Abstract
Lipid peroxidation in tissue and in tissue fractions represents a degradative process, which is the consequence of the production and the propagation of free radical reactions primarily involving membrane polyunsaturated fatty acids, and has been implicated in the pathogenesis of numerous diseases, including systemic lupus erythematosus (SLE). We have found that bovine serum albumin incubated with peroxidized polyunsaturated fatty acids significantly cross-reacted with the sera from MRL-lpr mice, a representative murine model of SLE. To identify the active substances responsible for the generation of autoantigenic epitopes recognized by the SLE sera, we performed the activity-guiding separation of a principal source from 13-hydroperoxy-9Z,11E-octadecadienoic acid and identified 4-oxo-2-nonenal (ONE), a highly reactive aldehyde originating from the peroxidation of ω6 polyunsaturated fatty acids, as the source of the autoantigenic epitopes. When the age-dependent change in the antibody titer against the ONE-modified protein was measured in the sera from MRL-lpr mice and control MRL-MpJ mice, all of the MRL-lpr mice developed an anti-ONE titer, which was comparable with the anti-DNA titer. Strikingly, a subset of the anti-DNA monoclonal antibodies generated from the SLE mice showing recognition specificity toward DNA cross-reacted with the ONE-specific epitopes. Furthermore, these dual-specific antibodies rapidly bound and internalized into living cells. These findings raised the possibility that the enhanced lipid peroxidation followed by the generation of ONE may be involved in the pathogenesis of autoimmune disorders.
Keywords: Antibodies, Fatty Acid Oxidation, Immunochemistry, Oxidative Stress, Post-translational Modification, Anti-DNA Autoantibody, Autoimmune Diseases, Lipid Peroxidation, Oxidation-specific Epitopes
Introduction
Several lines of evidence indicate that the nonenzymatic oxidative modification of proteins and the subsequent accumulation of the modified proteins have been found in cells during aging, during oxidative stress, and in various pathological states including premature diseases, muscular dystrophy, rheumatoid arthritis, and atherosclerosis (1, 2). It has also been suggested that many of the effects of cellular dysfunction under oxidative stress are mediated by the products of nonenzymatic reactions, such as the peroxidative degradation of polyunsaturated fatty acids (3, 4). Lipid peroxidation leads to the formation of a broad array of different products with diverse and powerful biological activities. Among them are a variety of different aldehydes. The primary products of lipid peroxidation, lipid hydroperoxides, can undergo carbon-carbon bond cleavage via alkoxyl radicals in the presence of transition metals, giving rise to the formation of short chain, unesterified aldehydes of three to nine carbons in length and a second class of aldehydes still esterified to the parent lipid (5). These aldehydes generated during the lipid peroxidation have been implicated as causative agents in cytotoxic processes initiated by the exposure of biological systems to oxidizing agents. Some of the lipid peroxidation products exhibit a facile reactivity with proteins, generating a variety of intra- and intermolecular covalent adducts. Binder and Silverman (6) proposed that such adducts, called oxidation-derived epitopes, generated on self-antigens, are important immunodominant targets of natural antibodies and suggested that these antibodies play an important function in the host response to the consequences of oxidative stress during oxidative events that occur when cells undergo apoptosis. The fact that the post-translational modification of proteins is enhanced in aging and stressed cells and arises under physiological conditions (1, 2) suggests the existence of an association between the formation of oxidation-specific epitopes and immune disorders.
Systemic lupus erythematosus (SLE)2 is a potentially fatal systemic autoimmune disease, characterized by the increased production of autoantibodies, immune complex deposition in the microvasculature, leukocyte infiltration, and, ultimately, tissue damage in a number of organs. Of the multiple autoantibodies described in this disease, antibodies against the native DNA are among the most characteristic, yet the triggering antigen in the disease is still unknown. The appearance of these antibodies in humans and in murine models of lupus correlates with the progression of the disease, and by comparison with all the other lupus autoantibodies, those against the double-stranded DNA are thought to be the most pathogenic and involved in the development of renal pathology (7). However, because of the systemic character and complexity of the disease, it still remains unclear what exactly are the primary stimuli that drive such autoantibody responses and the mechanisms that regulate the entire pathological process in lupus. Although some bacterial DNAs appear to be immunogenic in normal mouse strains, they do not elicit the production of antibodies that cross-react with the eukaryotic DNA (8, 9). Thus, it remains unclear whether DNA or nucleosomes induce the anti-DNA antibodies in lupus-prone mice or whether the anti-DNA antibodies are made in response to some other antigen, either itself or foreign.
There is increasing evidence that lipid peroxidation plays a role in SLE: (i) SLE patients have an enhanced urinary excretion of isoprostanes, the well established biomarkers of lipid peroxidation (10); (ii) the levels of the lipid peroxidation-derived short chain aldehydes are significantly elevated in children with a high disease activity of SLE (11); and (iii) there are elevated levels of the oxidized LDL together with elevated levels of autoantibodies related to the oxidized LDL in female patients with SLE (12). The involvement of lipid peroxidation was also suggested by the observation that lipid peroxidation-specific epitopes were detected in tissues from the SLE patients (13). These findings raised the possibility that the enhanced lipid peroxidation, through its pivotal role in oxidative stress, may be involved in the pathogenesis of autoimmune disorders. In light of the importance of lipid peroxidation-specific epitopes in autoimmune diseases, we sought to identify the source of the autoantigenic epitopes from the in vitro peroxidized polyunsaturated fatty acids. Following the identification of an active substance, we generated several anti-DNA mAbs from MRL-lpr mice and characterized their dual specificity toward DNA and lipid peroxidation-derived protein ligands. We also describe their genetic origins and ability to bind and internalize into live cells.
EXPERIMENTAL PROCEDURES
Materials
Soybean lipoxidase (type I-B) and calf thymus DNA were purchased from Sigma. Linoleic acid was obtained from Nu-Chek Prep, Inc. (Elysian, MN). 13-Hydroperoxy-9Z,11E-octadecadienoic acid was synthesized by a previously described method (14). Other aldehydes were purchased from the Cayman Chemical Co. (Ann Arbor, MI). The HRP-linked anti-rabbit IgG immunoglobulin, HRP-NeutrAvidin, and ECL Western blotting detection reagents were obtained from Amersham Biosciences.
Animals
Female MRL-lpr and MRL-MpJ mice were purchased from Chubu Kagaku Shizai Co., Ltd. (Nagoya, Japan). All of the animal protocols were approved by the Animal Experiment Committee in the Graduate School of Bioagricultural Sciences of Nagoya University.
