Abstract
Several streptococcal species are able to take up naked DNA from the environment and integrate it into their genomes by homologous recombination. This process is called natural transformation. In Streptococcus pneumoniae and related streptococcal species, competence for natural transformation is induced by a peptide pheromone through a quorum-sensing mechanism. Recently we showed that induction of the competent state initiates lysis and release of DNA from a subfraction of the bacterial population and that the efficiency of this process is influenced by cell density. Here we have further investigated the nature of this cell density-dependent release mechanism. Interestingly, we found that competence-induced pneumococci lysed competence-deficient cells of the same strain during cocultivation and that the efficiency of this heterolysis increased as the ratio of competent to noncompetent cells increased. Furthermore, our results indicate that the lysins made by competent pneumococci are not released into the growth medium. More likely, they are anchored to the surface of the competent cells by choline-binding domains and cause lysis of noncompetent pneumococci through cell-to-cell contact.
Natural transformation in streptococci can be divided into separate stages consisting of competence induction, double-stranded DNA binding, single-stranded DNA uptake, and homologous recombination (5, 11). In Streptococcus pneumoniae strain Rx, which has been used in this study, the process is regulated by a secreted 17-amino-acid-long peptide pheromone (CSP-1) which triggers competence development when it reaches an external concentration of 1 to 10 ng/ml (9). In a batch culture, this corresponds to about 107 cells/ml. CSP-1, which is encoded by comC, is secreted by an ABC transporter (ComAB) (12) and acts through a two-component regulatory system consisting of a histidine kinase receptor (ComD) and its cognate response regulator (ComE) (10, 18, 26). The alternative sigma factor ComX, which presumably is directly regulated by ComE, activates transcription of the late genes whose products are involved in DNA binding, DNA uptake, and recombination, etc. The late genes share a consensus promoter sequence called a cin-box (TACGAATA) (2, 19) that is recognized by ComX when this sigma factor is associated with the RNA polymerase holoenzyme (13, 15).
Although much has been learned about natural transformation in streptococci over the past decade, some important questions remain to be answered. One such question concerns the origin of the naked DNA taken up by competent cells, the so-called donor DNA. Traditionally it has been believed that DNA released from dead bacteria constitutes the only source of donor DNA and that donor DNA will always be present in the natural habitats of competent streptococci. In contrast to this view, evidence has recently been presented indicating that a DNA release mechanism is operating during competence to ensure that donor DNA is available at the appropriate time (25). When a population of S. pneumoniae is induced to competence, a subfraction of the cell population will lyse and release its contents to the surroundings. DNA release and DNA uptake reach their maxima at about the same time, demonstrating that release of DNA from the donor cells is coordinated in time and space with uptake by the recipients (25). So far, the DNA release mechanism has not been fully elucidated, but it is known to be influenced by cell density and to be under control of the same quorum-sensing system that regulates competence development (ComCDE) (25). In addition, evidence has been obtained indicating that LytA and maybe additional choline-binding proteins (CBPs) are involved (25). In the present work, experiments were carried out to discern between autolysis and heterolysis (we define heterolysis as the lysis of one bacterium brought about by another). Our results show that competent pneumococci are capable of lysing noncompetent pneumococci of the same strain during cocultivation. These results suggest that heterolysis rather than autolysis is the mechanism of DNA release in a competent population of S. pneumoniae. However, it cannot be ruled out that both mechanisms operate simultaneously.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study were derived from S. pneumoniae strain Rx (Table 1). Bacterial strains were grown in casein tryptone (CAT) medium containing (per liter) 167 mmol of K2HPO4, 5 mg of choline chloride, 5 g of tryptone, 10 g of enzymatic casein hydrolysate, 1 g of yeast extract, and 5 g of NaCl (16). After sterilization, glucose was added to a concentration of 0.2%. Unless otherwise specified, all incubations were carried out at 37°C and all density measurements of bacterial cultures were done spectrophotometrically at 550 nm.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| A1 | CP1415 but Rifr through spontaneous conversion | This work |
| CP1200 | Rx but malM511 str-1 | 24 |
| CP1415 | CP1200 but comA negative by insertion of the ermAM cassette into the ClaI site of comA | 17 |
| CP1500 | hex nov-r1 bry-r str-1 ery-r1 ery-r2 | 3 |
| EH1 | EK4166 but comE negative by replacement with a Kanr cassette via double-crossover recombinations | This work |
| EK100 | CP1415 but egb negative by transformation with genomic DNA form R262 | 25 |
| EK4166 | EK100 but hirL::pEVP3 by transformation with plasmid DNA | 25 |
| EK4167 | EK4166 but Novr by transformation with CP1500 genomic DNA | 25 |
| H1 | CP1415 but Novr by transformation with CP1500 genomic DNA | 25 |
| H2 | H1 but comE::pXF518 by transformation with R319 genomic DNA | 25 |
| H3 | H1 but lytA::pEVP3 by transformation with plasmid DNA | 25 |
| H4 | EK4166 but lytA::pFW13 by transformation with plasmid DNA | This work |
| H5 | EK100 but lytC::pEVP3 by transformation with plasmid DNA | This work |
| H6 | A1 but LytA− by transformation with genomic DNA from H3 | This work |
| R262 | R800 but negative for endogenous β-galactosidase activity (ebg negative) | 1 |
| R319 | R800 but comE::pXF518 by transformation with plasmid DNA | 1 |
| S1 | CP1415 but orf62-orf51::pEVP3 by transformation with plasmid DNA | This work |
| S. pneumoniae Rx | R36A derivative | 21 |
| Plasmids | ||
| pEVP3 | Nonreplicative vector (CmrlacZ) | 4 |
| pFW13 | Nonreplicative vector (Kmr) | 20 |
Construction of S. pneumoniae mutant strains.
