robinson E., Grieve D. J. (2009) Pharmacol. Ther. 122, 246–263 [DOI] [PubMed] [Google Scholar]
- 23.Ren S., Xin C., Beck K. F., Saleem M. A., Mathieson P., Pavenstädt H., Pfeilschifter J., Huwiler A. (2005) Biochem. Biophys. Res. Commun. 338, 1818–1824 [DOI] [PubMed] [Google Scholar]
- 24.Wang N. (2008) PPAR Res. 2008, 164163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida S., Ono M., Shono T., Izumi H., Ishibashi T., Suzuki H., Kuwano M. (1997) Mol. Cell. Biol. 17, 4015–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartels M., Schweda A. T., Dreikhausen U., Frank R., Resch K., Beil W., Nourbakhsh M. (2007) J. Immunol. 179, 7605–7613 [DOI] [PubMed] [Google Scholar]
- 27.Rochelson B., Dowling O., Schwartz N., Metz C. N. (2007) J. Reprod. Immunol. 73, 101–107 [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Calvo R., Serrano L., Coll T., Moullan N., Sánchez R. M., Merlos M., Palomer X., Laguna J. C., Michalik L., Wahli W., Vázquez-Carrera M. (2008) Diabetes 57, 2149–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bility M. T., Devlin-Durante M. K., Blazanin N., Glick A. B., Ward J. M., Kang B. H., Kennett M. J., Gonzalez F. J., Peters J. M. (2008) Carcinogenesis 29, 2406–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D. J., Prabhu K. S., Gonzalez F. J., Peters J. M. (2006) Carcinogenesis 27, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 31.Han S., Ritzenthaler J. D., Sun X., Zheng Y., Roman J. (2009) Am. J. Respir. Cell Mol. Biol. 40, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.He P., Borland M. G., Zhu B., Sharma A. K., Amin S., El-Bayoumy K., Gonzalez F. J., Peters J. M. (2008) Toxicology 254, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piqueras L., Reynolds A. R., Hodivala-Dilke K. M., Alfranca A., Redondo J. M., Hatae T., Tanabe T., Warner T. D., Bishop-Bailey D. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 63–69 [DOI] [PubMed] [Google Scholar]
- 34.Balakathiresan N. S., Bhattacharyya S., Gutti U., Long R. P., Jozwik C., Huang W., Srivastava M., Pollard H. B., Biswas R. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L1012–L1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J. W., Wang P., Kattah M. G., Youssef S., Steinman L., DeFea K., Straus D. S. (2008) J. Immunol. 181, 6536–6545 [DOI] [PubMed] [Google Scholar]
- 36.Chen W., Monick M. M., Carter A. B., Hunninghake G. W. (2000) Exp. Lung Res. 26, 13–26 [DOI] [PubMed] [Google Scholar]
- 37.Suzuki M., Tetsuka T., Yoshida S., Watanabe N., Kobayashi M., Matsui N., Okamoto T. (2000) FEBS Lett. 465, 23–27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.