General Experimental Procedures
The 1H NMR spectrum was recorded at 27 °C using an AMX400 (400 MHz) spectrometer (Bruker, Rheinstetten, Germany). The solvent peak served as an internal standard for reporting the chemical shifts, which are expressed as parts per million downfield from tetramethylsilane (δ scale). HPLC was conducted using a JASCO system (JASCO, Tokyo, Japan).
Modification of Protein by Lipid Peroxidation Products
Modification of the protein by the lipid peroxidation products was performed by incubating BSA (1.0 mg/ml) with linoleic acid (5.0 mm), FeSO4(NH4)2SO4·6H2O (0.5 mm), and ascorbic acid (2.0 mm) in 0.1 m HEPES buffer (pH 7.4) with 10% ethanol at 37 °C under atmospheric oxygen. After 2, 4, 8, 12, 24, 48, and 72 h, aliquots were collected, and the reaction was terminated by adding butylated hydroxytoluene (1.0 mm) and diethylene triamine pentaacetic acid (100 μm).
Identification of a Lipid Peroxidation-derived Source of Autoantigenic Epitopes
Decomposition of the lipid hydroperoxide was performed by incubating 13-hydroperoxy-9Z,11E-octadecadienoic acid (1.0 mm) with FeSO4(NH4)2SO4.6H2O (125 μm) and ascorbic acid (500 μm) in 0.1 m HEPES buffer (pH 7.4) with 10% ethanol. The reaction mixture was vortexed and then followed by incubation with continuous shaking at 37 °C. After 4 h, the reaction mixture was cooled in an ice bath and extracted with diethylether. The diethylether layer was dried (Na2SO4) and concentrated in vacuo, and the residue was dissolved in acetonitrile and subjected to reverse phase HPLC on a Develosil ODS-HG-5 column (8.0-mm inner diameter × 250 mm; Nomura Chemicals, Aichi, Japan) eluted with a gradient of water containing 0.1% TFA (solvent A) with acetonitrile (solvent B) (0–20 min, 40% B; 40–50 min, 100% B) at a flow rate of 2.0 ml/min. The elution profiles were monitored by a photodiode array. The eluate was collected at 2-min intervals. The collected products were evaporated from acetonitrile and extracted using a 100-mg C18 Sep-Pak cartridge (Waters) eluted with diethylether. After concentration in vacuo, all of the fractions were incubated with 0.05 mg/ml BSA at 37 °C for 24 h as the coating agents in ELISA. The ELISA-positive fraction was further separated by reverse phase HPLC on a ChromaNik sunrise PhE column (4.6-mm inner diameter × 250 mm; ChromaNik, Osaka, Japan) eluted with a 30% acetonitrile containing 0.1% TFA at the flow rate of 1.2 ml/min. The elution profiles were monitored by a photodiode array. The eluate was collected at 2-min intervals. The collected products were evaporated from acetonitrile and extracted using a 100-mg C18 Sep-Pak cartridge (Waters) eluted with diethylether. After concentration in vacuo, all of the fractions were incubated with 0.05 mg/ml BSA at 37 °C for 24 h as the coating agents in ELISA. The semicarbazone derivative of the aldehydes was prepared as described previously (15). The semicarbazone derivatives were analyzed by ultra performance liquid chromatography-electrospray tandem mass spectrometry.
ELISA
A 100-μl aliquot of the antigen solution was added to each well of a 96-well microtiter plate and incubated for 20 h at 4 °C. The antigen solution was then removed, and the plate was washed with PBS containing 0.5% Tween 20 (PBS/Tween). Each well was incubated with 200 μl of 4% Blockace (Yukijirushi, Sapporo, Japan) in PBS/Tween for 60 min at 37 °C to block the unsaturated plastic surface. The plate was then washed three times with PBS/Tween. A 100-μl aliquot of a 1 × 103 dilution of serum was added to each well and incubated for 2 h at 37 °C. After discarding the supernatants and washing three times with PBS/Tween, 100 μl of a 5 × 103 dilution of goat anti-mouse IgG conjugated to horseradish peroxidase in PBS/Tween was added. After 1 h at 37 °C, the supernatant was discarded, and the plates were washed three times with PBS/Tween. The enzyme-linked antibody bound to the well was revealed by adding 100 μl/well of 1,2-phenylenediamine (0.5 mg/ml) in a 0.1 m citrate/phosphate buffer (pH 5.5) containing 0.003% hydrogen peroxide. The reaction was terminated by the addition of 2 m sulfuric acid (50 μl/well), and the absorbance at 490 nm was read using a micro-ELISA plate reader.
Preparation of mAbs from MRL-lpr Mice
Spleen cells from 14-week-old MRL-lpr mice were fused with P3/U1 murine myeloma cells and cultured in hypoxantine/aminopterin/thymidine selection medium. The culture supernatants of the hybridoma were screened using an ELISA, employing pairs of wells in microtiter plates on which were absorbed calf thymus DNA and ONE-treated BSA as antigens (0.5 μg of protein or DNA/well). After incubation with 100 μl of the hybridoma supernatants and with intervening washes with Tris-buffered saline (pH 7.8) containing 0.05% Tween 20 (TBS-Tween), the wells were incubated with alkaline phosphatase-conjugated goat antimouse IgG, followed by a substrate solution containing 1 mg/ml p-nitrophenyl phosphate. Hybridoma cells, corresponding to the supernatants that were positive on either DNA or ONE-modified BSA and negative on native BSA, were then cloned by limited dilution. After repeated screenings, three clones showing the most distinctive recognition of DNA and two clones showing the most distinctive recognition of both ONE-modified BSAs were obtained.
Antibody Sequence Analysis
Immunogloblin variable region genes were cloned and sequenced following amplification by PCR. The total RNA was prepared from 5 × 106 hybridoma cells by the phenol-guanidine isothiocyanate method (TRIzol; Invitrogen) according to the manufacturer's protocol. The first strand cDNA synthesis was performed with recombinant Moloney murine leukemia virus reverse transcriptase (Superscript II; Invitrogen) using the manufacturer's protocol. A 5-μg sample of the total RNA was primed with 10 pmol of random primers. Variable region genes were amplified using degenerate sense primers homologous to the mouse heavy and light chain leader sequences and antisense constant primers (Novagen), as described previously (16). The amplification products were ligated into the pGEM-T Easy Vector (Promega) using standard protocols, and both strands of inserts were sequenced using an automated dye chain termination DNA sequencer. The obtained sequences were analyzed using the DNASTAR software (DNASTAR, Madison, WI). The BLAST protocol was used to search the GenBankTM database to determine the homology with the V regions of other murine Abs that have been sequenced (17).