The H4 mutant was constructed by insertion-duplication mutagenesis. The primers LytA.1 (5′-ATATTGGTACCGCGGTTGGAATGCTGAGACCTATGCAGC-3′) and LytA.2 (5′-ATATTGGATCCGTACCAGTAGCCAGTGTCATTCTTCTGCC-3′) were used to amplify a ∼350-bp internal region of the lytA gene. Purified genomic DNA from strain CP1415 was used as a template. The resulting PCR fragment was cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's recommendations. The DNA fragment was excised by SpeI and ApaI and ligated into multiple cloning site II (MCS-II) of the vector pFW13 (20), precleaved with the same enzymes. The ligation reaction was used to transform chemically competent TOP10 cells (Invitrogen), and positive clones were selected on agar plates containing 50 μg of kanamycin/ml. The insertion-duplication mutant was made by natural transformation of EK4166 by using purified pFW13 containing the amplified 350-bp fragment. To verify that the correct mutant had been made, a PCR was performed using primer LYTA.3 (5′-GAACAGATTTGCCTCAAGTCGGCG-3′), located upstream of primer LYTA.1, and primer PFW13.4 (5′-TAACCGTATTACCGCCTTTG-3′), located downstream of MCS-II in pFW13. A PCR product of the expected size was obtained, demonstrating that the pFW13 plasmid had integrated into the lytA gene. The H4 mutant also had the expected phenotype. It did not autolyze in the stationary phase even in the presence of 0.1% Triton X-100.
The H5 mutant was constructed in the same way as the H4 mutant. PCR was performed with the primers LytC.1 (5′-TTAAGCATGCGAATGTCGGCTGGGTTCACAGAGATGG-3′) and LytC.2 (5′-TTAAGGATCCCTCAACATCATAATAGATAGGGTAAGACAGG-3′) containing SphI and BamHI sites at their 5′ ends, respectively. The resulting ∼380-bp internal fragment of the lytC gene was then cloned into the pCR2.1-TOPO vector. The cloned PCR fragment was excised from the pCR2.1-TOPO vector with SphI and BamHI and ligated into the pEVP3 vector (4), predigested with the same restriction enzymes. Finally, the H5 mutant was constructed by transforming competent EK100 cells with approximately 1 μg of purified pEVP3 containing the 380-bp lytC fragment. Insertion-duplication mutants were selected on agar plates containing 2.5 μg of chloramphenicol/ml. To confirm that the correct LytC− mutant had been made, control PCRs were carried out using genomic DNA from selected transformants as templates. LytC.3 (5′-CAGATGGTCGTTACTCGCA-GAATG-3′), corresponding to a sequence in the lytC gene upstream of the site of integration, was used in combination with pEVP3.3 (5′-GTAACTTCCACAGTAGTTCACCACC-3′) or PEVP3.4 (5′-CCCGGTCGACCCGTAATCTTACGTC-3′). These primers are complementary to sequences in the pEVP3 vector. The resulting PCR fragments were of the expected sizes, demonstrating that the lytC gene had been disrupted and that pEVP3 had been inserted in the correct orientation, placing the promoterless lacZ gene of the pEVP3 vector under control of the lytC promoter.
Rimini et al. (23) have shown that the rate of transcription of two small open reading frames termed orf62 and orf51 increases about 12-fold upon competence development. There is also a cin-box located immediately upstream of the start codon of orf62, showing that orf62 and orf51 belong to the ComX regulon. To be able to monitor expression of the late genes under various growth conditions, we wanted to construct a mutant containing a β-galactosidase reporter system controlled by the orf62-orf51 promoter. This was carried out by amplifying a ∼270-bp DNA fragment corresponding to about half of orf62 and the complete orf51 by PCR using the primers orf62.1 (5′-CTCGCTCCCTTGGTTATCTTTGGAG-3′) and orf51.2 (5′-CTAGCATGACTTACCAAACTTTTTACGAAGG-3′). The DNA fragment was first cloned into the pCR2.1-TOPO vector, then excised using the pCR2.1-TOPO polylinker restriction enzymes BamHI and NsiI, and finally ligated into the pEVP3 vector, precleaved with BamHI and NsiI. Purified pEVP3 vector harboring the 270-bp fragment was then used to transform the EK100 strain by natural transformation. Colonies growing on agar plates containing 2.5 μg of chloramphenicol/ml were isolated and tested for β-galactosidase activity in the presence and absence of CSP-1. A mutant termed S1 that had the expected phenotype was isolated. It produced no β-galactosidase in the absence of CSP-1, but the addition of 250 ng of this peptide pheromone/ml induced strong expression of the β-galactosidase enzyme.