Glomerular Immunoglobulin Deposition
For light microscopy, kidney tissues were fixed and embedded in paraffin, and 4-μm sections were stained with hematoxylin and eosin. For immunofluorescence, paraffin sections fixed in 4% paraformaldehyde were stained with the Alexa Fluor 488-conjugated antibody against mouse IgG (Molecular Probes; Invitrogen). The images were digitally captured using a laser scanning confocal microscope and Pascal LSM software (LSM5 Pascal; Zeiss, Jena, Germany).
Antibody Elution Study
The kidneys were thawed, minced, and homogenized in a high speed blender for 1 min in 4 volumes of cold PBS and then repeatedly washed in PBS. The washed homogenates were then incubated with 1 ml of glycine HCl buffer (20 mm, pH 3.0) at room temperature for 10 min and centrifuged at 1,500 rpm, and the pH of the supernatant was adjusted to pH 7.4. The eluates were dialyzed overnight against PBS at 4 °C. The samples were assessed for anti-DNA and anti-ONE titers.
Antibody Internalization Assay
Jurkat cells were incubated in the complete culture medium with mAb DSO, anti-DNA mAb, or control murine IgG. At different time points, the cells were fixed with 1% paraformaldehyde for 20 min. After extensive washing and treating with or without 0.1% Triton X-100 for 5 min, the cells were stained with the Alexa Fluor 488-conjugated antibody against mouse IgG for 1 h. Cell penetration was analyzed using a flow cytometer JSAN (Bay Bioscience, Kobe, Japan). A total of 10,000 events/sample were counted and analyzed. For confocal laser scanning microscopy, the cells were handled as above, and the nucleus was then stained with propidium iodide (160 nm) (Sigma) for 10 min after incubation with the Alexa Fluor 488-conjugated anti-IgG. The images were digitally captured using a laser scanning confocal microscope and Pascal LSM software (LSM5 Pascal; Zeiss).
RESULTS
Lipid Peroxidation Modification of Serum Albumin Generates Autoantigenic Epitopes Recognized by SLE Sera
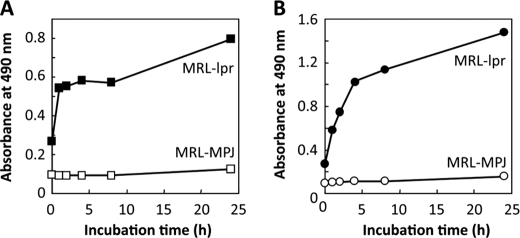
These experiments were done to determine whether reactive products derived from peroxidized polyunsaturated fatty acids could modify protein and lead to the formation of autoantigenic epitopes recognized by the sera from MRL-lpr mice, a representative murine model of SLE, and from control MRL-MpJ mice. To this end, BSA was incubated with a polyunsaturated fatty acid (arachidonic acid or linoleic acid) in the presence of the Fe2+/ascorbate free radical-generating system and evaluated the formation of the autoantigenic epitopes. Strikingly, along with the progress in the peroxidation of linoleate, the protein showed a significant cross-reactivity with the sera from the MRL-lpr mice, whereas no cross-reactivity with the sera from the MRL-MpJ mice was observed (Fig. 1A). A similar increase in the cross-reactivity with the SLE sera was obtained when the protein was incubated with peroxidized arachidonate (Fig. 1B). Thus, the peroxidation of these polyunsaturated fatty acids is likely to generate product(s) that may have the ability to cause formation of autoantigenic epitopes that cross-react with the SLE sera.
FIGURE 1.
Lipid peroxidation modification of serum albumin generates autoantigenic epitopes recognized by the sera from MRL-lpr mice. BSA was incubated with a polyunsaturated fatty acid (arachidonic acid or linoleic acid) in the presence of the Fe2+/ascorbate free radical-generating system. Cross-reactivity of modified proteins with the sera from MRL-lpr and MRL-MpJ mice was examined by ELISA. A, formation of autoantigenic epitopes upon reaction of BSA with peroxidized linoleic acid. Closed square, MRL-lpr mice; open square, MRL-MpJ mice. B, formation of autoantigenic epitopes upon reaction of BSA with peroxidized arachidonic acid. Closed circle, MRL-lpr mice; open circle, MRL-MpJ mice.
Identification of a Lipid Peroxidation-derived Active Molecule
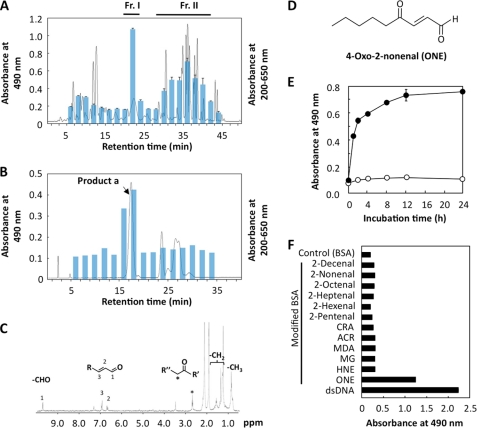
To define the lipid peroxidation product responsible for the formation of autoantigenic epitopes in the protein, we prepared the oxidized products by incubation of 13-hydroperoxy-9Z,11E-octadecadienoic acid with 125 μm Fe2+ and 0.5 mm ascorbic acid in 0.1 m HEPES buffer (pH 7.4) at 37 °C for 4 h. The diethyl ether extract of the reaction mixture was separated and fractionated every 2 min by reverse phase HPLC (Fig. 2A). The aliquots of each fraction were then incubated with BSA in 0.1 m HEPES buffer (pH 7.4) at 37 °C for 24 h and evaluated for its cross-reactivity with the sera from MRL-lpr mice. The ELISA analysis showed that the two fractions (fractions I and II), upon incubation with BSA, had a significant cross-reactivity with the autoantibodies. Because fraction I was expected to contain a major source of autoantigenic epitopes, we tried to isolate the product from fraction I. Using reverse phase HPLC on a phenethyl column, fraction I was further separated from other products, yielding the main product (product a) eluted from 16 to 18 min, which upon reaction with BSA formed autoantigenic epitopes cross-reactive with the SLE serum (Fig. 2B). After purification, the chemical structure of this product was characterized by NMR and ultra performance liquid chromatography-electrospray tandem mass spectrometry analyses. The 1H NMR spectrum of product a showed the presence of an aldehydic proton at δ 9.73, two mutually coupled signals, and δ 6.97 (doublet) and δ 6.70 (double doublet) of the α,β-unsaturated aldehyde moiety. Moreover, the product has a carbonyl group; this was supported by the chemical shift in the methylene proton at δ 2.72. These data suggested the presence of a γ-keto-α,β-unsaturated aldehyde structure (Fig. 2C). The ultra performance liquid chromatography-electrospray tandem mass spectrometry analysis of product a after semicarbazone derivatization showed that the product has a precursor ion at m/z 269 [M + H]+ and collision-induced dissociation on m/z 269, giving rise to peaks at m/z 209 [M - NHCONH2] and m/z 151 [M + 1 − 2(NHCONH2)]. Thus, the most likely structure of this product was 4-oxo-2-nonenal (ONE) (Fig. 2D), a recently identified aldehyde originating from the peroxidation of ω6 polyunsaturated fatty acids (18, 19). Indeed, the semicarbazone derivative of the authentic ONE gave the same fragmentation pattern as product a (supplemental Fig. S1). Furthermore, we observed that (i) ONE, upon incubation with BSA, generated autoantigenic epitopes that cross-reacted with the sera from MRL-lpr mice (Fig. 2E); and (ii) among several unsaturated aldehydes of chain lengths varying from three to nine carbons, the ONE-modified protein exclusively cross-reacted with the MRL-lpr mice sera (Fig. 2F). Thus, the active molecule generated from lipid peroxidation was identified to be ONE.
FIGURE 2.
Identification of a source of autoantigenic epitopes recognized by the sera from MRL-lpr mice. A, lipid peroxidation products were fractionated by reverse phase HPLC on an ODS column, and the fractions were collected every 2 min. All of the fractions were incubated with serum albumin at 37 °C for 24 h as the coating agent in ELISA. Solid line, profile of UV absorbance at 200–650 nm. Bar, ELISA analysis. B, fraction I was analyzed by reverse phase HPLC on a phenethyl column, and the fractions were collected every 2 min. All of the fractions were incubated with serum albumin at 37 °C for 24 h, as the coating agent in ELISA. Solid line, profile of UV absorbance at 200–650 nm. Bar, ELISA analysis. C, 1H NMR spectrum of the product a. D, chemical structure of ONE. E, cross-reactivity of the MRL-lpr mice sera with ONE-treated proteins. Affinity of the MRL-lpr mice sera was determined by a direct antigen ELISA using ONE-treated BSA as the absorbed antigens. F, cross-reactivity of the MRL-lpr mice sera with aldehydes-treated proteins. Affinity of the MRL-lpr mice sera was determined by a direct antigen ELISA using dsDNA and native and aldehyde-treated BSA as the absorbed antigens.
Elevation of the Anti-ONE Titer in Autoimmune Diseases
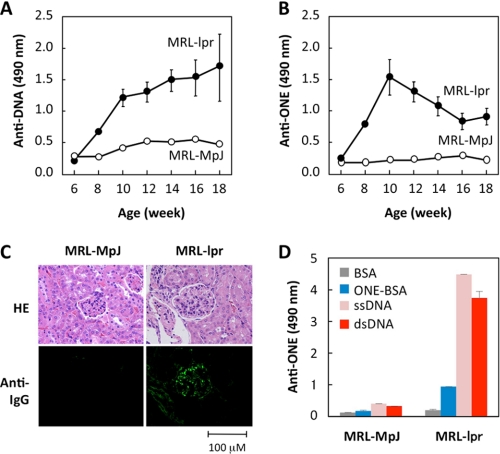
Following the identification of ONE, the age-dependent change in the antibody titer against the ONE-specific epitopes was measured in the sera from the MRL-lpr mice and control MRL-MpJ mice. The control MRL-MpJ mice did not show any significant anti-DNA and anti-ONE titers. In contrast, the MRL-lpr mice displayed a spontaneous age-dependent elevation of the anti-DNA titer (Fig. 3A). Importantly, the mice also displayed a significant increase in the anti-ONE titers (Fig. 3B). Of note, the anti-DNA titer progressively increased up to the age of 18 weeks, whereas the anti-ONE response showed different kinetics; it peaked at the age of 10 weeks and then decreased. These data suggest that, although the details remain unknown, the anti-DNA and anti-ONE responses might be differentially regulated in the MRL-lpr mice.
FIGURE 3.
Anti-ONE response in MRL-lpr mice. A, anti-DNA response in MRL-MpJ (open circle) and MRL-lpr mice (closed circle). The antibody response was examined by an ELISA employing pairs of wells in microtiter plates on which were absorbed calf thymus DNA as the antigen. B, anti-ONE response in MRL-MpJ (open circle) and MRL-lpr mice (closed circle). The antibody response was examined by an ELISA employing pairs of wells in microtiter plates on which were absorbed ONE-treated BSA as the antigen. C, glomerular immunoglobulin deposition in 20-week-old MRL-MpJ and MRL-lpr mice. Kidney sections were stained with Alexa Fluor 488-conjugated antibody against mouse IgG. HE, hematoxylin and eosin. D, immunoreactivity of antibodies eluted from the kidneys of MRL-MpJ and MRL-lpr mice. Affinity of the antibodies was determined by a direct antigen ELISA using ssDNA, dsDNA, BSA, and ONE-treated BSA as the absorbed antigens.
Glomerulonephritis is a major cause of morbidity and mortality in patients with SLE. The deposition of autoantibodies in the glomeruli plays a key role in the development of lupus nephritis. The MRL-lpr mice indeed had a moderate immunoglobulin deposition, mostly localized in the glomeruli, whereas no glomerular immunoglobulin deposition was present in the control MRL-MpJ mice (Fig. 3C). We also assessed the specificity of the antibody that deposited in glomeruli of the MRL-lpr mice and observed that the titers of the anti-ONE antibodies were significantly higher in the MRL-lpr mice than in the MRL-MpJ mice (Fig. 3D). These data suggest that the pathogenic autoantibodies recognize the ONE-specific epitopes as autoantigenic epitopes.
Elevation of the anti-ONE titers was also observed in the autoimmune hemolytic anemia-prone New Zealand Black mice (supplemental Fig. S2). In addition, we observed enhanced anti-ONE antibody levels in the superoxide dismutase 1 (SOD1)-deficient (SOD1−/−) mice and that the anti-ONE titer was significantly suppressed by the transgenic expression of SOD1 (supplemental Fig. S3). These data were consistent with our previous observations that the SOD1 deficiency causes anemia and autoimmune responses against red blood cells and that the transgenic expression of human SOD1 in erythroid cells rescues them (20). Moreover, we evaluated the presence of the anti-ONE antibodies in SLE patients and observed pronounced increases in them (supplemental Fig. S4). These data suggest that the antibody response against the ONE-specific epitopes may be an immunological characteristic common to various types of autoimmune diseases.