The EH1 strain is identical to EK4166, except that its comE gene is replaced by a Kanr cassette via double-crossover recombinations. To make the EH1 strain, we took advantage of the pFW13 vector, which contains a kanamycin resistance gene flanked by two MCSs, MCS-I and MCS-II. An ∼880-bp DNA fragment, corresponding to the region immediately upstream of the comE gene, was amplified by PCR using the primers ComE.56 (5′-TTAAGCTAGCATCTTTCGTTTCAGATATGGTAAGTACG-3′) and ComE.57 (5′-TTAAGACGTCCATCCAATATTCTCTCTAGTCTCACTTGATG-3′) and ligated into the pCR2.1-TOPO vector as described above. The DNA fragment was excised by AatII and NheI and cloned into MCS-I of pFW13. Next, a ∼750-bp DNA fragment, corresponding to the region immediately downstream of the comE gene, was amplified by PCR using the primers comE.53 (5′-ATTACCATGGTCTCAAAAGTGATTGACAATTAGCAAG-3′) and comE.55 (5′-ATTACATATGGCTATGGTACAATTACTGATGGAACAGCC-3′) and cloned into the pCR2.1-TOPO vector. The fragment was excised from the pCR2.1-TOPO vector with NcoI and NdeI and ligated into MCS-II of the pFW13 vector harboring the 880-bp fragment in MCS-I. Using this construct as a template and comE.55 and comE.56 as primers, we amplified a ∼3,500-bp fragment consisting of the Kanr gene flanked with the two cloned fragments described above. This linear DNA fragment was then used to transform the EK4166 strain by natural transformation. A transformant growing on agar plates containing 150 μg of kanamycin/ml was isolated and assayed for transformability by using genomic DNA from the novobiocin-resistant strain CP1500. The transformant, termed EH1, turned out to be completely noncompetent, demonstrating that the Kanr cassette most likely had replaced the comE gene. To further verify that this was the case, the site of integration of the Kanr cassette was examined by PCR. The reaction was carried out with genomic DNA from EH1, the primer tArg2 (5′-CATAGCTCAGCTGGATAGAGCATTCGCCTTC-3) that is complementary to the Arg-tRNA gene upstream of the comCDE operon, and the primer pFW13.6 (CATTTATTTACCTCCTTTT-GGTTACCTCAC-3) that is complementary to the promoter region of the Kanr gene. Analysis of the PCR by agarose gel electrophoresis revealed a single band of the expected size, demonstrating that the Kanr cassette had integrated at the correct location.
β-galactosidase release assay.
An overnight culture of the bacterial strain was diluted to an optical density at 550 nm (OD550) of 0.1 in prewarmed (37°C) CAT medium and incubated until it reached an OD of 0.3. To ensure vigorous growth, it was rediluted to an OD550 of 0.05 and incubated further. When the culture reached an OD of 0.1, two 10-ml samples were withdrawn and induced to competence by addition of 250 ng of CSP-1 (NH2-EMRLSKFFRDFILQRKK-COOH)/ml, incubated for 30 min, and placed on ice. The cells in the first sample were immediately removed by centrifugation at 2,500 × g for 10 min at 4°C. Then the supernatant was sterile filtered using a 0.2-μm-pore-size filter and stored on ice until assayed. In order to measure the total β-galactosidase activity in the culture, the cells in the second sample were lysed by incubating the culture for 10 min at 37°C with 0.1% Triton X-100. Following lysis, the culture was kept on ice until assayed. Samples were collected at regular intervals during logarithmic growth (OD550s of 0.1 and 0.2, etc.). Uninduced samples were always run in parallel as negative controls. All samples were assayed for β-galactosidase activity as described previously (25). Cocultivation experiments were performed in the same way, except that 5 ml of each strain was mixed when the OD550 of each culture reached 0.1 and 0.2, etc. In the experiment in which the effect of temperature on β-galactosidase release was examined, the cultures were placed at their respective temperatures 30 min before competence was induced by the addition of 250 ng of CSP-1/ml. At this stage, the OD550s of the cultures had reached approximately 0.35. Apart from this, the assay was carried out as described above.
Cocultivation transformation assay.
To measure the efficiency of gene exchange between two strains growing in liquid medium, a cocultivation assay was developed. Overnight cultures of the two strains H2 (ComA− ComE− Novr) and A1 (ComA− Rifr) were diluted to OD550s of 0.05 and incubated at 37°C until they reached OD550s of 0.2. To ensure vigorous growth, both cultures were diluted once more to OD550s of 0.05 and incubated further at 37°C. When they reached OD550s of 0.1, 0.2, and 0.4, 0.5 ml from each culture (H2 and A1) was withdrawn and mixed in a 15-ml plastic tube containing 250 ng of CSP-1. The mixed strains were incubated at 37°C for 30 min, diluted four times with prewarmed CAT medium, and then further incubated at 37°C for 60 min before being plating onto CAT agar plates containing 2 μg of rifampin/ml and 5 μg of novobiocin/ml. After incubation of the plates at 37°C for 24 to 48 h, the number of transformants resistant to both antibiotics was determined. Uninduced samples were run in parallel as negative controls. Identical gene exchange assays were carried out by cocultivating strains H3 and H6 and strains A1 and H1.