Isolation and Specificity of Autoantibodies
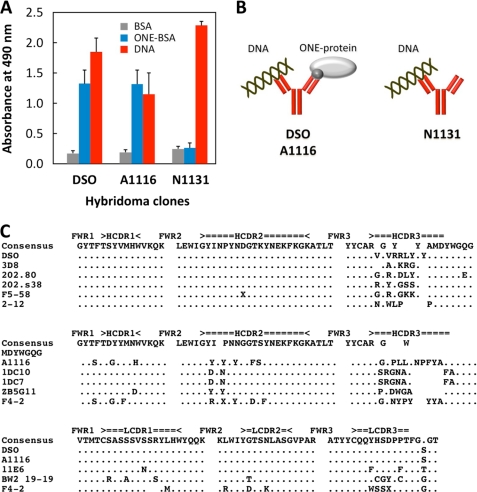
Based on the observations that the MRL-lpr mice spontaneously developed the anti-DNA and the anti-ONE titers, we tried to isolate the hybridoma clones producing the antibodies specific for DNA and/or ONE-specific epitopes from the MRL-lpr mice and characterize their specificity in detail. Spleen cells from female MRL-lpr mice at 14 weeks of age were fused with P3/U1 murine myeloma cells, and after screening based on specific binding to the DNA, we established three hybridoma clones, DSO, A1116, and N1131, producing the antibodies specific for the DNA. Two of these cloned autoantibodies, DSO and A1116, were IgGs, whereas N1131 was an IgM. Strikingly, the anti-DNA antibodies produced from these hybridoma clones, DSO and A1116, cross-reacted not only with the single- and double-stranded DNA but also with the ONE-specific epitopes (Fig. 4A), suggesting that the anti-DNA IgGs originating from the MRL-lpr mice may have a common specificity toward the ONE-specific epitopes (Fig. 4B). In agreement with this hypothesis, hybridoma clones, LA6, LG1, and LA23, isolated from the MRL-lpr mice after screening based on the binding to the ONE-specific epitopes produced the anti-ONE mAbs that cross-reacted with the double-stranded DNA (supplemental Fig. S5).
FIGURE 4.
Isolation and specificity of the dual-specific autoantibodies. A, immunoreactivity of the anti-DNA mAbs originating from MRL-lpr mice. The affinity of the antibodies was determined by a direct antigen ELISA using dsDNA, BSA, and ONE-treated BSA as the absorbed antigens. B, specificity of the anti-DNA IgGs originated from MRL-lpr mice. C, sequence alignments of the VH and VL domains of anti-DNA mAbs and homologous antibodies. A dot denotes sequence identity and a space denotes a gap in the sequence. The characters above the alignment represent the IgBLAST complementarity-determined regions (CDR1, CDR2, and CDR3) and framework regions (FWR1, FWR2, and FWR3). The sequences were aligned using the program ClustalW 1.82 and were manually modified. Accession numbers for the sequences are as follows. 202.80, CAA80103; 202.s38, CAA80108; 2-12, AAA97369; F5–58, AAB03597; 1DC7, AAS00741; 1DC10, AAS00737; ZB5G11, AY436953; F4-2 (VH), AAB03592; F4-2 (VL), AAB03601; 3D8, AF232220; 11E6, AAA96774; and BW2 19-19, AAL92956.
Sequence Analysis of Dual-specific Autoantibodies
The VH and VL nucleotide sequences were determined for the dual-specific mAbs DSO and A1116 isolated from the MRL-lpr mice, and the identity of their V region genes was determined by searching the GenBankTM database for homologies to the known V region genes using the BLAST protocol (17). To confirm that these mAbs represent the anti-DNA antibodies, the V region sequences were compared. A homology search revealed that the mAb DSO shared a high degree of sequence identity with the anti-DNA antibodies. The VH domain of the mAb DSO was almost identical, except their CDR3 regions (98–100% homology) to the mAbs 3D8 (anti-ssDNA/dsDNA), 202.80 (anti-ssDNA), F5–58 (anti-ssDNA/dsDNA), 202.s38 (anti-cardiolipin/ssDNA), and 2-12 (anti-Sm/anti-ssDNA) (Fig. 4C). In addition, the sequence identity of 89–99% for the VL domain of mAb DSO was shared with mAbs A1116, 11E6 (anti-nucleosome), F4-2 (anti-ssDNA), and BW2 19-19 (anti-DNA). Of interest, the presence of an arginine-arginine doublet, which may be responsible for anti-DNA reactivity (21), was noted in the hypervariable region of mAb DSO. The VH gene of A1116 also revealed a sequence similarity with the anti-DNA mAbs, ZB5G11 (anti-ssDNA), 1DC7 (anti-nuclear), 1DC10 (anti-nuclear), 11E6 (anti-nucleosome), and F4-2 (anti-ssDNA), whereas their identities were significantly lower (87–88% identity, except for CDR3 regions) than the identity of mAb DSO to the anti-DNA mAbs.
Cell Internalization of Dual-specific Autoantibodies
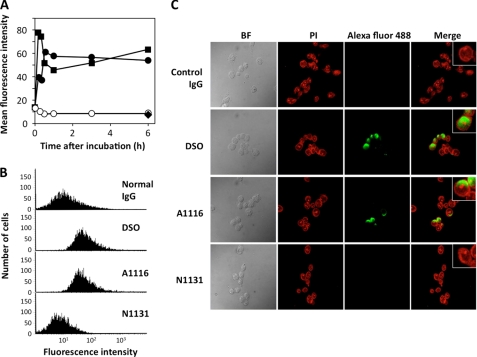
One of the mechanisms proposed to explain the pathogenicity of the autoantibodies relies on their ability to either remain on the cell surface and participate in the complement-mediated cytotoxicity or be internalized to traverse the cytoplasm and localize to the nucleus (22). To determine the penetration of the autoantibodies into live cells, Jurkat T cells were incubated with the dual-specific mAbs isolated from the MRL-lpr mice for the indicated times. The nonimmune control murine IgG was also used for comparison. After washing, fixation, and permeabilization, the intracellular mAb was stained with the Alexa Fluor 488-conjugated antibody against the mouse IgG and analyzed by flow cytometry. The dual-specific mAb DSO entered the cells in a very rapid manner (Fig. 5A). Penetration of the mAb reached a plateau within 1 h. Another dual-specific mAb A1116 was also internalized into the cells, whereas the internalization of the DNA-specific mAb N1131 was significantly less efficient than the mAbs DSO and A1116. The control IgG did not bind above background levels to the viable cells. After incubation with the mAb DSO, no positive staining was detected in the cells treated with the Alexa Fluor 488-conjugated antibody against the mouse IgG but without the Triton permeabilization (Fig. 5B), suggesting no binding of the antibody to the cell surface.
FIGURE 5.