RESULTS
Competence-induced heterolysis.
During competence development in a population of S. pneumoniae cells, a subfraction of the population was lysed, causing release of DNA to the surroundings (25). Addition of CSP-1 did not elicit a corresponding response in a competence-deficient mutant in which the response regulator ComE had been disrupted. ComE, which is part of the two-component regulatory system (ComDE) that monitors the external concentration of the competence pheromone, is a key component in the signal transduction pathway triggering competence development in S. pneumoniae. The absence of cell lysis in a ComE− mutant clearly shows that the signal transduction pathway encoded by comDE also controls the expression of genes involved in cell lysis and strongly suggests that this phenomenon is an integral part of natural transformation (25).
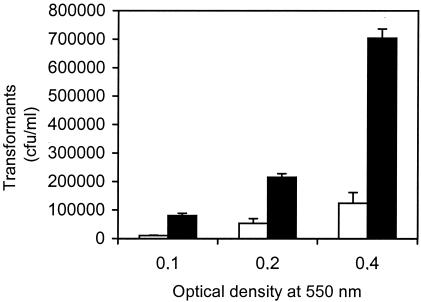
To further elucidate the mechanism behind competence-induced cell lysis, we asked ourselves whether transfer of DNA can take place between a competence-inducible strain and a competence-deficient strain of S. pneumoniae. In order to answer this question, a cocultivation experiment was carried out with the two S. pneumoniae strains A1 (ComA− Rifr) and H2 (ComA− ComE− Novr). Vigorously growing cultures of A1 and H2 were mixed in equal amounts at OD550s of 0.1, 0.2, and 0.4, subjected to 250 ng of CSP-1/ml, incubated for 1.5 h at 37°C, and spread onto agar plates containing 2 μg of rifampin/ml and 5 μg of novobiocin/ml (see Materials and Methods for details). The plates were incubated at 37°C, and the number of transformants resistant to both antibiotics was determined the next day. The results presented in Table 2 show that competence-induced A1 cells must have taken up DNA released from the competence-deficient H2 strain. In a negative control experiment run in parallel, CSP-1 was omitted. As expected, no transformant was obtained in this experiment. The Novr DNA taken up by the A1 cells may have originated from dead H2 cells and been present in the growth medium of the H2 cells before they were mixed with the A1 cells. Alternatively, release of Novr DNA may have been an active process mediated by the A1 cells after they had been induced to competence by addition of CSP-1. To discern between these two mechanisms, we set up a cocultivation experiment in which the competence-deficient (ComE−) strain EH1 expressing Escherichia coli β-galactosidase from a constitutive promoter was mixed with an equal amount of the competence-inducible strain EK100 lacking this enzyme. When the EK100 strain was induced to competence by addition of CSP-1, cytoplasmic β-galactosidase from the EH1 strain was released to the growth medium. However, very little β-galactosidase activity was detected in the medium of the negative control, i.e., when no CSP-1 had been added to the mixed culture (Fig. 1). Furthermore, when CSP-1 was added to a culture consisting only of the EH1 strain, no release of β-galactosidase was detected (results not shown). These results can only be explained by a release mechanism that involves heterolysis, i.e., lysis of the competence-deficient EH1 strain by the competent EK100 strain.
TABLE 2.
Cocultivation of a competence-inducible strain (A1) and a competence-deficient strain at different densities during logarithmic growth
| Deficient strain or DNA fragment | Pheromone added | No. of CFU of transformants/ml at OD550 of:
|
||
|---|---|---|---|---|
| 0.1 | 0.2 | 0.4 | ||
| H2a | Yes | 1.8 × 104 | 4.8 × 104 | 9.7 × 104 |
| No | 2.0 × 101 | 0 | 1.0 × 101 | |
| Novr DNA from CP1500b | Yes | 5.9 × 105 | 3.4 × 105 | 2.4 × 105 |
| No | 0 | 0 | 1.0 × 101 | |
Competence-deficient mutant lacking a functional comE gene.
One microgram of purified DNA per milliliter from novobiocin-resistant strain CP1500 was used to transform strain A1.
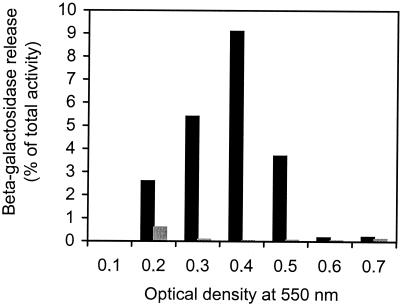
FIG. 1.
Competence-induced release of intracellular β-galactosidase into the growth medium during cocultivation of S. pneumoniae strains EH1 and EK100. The EH1 strain is competence deficient and expresses β-galactosidase from a constitutive promoter. In contrast, the EK100 strain is competence inducible but β-galactosidase deficient. Cultures of EH1 and EK100 growing in parallel were mixed at a ratio of 1:1 when they reached OD550s of 0.1 and 0.2, etc. After 30 min with or without CSP-1, samples were collected for analysis of β-galactosidase activity. Both cell-free growth medium (supernatants) and cell lysates (supernatants and lysed cells) were assayed. The amount of β-galactosidase present in the supernatants (estimated in Miller units) is given as a percentage of the total activity present in the cell lysates. Black columns represent cocultivation in the presence of 250 ng of CSP-1/ml. Grey columns represent cocultivation in the absence of CSP-1. The results shown are representative of results from three separate experiments.
Do competent cells release a lysin?
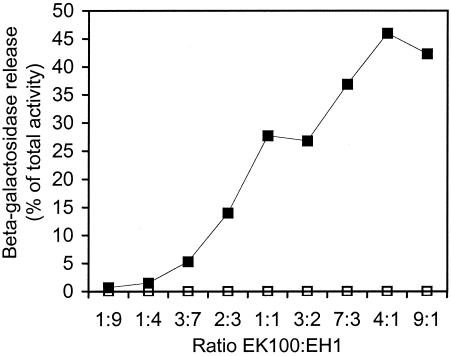
Previously it was shown that the fraction of cells lysed when cultures of S. pneumoniae are induced to competence at different cell densities increases with OD550s of up to 0.3 to 0.4 and then declines (25). The mechanism behind this cell density dependency is not understood, but it is likely that the decline in release can be explained, at least in part, by loss of competence induction towards the end of the logarithmic growth phase (9). Our finding that competent cells can lyse competence-deficient cells suggests that competent cells may secrete a lysin. If this is the case, it would be expected that increasing the ratio of competence-inducible cells to competence-deficient cells would result in more-efficient lysis. In cocultivation experiments similar to the one described above, different ratios of strains EH1 and EK100 were used (Fig. 2). The results of these experiments clearly showed that the percentage of EH1 cells lysed increased when the ratio of competent EK100 cells to competence-deficient EH1 cells increased. Thus, the competent cells must produce one or more lysins that attack noncompetent cells and lyse them. If the lysin(s) is secreted into the external medium, it should be possible to induce lysis of noncompetent cells by adding cell-free medium from a culture of competent cells. A vigorously growing culture of EK100 was induced to competence by addition of 250 ng of CSP-1/ml at an OD550 of 0.4 and incubated at 37°C for 30 min. The cells were removed by centrifugation, and the supernatants were sterile filtered. Uninduced samples were run in parallel as negative controls. The temperature of the sterile-filtered supernatants was adjusted to 37°C, and then the supernatants were mixed immediately with equal volumes of vigorously growing cultures of EH1 cells. These mixtures were incubated for another 30 min, and the supernatants were harvested and assayed for β-galactosidase activity as described before. We did not detect any release of β-galactosidase into the culture sample in which EH1 cells had been mixed with growth medium from competence-induced EK100 cells (results not shown). To ensure that the EH1 cells employed in these experiments produced normal amounts of β-galactosidase, parallel samples were lysed by addition of 0.1% Triton X-100 and their total enzyme contents were measured. The levels of total β-galactosidase activity present in these samples were as expected. In conclusion, it seems that the lysin produced by competent S. pneumoniae cells is not released into the growth medium. Rather, our results suggest that the lysin is attached to the cell surface and that cell-cell contact is necessary for heterolysis to take place.
FIG. 2.
Release of cytoplasmic β-galactosidase from the competence-deficient EH1 strain during cocultivation with strain EK100. The EK100 strain is competence inducible when CSP-1 is added but is completely without endogenous β-galactosidase activity. When cultures of the two strains both reached an OD550 of 0.4, they were mixed at different ratios and 250 ng of CSP-1/ml was added immediately. After 30 min, samples were harvested and cell-free supernatants and cell lysates (supernatants and lysed cells) were prepared and assayed for β-galactosidase activity. Uninduced samples were run in parallel as negative controls. The amount of β-galactosidase present in the supernatants (estimated in Miller units) is given as a percentage of the total activity present in the cell lysates. Closed squares represent results for supernatants from CSP-1-induced cells, and open squares represent results for supernatants from uninduced cells. The experiment was repeated several times with similar results.
Role of LytA.
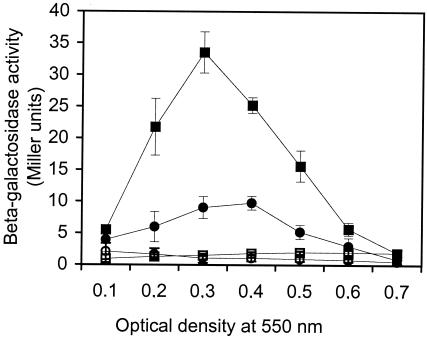
A good candidate for the unidentified lysin discussed above is the murein hydrolase LytA, which is anchored noncovalently to the pneumococcal cell wall through six choline-binding domains (14). It is expressed in noncompetent cells, but its expression is upregulated about sixfold during competence development (23). It has previously been shown that a LytA− mutant releases less DNA when induced to competence than a wild-type strain (25). Here we decided to make a LytA-deficient mutant that expresses E. coli β-galactosidase constitutively (H4) and to measure CSP-1-induced release of β-galactosidase from this mutant at different cell densities. It turned out that significantly less β-galactosidase is released from the LytA− mutant during competence development than from the positive control. The difference is two- to fourfold depending on the cell density of the culture (Fig. 3). However, at OD550s of 0.3 and 0.4, the amount of β-galactosidase released from the LytA− mutant is still almost 10-fold higher than that released from the uninduced control (Fig. 3). This indicates that more than one lysin is involved in the release mechanism. This view is supported by the results of the DNA exchange experiment presented in Fig. 4. Strains resistant to novobiocin (H1) and rifampin (A1) were cocultivated at OD550s of 0.1, 0.2, and 0.4 as described above, and transformants were selected on agar plates containing 2 μg of rifampin/ml and 5 μg of novobiocin/ml. In parallel, the two LytA− mutants H3 and H6, resistant to novobiocin and rifampin, respectively, were cocultivated and the number of transformants resistant to both antibiotics was determined. The results show that the numbers of transformants obtained with the LytA− mutants are four- to eightfold lower (depending on cell density) than the corresponding numbers obtained with the positive control.