Internalization of the dual-specific antibodies into Jurkat cells. A, time-dependent internalization of dual-specific mAbs. Internalization was analyzed by flow cytometry after treating Jurkat cells with the mAbs for different lengths of time. Closed square, DSO; closed circle, A1116; closed diamond, N1131; open circle, normal IgG. B, internalization, not cell surface binding, of dual-specific mAb into the cells. The Jurkat cells were treated with mAb DSO for 1 h. The cells were immediately fixed, extensively washed, and incubated with or without 0.1% Triton X-100. The penetration of the mAb was analyzed by flow cytometry. C, detection of internalized antibodies by confocal microscopy. The Jurkat cells were treated with the mAbs for 6 h. The cells were then stained with propidium iodide (PI) or FITC-labeled Alexa Flour 488-labeled anti-IgG and analyzed by confocal microscopy. Each figure is representative of three independent experiments.
To confirm the intracellular localization of the dual-specific mAbs, the Jurkat cells treated with the mAbs were stained with the propidium iodide or Alexa Fluor 488-conjugated antibody against the mouse IgG and analyzed by confocal laser scanning microscopy (Fig. 5C). The intracellular penetration of mAb was examined on live Jurkat cells incubated for 6 h with the antibody. Both mAb DSO and A1116 generated intense signals mostly localized in the nucleus of the cells. The cells treated with mAb N1131 and nonimmune control murine IgG did not show any positive staining even at 6 h.
DISCUSSION
A large number of oxygenated products have been identified from nonenzymatically oxidized unsaturated fatty acids. The primary and secondary nonvolatile products include hydroperoxides, epoxides, dihydroperoxides, and hydroperoxy cyclic peroxides (23). Many of these can undergo decomposition reactions to yield a large number of volatile products including unsaturated aldehydes, epoxyaldehydes, alkanes, and short chain fatty acids (23). In this study, to investigate whether lipid peroxidation is associated with the formation of autoantigenic epitopes, we examined the cross-reactivity of protein-bound lipid peroxidation products with the sera from MRL-lpr mice, which carry a defective Fas gene and develop a spontaneous lupus-like disease as they age, and identified ONE as a potential source of autoantigenic epitopes. The peroxidation of polyunsaturated fatty acids is known to generate a number of α,β-unsaturated aldehydes, such as nonenal, heptenal, pentenal, and crotonaldehyde; however, even extensive modification of a protein with these aldehydes failed to cause recognition by the sera. Modification of the protein with 4-hydroxy-2-nonenal, a peroxidation product that is relatively abundant in lipid peroxidation, was also found not to lead to recognition by the sera. ELISA analysis of the reactive HPLC fractions (Fig. 2) suggested the presence of other oxygenated compounds that may be fatty acid epoxides, hydroxides, or peroxides, but it was not established whether they are the substances that are responsible for recognition by the sera. Therefore, it is apparent from these data that the SLE sera exhibit a remarkable specificity for certain ONE-specific epitopes generated on protein molecules.
Following the identification of ONE as the source of the autoantigenic epitopes, we also measured the change in the antibody titer against the ONE-specific epitopes in the sera from MRL-lpr mice and observed the spontaneous age-dependent increase in the anti-DNA and anti-ONE responses (Fig. 3). In addition to the SLE-prone MRL-lpr mice, the anti-ONE response was elevated in the autoimmune hemolytic anemia-prone New Zealand Black mice, and an antioxidant, N-acetylcysteine, significantly suppressed the inflammatory response (supplemental Fig. S2). More intriguingly, utilizing the SOD1−/− mice, we showed that a SOD1 deficiency was associated with a significant increase in the levels of the anti-ONE titer and that the transgenic expression of SOD1 in the SOD1−/− mice resulted in a significant suppression of the anti-ONE response (supplemental Fig. S3). The increase in the levels of the anti-ONE antibodies was also observed in the SLE patients (supplemental Fig. S4). Thus, the antibody response against the ONE-specific epitopes may be an immunological characteristic common to various types of autoimmune diseases, and the serum anti-ONE titer may have a potentially important diagnostic value in human autoimmune diseases. Moreover, these observations suggest the possibility that, although the potential role in pathogenesis needs to be further explored, lipid peroxidation may not simply be an associated side effect of the disease progression but a possible etiology of SLE and other autoimmune diseases.
Based on the observations that the MRL-lpr mice spontaneously developed the anti-DNA and the anti-ONE titers, we tried to isolate the hybridoma clones, producing the antibodies specific for DNA and/or ONE-specific epitopes, from the MRL-lpr mice. In addition to the anti-DNA mAb N1131 specific for the native DNA, we isolated the anti-DNA antibodies, DSO and A1116, that cross-reacted not only with the single- and double-stranded DNA, but also with the ONE-specific epitopes (Fig. 4A). These data suggest the existence of at least two distinct populations of B cell clones in MRL-lpr mice; one clone produces the antibodies specific to both ONE-modified proteins and native DNA; the other clone produces the antibodies specific to native DNA (Fig. 4B). Dual specificity toward DNA and the ONE-modified protein is not unique to these antibodies raised from the MRL-lpr mice. We have indeed observed that the anti-DNA mAb BV 16-13, which has been characterized in detail because of its specificity for both DNA and the hinge region (24), also cross-reacts with the ONE-modified protein. In addition, we have examined the affinity of the anti-DNA mAbs D431 and D466 obtained from the 56R and CD40L-double transgenic mice (25, 26), a spontaneous murine model of SLE, to the ONE-modified protein and found that, of the two anti-DNA mAbs, mAb D466 cross-reacted with the ONE-modified BSA.3 Of interest, the anti-DNA mAb raised from the BALB/c mice immunized with the 4-hydroxy-2-nonenal-modified protein has been shown to cross-react with the ONE-modified protein (13). Several different antigenic cross-reactivities have been identified for the anti-DNA antibodies (27). These antibodies share structural similarities with antibodies against bacterial polysaccharide, and some cross-react with the bacterial polysaccharide and protect mice against a lethal bacterial infection (28, 29). Other studies have also demonstrated a cross-reactivity of the anti-DNA antibodies with microbial protein antigens, non-nucleic acid autoantigens, cell membranes, and extracellular matrix components (30–34). Thus, at least some of the anti-DNA antibodies seen in autoimmune disease are likely to arise from antigens associated with the modification by ONE, whereas some related adducts arising from other lipid oxidation products or possibly some ligands that are of quite distinct origin are also likely to be involved. Further study is required to establish the mechanism for the cross-reactivities of the anti-DNA mAbs toward covalently modified proteins with the lipid peroxidation products.