FIG. 3.
Comparison of strains EK4166 and H4 with respect to the level of competence-induced release of cytoplasmic β-galactosidase. The lytA gene of strain H4 has been disrupted, but otherwise the strain is identical to EK4166. At different cell densities (OD550s of 0.1 to 0.7), samples were withdrawn and induced to competence by adding 250 ng of synthetic CSP-1/ml. After 30 min at 37°C, cell-free supernatants were prepared and assayed for β-galactosidase activity. Corresponding uninduced samples were run in parallel as negative controls. Closed squares, activity in supernatants from CSP-1-induced EK4166 cells; open squares, activity in supernatants from uninduced EK4166 cells; closed circles, activity in supernatants from CSP-1-induced H4 cells; open circles, activity in supernatants from uninduced H4 cells. The β-galactosidase activity is given in Miller units, and amounts shown are the means ± standard errors of results for triplicate samples.
FIG. 4.
Influence of LytA on efficiency of gene exchange in vitro. Two LytA-deficient S. pneumoniae strains, H3 (Novr) and H6 (Rifr), were mixed at three different densities (OD550s of 0.1, 0.2, and 0.4) and induced to competence by adding 250 ng of CSP-1/ml. After cocultivation for 1.5 h, cells were plated onto agar plates containing rifampin and novobiocin and incubated at 37°C. Transformants were scored the next day. The two strains H1 (Novr) and A1 (Rifr), both containing intact lytA genes, were treated in the same way. Black columns represent numbers of transformants obtained by cocultivation of H1 and A1, while white columns represent numbers of transformants obtained by cocultivation of the LytA-deficient strains H3 and H6. Values shown are numbers of CFU of transformants per milliliter and are the means ± standard errors of results for triplicate samples. Uninduced samples, receiving no peptide pheromone, were run in parallel as negative controls. The numbers of transformants observed in uninduced samples ranged from 0 to 200 CFU/ml.
LytC is not the missing lysin.
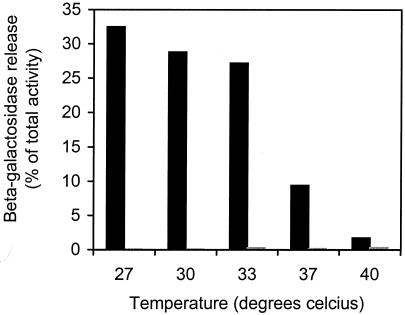
Evidently, LytA plays an important role in competence-induced DNA release in S. pneumoniae. However, as demonstrated above, there must be at least one additional lysin produced by competent pneumococci. The release mechanism is sensitive to the presence of choline in the growth medium, demonstrating that one or more CBPs are involved (25). Without affecting transformability, the presence of 0.02% choline reduces release of β-galactosidase by about 50% and 0.2% reduces release by up to 90% (results not shown). We therefore hypothesized that another choline-binding murein hydrolase could be involved. The most promising candidate was the choline-binding lysozyme LytC (7, 14), which had been reported to be competence induced (23). LytC has optimum activity at 30°C (7), and we therefore first investigated whether the competence-induced lysis mechanism is more efficient at this temperature. The results given in Fig. 5 show that this indeed is the case. Release of β-galactosidase at 30°C increases two- to threefold compared to the corresponding release at 37°C. Interestingly, we also found that release is practically abolished at 40°C. In principle, high temperatures may influence release of β-galactosidase indirectly by suppressing competence induction or they may act directly on the cell lysis mechanism. In order to address this question, we used a mutant (S1) that harbors a chromosomally located lacZ reporter gene under control of the orf62-orf51 promoter (23). orf62 and orf51 are two small bacteriocin-like reading frames that are part of the ComX regulon. Two cultures of the S1 strain (OD550 = 0.35) growing at 37and 40°C were induced to competence by addition of CSP-1, incubated for 30 min, lysed by addition of 0.1% (vol/vol) Triton X-100, and assayed for β-galactosidase activity. Uninduced samples were run in parallel. No β-galactosidase activity was detected in the negative controls (results not shown). The activity measured in the induced S1 culture grown at 40°C (153 Miller units) was somewhat reduced compared to the activity measured in the corresponding culture grown at 37°C (207 Miller units). However, the reduction does not seem large enough to explain the nearly complete loss of competence-induced cell lysis at 40°C. It must therefore be concluded that the observed effect is due to temperature sensitivity of the release mechanism itself.