ONE reacts with biological molecules, such as protein and DNA, at rates significantly greater than other lipid peroxidation-derived aldehydes (18, 35–39). Upon reaction with proteins, ONE selectively modifies the nucleophilic side chains of lysine, histidine, cysteine, and arginine (40). The predominant initial reaction appears to involve the Michael addition to the central ONE double bond, more at C3 than at C2, to produce substituted 4-oxononanals. These adducts are relatively unstable and could be further converted into stable products including dihydrofuran, dihydropyrrole, and isomeric 4-ketoamide derivatives originating from the reaction of ONE with lysine (41) and a substituted imidazole derivative with arginine (42). ONE also forms furan derivatives upon its reaction with the cysteine and histidine derivatives (43, 44). In our recent study, we carried out a comprehensive study on the identification of the ONE-N-acetyl-l-cysteine adducts and identified several advanced reaction products that originated from the initial Michael adducts (45). We tried to elucidate the structure of the autoantigenic epitopes recognized by the MRL-lpr-derived mAb DSO and identified the ONE-cysteine adduct as a candidate epitope.4 Although the detailed mechanisms for the dual cross-reactivity of the mAb toward the native DNA and the ONE-cysteine adduct remain unclear, there are several possible explanations for the antibody multispecificity. The antibody-combining site may have more than one contact region for unrelated ligands (46). An intriguing possibility is an antibody conformational isomerism, in which the antibody has two structurally dissimilar binding site conformations and can bind to two structurally distinct antigens; one site has a deep hole that binds to aromatic haptens, and the other binds to an unrelated protein or DNA antigen (47, 48). Molecular mimicry, which is characterized by an immune response to an environmental agent that cross-reacts with a host antigen, may be unlikely, because of the structural dissimilarity between the DNA and ONE-cysteine Michael adduct. Even so, the ONE-associated dual cross-reactivity of the antibodies may be ascribed, at least in part, to the highest reactivity of this major lipid hydroperoxide-derived bifunctional electrophile toward biomacromolecules, such as protein and DNA.
Multiple studies have shown that the presence of basic residues in the antibody hypervariable regions, mostly arginine residues in the HCDR3, may be responsible for the anti-DNA reactivity (21) and that cationization of otherwise irrelevant antibodies also appears to induce cellular uptake (49). Some distinct features of potential significance were found in the HCDR3 of the dual-specific mAb DSO (Fig. 4C). The HCDR3 displays an arginine-arginine doublet at the 5′ end where the second arginine appears to have been generated by junctional diversity and presumably selected by the antigen. It has been hypothesized that the positively charged amino acids, especially the arginine residues, in the HCDR3 of the anti-dsDNA are important sites involved in interaction with negatively charged macromolecules on the cell surface, thus facilitating the anti-dsDNA internalization (50). These features were absent in the mAb A1116 whose HCDR3 global charge was significantly less positive. However, the relatively long and hydrophobic sequence of the HCDR3, both ends of which are flexible residues (glycines and alanine), indicatesthat multiple conformational states and/or multiple dissimilar binding site conformations of HCDR3 enable the cross-reactivites of A1116 toward the ONE-modified proteins. It was also noted that both mAbs DSO and A1116 are encoded by VH genes that are similar to those found in the anti-DNA obtained from normal mice immunized with a protein-bound lipid peroxidation product (13). The similarity between the autoimmune and immunization-induced anti-DNA has been previously noted and supports the hypothesis that the development of the anti-DNA in normal and lupus-prone mice may occur by a common mechanism in which DNA or a protein-bound ligand stimulates B cells to differentiate in a receptor-mediated response.
It has been suggested that pathogenic autoantibodies should be able to enter live cells in multiple tissues, reach the appropriate intracellular compartment and interfere with the cell function. We indeed observed that the dual-specific mAb DSO entered human Jurkat leukemia T lymphocytes (Fig. 5). The functional consequence of the antibody internalization into the cells remains obscure; however, there are data to suggest that the anti-DNA antibodies either can remain on the cell surface (nonpenetrating) and participate in the complement-mediated cytotoxicity or can be internalized to traverse the cytoplasm and localize in the nucleus (22). In addition, a number of anti-DNA antibodies have been shown to mediate cellular response, such as a proliferative response, decreased cell viability, and increased synthesis of extracellular matrix components and pro-inflammatory cytokines (51–53). Thus, the preferential binding of dual-specific mAbs to living Jurkat cells may represent a potential mechanism for T cell activation, thereby further amplifying the inflammatory process.
Supplementary Material
This work was supported by a research grant from the Ministry of Education, Culture, Sports, Science, and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
N. Otaki and K. Uchida, unpublished data.
Y. Shimozu, N. Otaki, and K. Uchida, unpublished data.
- SLE
- systemic lupus erythematosus
- ONE
- 4-oxo-2-nonenal
- ssDNA
- single-stranded DNA.
REFERENCES
- 1.Stadtman E. R. (1992) Science 257, 1220–1224 [DOI] [PubMed] [Google Scholar]
- 2.Stadtman E. R., Levine R. L. (2000) Ann. N.Y. Acad. Sci. 899, 191–208 [DOI] [PubMed] [Google Scholar]
- 3.Uchida K. (2000) Free Radic. Biol. Med. 28, 1685–1696 [DOI] [PubMed] [Google Scholar]
- 4.Uchida K. (2003) Prog. Lipid Res. 42, 318–343 [DOI] [PubMed] [Google Scholar]