FIG. 5.
Effect of temperature on competence-induced release of cytoplasmic β-galactosidase from S. pneumoniae. Strain EK4166 was induced to competence at five different temperatures by addition of 250 ng of CSP-1/ml (black columns). Both cell-free supernatants and cell lysates (supernatants and lysed cells) were assayed for β-galactosidase activity. The amount of β-galactosidase present in the supernatants (estimated in Miller units) is given as a percentage of the total activity present in the cell lysates. Corresponding uninduced samples were run in parallel as negative controls (grey columns). This experiment has been repeated several times with similar results.
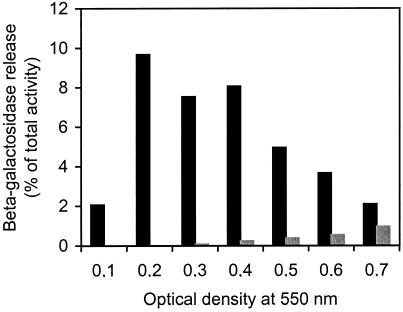
Together, the various data discussed above suggested to us that LytC could be involved in competence-induced cell lysis. We therefore decided to make a lytC knockout mutant. An internal fragment of lytC was cloned into the pEVP3 vector, and this construct was used to transform strain EK100. The pEVP3 vector contains a promoterless E. coli lacZ reporter gene immediately downstream of the polylinker. Transforming strain EK100 with this construct resulted in integration of the recombinant plasmid into the lytC gene by an insertion-duplication mechanism, giving rise to a chromosomally located transcriptional fusion between lytC and lacZ. The strain (H5) carrying this reporter construct was used to control the results of a previously published microarray analysis indicating that transcription of lytC is upregulated during competence development (23). Unexpectedly, our results showed that rates of transcription from the lytC promoter remained the same in induced and uninduced cultures, demonstrating that lytC is not a competence-regulated gene. However, as the lacZ gene was constitutively transcribed at a high rate, the H5 strain could be used directly in a β-galactosidase release assay. The results of this assay (Fig. 6) showed that the level of β-galactosidase released into the growth medium when the LytC− mutant was induced to competence was not reduced compared to the level released by the wild type. We therefore concluded that LytC is not involved in the competence-activated lysis mechanism.
FIG. 6.
Competence-induced release of β-galactosidase from the LytC-deficient H5 strain during logarithmic growth. Samples were collected at different cell densities (OD550s of 0.1 to 0.7) and induced to competence by adding 250 ng of CSP-1/ml (black columns). Both cell-free supernatants and cell lysates (cells and supernatants) were assayed for β-galactosidase activity. The amount of β-galactosidase present in the supernatants (estimated in Miller units) is given as a percentage of the total activity present in the cell lysates. Corresponding uninduced samples were run in parallel as negative controls (grey columns). The presented data are representative of results from three independent experiments.
DISCUSSION
The data presented here and in a recent paper by Steinmoen et al. (25) clearly show that S. pneumoniae cells grown under laboratory conditions actively release DNA when induced to competence. This coordination between DNA release and uptake makes efficient gene exchange possible in a batch culture, as demonstrated by our cocultivation experiments with Novr and Rifr mutants of S. pneumoniae strain Rx (Table 2 and Fig. 4). Since both DNA uptake and release are controlled by the same quorum-sensing system (ComABCDE), these processes must be coordinated also under natural conditions. From an evolutionary perspective, this findingadds a new dimension to natural genetic transformation and makes this phenomenon a biologically significant event. In order to better understand how gene exchange takes place under natural conditions, it is essential to elucidate fully the DNA release mechanism. All evidence gathered so far indicates that DNA is released by cell lysis and not by DNA export machinery. In the present work, we wanted to establish that release of DNA in a competence-induced population of S. pneumoniae takes place by autolysis. However, to our surprise we found that competence-deficient cells (ComE−) are lysed when cocultivated with competence-induced cells, demonstrating that heterolysis must take place. This finding does not exclude autolysis, but the high levels of β-galactosidase released in the experiments whose results are presented in Fig. 1 and 2 suggest that heterolysis contributes substantially to the process. This made us wonder whether the observed heterolysis is due to secretion of a lysin by the competent cells or whether it depends on cell-to-cell contact between competent and competence-deficient cells. We were unable to detect an active agent in the supernatant harvested from competence-induced S. pneumoniae cultures, indicating that heterolysis involves lytic substances attached to the surface of competent cells. This model is in agreement with the fact that in most experiments release is more efficient at OD550s of 0.3 to 0.4 than at lower cell densities. In a batch culture, cell-to-cell contact will be more extensive at higher cell densities, leading to more-efficient lysis. The drop in release seen at even higher cell densities (OD550s of 0.6 to 0.7) is probably due to the loss of competence inducibility (insensitivity to CSP-1) seen at high cell densities (9). Our results show that the presence of 0.2% choline in the growth medium almost completely abolishes competence-induced lysis without affecting transformability. Previous studies have demonstrated that CBPs can be eluted from the pneumococcal surface with soluble choline (6, 8). This suggested that a cell wall-located CBP is involved in the observed lysis (25). The obvious candidate for this role was the competence-induced amidase LytA, which is anchored to the cell wall through a choline-binding domain consisting of six repeats. It turned out that DNA release in a LytA− mutant is significantly reduced, but not abolished, compared to that in the wild type (25). This finding was confirmed by the results of the β-galactosidase release assay shown in Fig. 3. Consequently, an additional choline-binding murein hydrolase seems to be involved. The pneumococcal CBP family consists of 12 members, which include the three murein hydrolases LytA, LytB, and LytC (8, 14). LytB is involved in the separation of daughter cells (6), whereas the exact biological role of LytC is unknown. However, LytC possesses lysozyme-like activity with a temperature optimum of 30°C (6, 7). In addition, it has been reported that transcription of the lytC gene is upregulated during competence development (23). To determine whether LytC could be the missing lysin, we cocultivated competent and competence-deficient strains at different temperatures. The results (Fig. 5) showed that lysis is more efficient at temperatures around the reported optimum temperature for LytC. To further investigate the possible role of LytC in competence-induced cell lysis, we constructed a LytC knockout mutant. In this mutant, a lacZ reporter gene was placed under control of the lytC promoter. In contrast to the report by Rimini et al. (23), we found that the lytC gene was constitutively highly expressed, but its rate of transcription did not increase during competence development. In addition, no reduction in the release of β-galactosidase was detected (Fig. 6), demonstrating that LytC is not involved in competence-induced cell lysis.