- 5.Esterbauer H., Schaur R. J., Zollner H. (1991) Free Radic. Biol. Med. 11, 81–128 [DOI] [PubMed] [Google Scholar]
- 6.Binder C. J., Silverman G. J. (2005) Springer Semin. Immunopathol. 26, 385–404 [DOI] [PubMed] [Google Scholar]
- 7.Koffler D., Carr R. I., Agnello V., Thoburn R., Kunkel H. G. (1971) J. Exp. Med. 134, 294–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pisetsky D. S., Grudier J. P., Gilkeson G. S. (1990) Arthritis Rheum. 33, 153–159 [DOI] [PubMed] [Google Scholar]
- 9.Gilkeson G. S., Pippen A. M., Pisetsky D. S. (1995) J. Clin. Invest. 95, 1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurz E. U., Lees-Miller S. P. (2004) DNA Repair 3, 889–900 [DOI] [PubMed] [Google Scholar]
- 11.Oren M. (1999) J. Biol. Chem. 274, 36031–36034 [DOI] [PubMed] [Google Scholar]
- 12.Brown J. M., Wouters B. G. (1999) Cancer Res. 59, 1391–1399 [PubMed] [Google Scholar]
- 13.Toyoda K., Nagae R., Akagawa M., Ishino K., Shibata T., Ito S., Shibata N., Yamamoto T., Kobayashi M., Takasaki Y., Matsuda T., Uchida K. (2007) J. Biol. Chem. 282, 25769–25778 [DOI] [PubMed] [Google Scholar]
- 14.Brash A. R., Song W. (1996) Methods Enzymol. 272, 250–259 [DOI] [PubMed] [Google Scholar]
- 15.Chen L. J., Hecht S. S., Peterson L. A. (1995) Chem. Res. Toxicol. 8, 903–906 [DOI] [PubMed] [Google Scholar]
- 16.Antone S. M., Adderson E. E., Mertens N. M., Cunningham M. W. (1997) J. Immunol. 159, 5422–5430 [PubMed] [Google Scholar]
- 17.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rindgen D., Nakajima M., Wehrli S., Xu K., Blair I. A. (1999) Chem. Res. Toxicol. 12, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 19.Lee S. H., Blair I. A. (2000) Chem. Res. Toxicol. 13, 698–702 [DOI] [PubMed] [Google Scholar]
- 20.Iuchi Y., Okada F., Takamiya R., Kibe N., Tsunoda S., Nakajima O., Toyoda K., Nagae R., Suematsu M., Soga T., Uchida K., Fujii J. (2009) Biochem. J. 422, 313–320 [DOI] [PubMed] [Google Scholar]
- 21.Radic M. Z., Mackle J., Erikson J., Mol C., Anderson W. F., Weigert M. (1993) J. Immunol. 150, 4966–4977 [PubMed] [Google Scholar]
- 22.Deshmukh U. S., Bagavant H., Fu S. M. (2006) Autoimmun. Rev. 5, 414–418 [DOI] [PubMed] [Google Scholar]
- 23.Frankel E. N. (1983) Prog. Lipid Res. 22, 1–33 [DOI] [PubMed] [Google Scholar]
- 24.Ballard D. W., Voss E. W., Jr. (1985) J. Immunol. 135, 3372–3380 [PubMed] [Google Scholar]
- 25.Chen C., Radic M. Z., Erikson J., Camper S. A., Litwin S., Hardy R. R., Weigert M. (1994) J. Immunol. 152, 1970–1982 [PubMed] [Google Scholar]
- 26.Higuchi T., Aiba Y., Nomura T., Matsuda J., Mochida K., Suzuki M., Kikutani H., Honjo T., Nishioka K., Tsubata T. (2002) J. Immunol. 168, 9–12 [DOI] [PubMed] [Google Scholar]
- 27.Spatz L., Iliev A., Saenko V., Jones L., Irigoyen M., Manheimer-Lory A., Gaynor B., Putterman C., Bynoe M., Kowal C., Kuo P., Newman J., Diamond B. (1997) Methods 11, 70–78 [DOI] [PubMed] [Google Scholar]
- 28.Kowal C., Weinstein A., Diamond B. (1999) Eur. J. Immunol. 29, 1901–1911 [DOI] [PubMed] [Google Scholar]
- 29.Limpanasithikul W., Ray S., Diamond B. (1995) J. Immunol. 155, 967–973 [PubMed] [Google Scholar]
- 30.Chan T. M., Yu P. M., Tsang K. L., Cheng I. K. (1995) Clin. Exp. Immunol. 100, 506–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob L., Lety M. A., Choquette D., Viard J. P., Jacob F., Louvard D., Bach J. F. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2956–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen C., Otto E., Kuhlemann K., Forster G., Kahaly G. J. (1996) Clin. Exp. Rheumatol. 15, 559–567 [Google Scholar]
- 33.Katz J. B., Limpanasithikul W., Diamond B. (1994) J. Exp. Med. 180, 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budhai L., Oh K., Davidson A. (1996) J. Clin. Invest. 98, 1585–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin D., Lee H. G., Liu Q., Perry G., Smith M. A., Sayre L. M. (2005) Chem. Res. Toxicol. 18, 1219–1231 [DOI] [PubMed] [Google Scholar]
- 36.Rindgen D., Lee S. H., Nakajima M., Blair I. A. (2000) Chem. Res. Toxicol. 13, 846–852 [DOI] [PubMed] [Google Scholar]
- 37.Kawai Y., Kato Y., Nakae D., Kusuoka O., Konishi Y., Uchida K., Osawa T. (2002) Carcinogenesis 23, 485–489 [DOI] [PubMed] [Google Scholar]
- 38.Kawai Y., Uchida K., Osawa T. (2004) Free. Radic. Biol. Med. 36, 529–541 [DOI] [PubMed] [Google Scholar]
- 39.Pollack M., Oe T., Lee S. H., Silva Elipe M. V., Arison B. H., Blair I. A. (2003) Chem. Res. Toxicol. 16, 893–900 [DOI] [PubMed] [Google Scholar]
- 40.Doorn J. A., Petersen D. R. (2002) Chem. Res. Toxicol. 15, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 41.Zhu X., Sayre L. M. (2007) Chem. Res. Toxicol. 20, 165–170 [DOI] [PubMed] [Google Scholar]
- 42.Oe T., Lee S. H., Silva Elipe M. V., Arison B. H., Blair I. A. (2003) Chem. Res. Toxicol. 16, 1598–1605 [DOI] [PubMed] [Google Scholar]
- 43.Zhang W. H., Liu J., Xu G., Yuan Q., Sayre L. M. (2003) Chem. Res. Toxicol. 16, 512–523 [DOI] [PubMed] [Google Scholar]
- 44.Yocum A. K., Oe T., Yergey A. L., Blair I. A. (2005) J. Mass Spectrom. 40, 754–764 [DOI] [PubMed] [Google Scholar]
- 45.Shimozu Y., Shibata T., Ojika M., Uchida K. (2009) Chem. Res. Toxicol. 22, 957–964 [DOI] [PubMed] [Google Scholar]
- 46.Schwartz R. S., Stollar B. D. (1985) J. Clin. Invest. 75, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James L. C., Roversi P., Tawfik D. S. (2003) Science 299, 1362–1367 [DOI] [PubMed] [Google Scholar]
- 48.Foote J. (2003) Science 299, 1327–1328 [DOI] [PubMed] [Google Scholar]
- 49.Pardridge W. M., Buciak J., Yang J., Wu D. (1998) J. Pharmacol. Exp. Ther. 286, 548–554 [PubMed] [Google Scholar]
- 50.Song Y. C., Sun G. H., Lee T. P., Huang J. C., Yu C. L., Chen C. H., Tang S. J., Sun K. H. (2008) Eur. J. Immunol. 38, 3178–3190 [DOI] [PubMed] [Google Scholar]
- 51.Yung S., Tsang R. C., Sun Y., Leung J. K., Chan T. M. (2005) J. Am. Soc. Nephrol. 16, 3281–3294 [DOI] [PubMed] [Google Scholar]
- 52.Chan T. M., Leung J. K., Ho S. K., Yung S. (2002) J. Am. Soc. Nephrol. 13, 1219–1229 [DOI] [PubMed] [Google Scholar]
- 53.Yung S., Tsang R. C., Leung J. K., Chan T. M. (2006) Kidney Int. 69, 272–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.