Unexpectedly, the lysis mechanism works very poorly, if at all, at 40°C (Fig. 5). Our results indicate that the temperature-sensitive step is downstream of comX, since transcriptional activation of the late genes functions normally at 40°C. Presumably, a late-gene product which constitutes a critical part of the lysis machinery does not function at this temperature. Another possibility is that Brownian movements, which are more intense at 40°C, disrupt stable cell-to-cell contact, preventing heterolysis.
Most likely, gene exchange under natural conditions takes place in complex multispecies biofilms, where there is close contact between individual cells. In oral streptococci such as S. mitis and S. gordonii, this contact is mediated by surface-exposed proteins called coaggregation adhesins (22). Presumably, pneumococci rely on similar surface proteins to make contact with one another and other species in the biofilm community. To better understand how gene exchange takes place under natural conditions, it is important to understand exactly how DNA is released from the donor cells and how this process is regulated. In a batch culture, competence is triggered when the concentration of CSP-1 reaches 1 to 10 ng/ml, corresponding to about 107 cells/ml. Within a biofilm community, diffusion of the CSP-1 peptide is probably much more restricted and competence most likely develops among small clusters of cells belonging to the same pherotype. Our results suggest that competent cells in such clusters can lyse neighboring cells by expressing cell wall-degrading enzymes such as LytA on their surfaces. We therefore postulate that competent streptococci actively acquire DNA by killing their neighbors. This contrasts with the traditional view in which it is assumed that DNA taken up by competent bacteria originates from donor bacteria that have died and fallen apart from natural causes.
Acknowledgments
We thank Andreas Podbielski (University Hospital Rostock, Rostock, Germany) for kindly providing the pFW13 vector.
This work was supported by a grant from the Research Council of Norway.
REFERENCES
- 1.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 3.Cato, A., and W. R. Guild. 1968. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double crossover model. J. Mol. Biol. 37:157-178. [DOI] [PubMed] [Google Scholar]
- 4.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 5.Claverys, J. P., and L. S. Håvarstein. 2002. Extracellular peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-d1814. [DOI] [PubMed] [Google Scholar]
- 6.García, P., M. González, R. López, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1281. [DOI] [PubMed] [Google Scholar]
- 7.García, P., M. P. González, E. García, J. L. García, and R. López. 1999. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol. Microbiol. 33:128-138. [DOI] [PubMed] [Google Scholar]
- 8.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Håvarstein, L. S., G. Coomaraswamy, and D. A Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 11.Håvarstein, L. S., and D. A. Morrison. 1999. Quorum-sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-26. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 12.Hui, F. M., and D. A. Morrison. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López, R., M. P. González, E. García, J. L. García, and P. García. 2000. Biological roles of two new murein hydrolases of Streptococcus pneumoniae representing examples of module shuffling. Res. Microbiol. 151:437-443. [DOI] [PubMed] [Google Scholar]
- 15.Lou, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison, D. A., S. A. Lacks, W. R. Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison, D. A., M. C. Trombe, M. K. Hayden, G. A. Waszak, and J. D. Chen. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAMβ1. J. Bacteriol. 159:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an autoinduced peptide pheromone and a two component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 19.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lütticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 21.Ravin, A. W. 1959. Reciprocal capsular transformations of pneumococci. J. Bacteriol. 77:296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 23.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Fransesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polizzi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 24.Shoemaker, N. B., and W. R. Guild. 1974. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol. Gen. Genet. 128:283-290. [DOI] [PubMed] [Google Scholar]
- 25.Steinmoen, H., E. Knutsen, and L. S. Håvarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ween, O., P. Gaustad, and L. S. Håvarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]