Abstract
Many bacterial pathogens present adhesins at the tips of long macromolecular filaments known as pili that are often important virulence determinants. Very little is known about how pili presented by Gram-positive pathogens mediate host cell binding. The crystal structure of a pilus adhesin from the important human pathogen Streptococcus pyogenes reveals an internal thioester bond formed between the side chains of a cysteine and a glutamine residue. The presence of the thioester was verified using UV-visible spectroscopy and mass spectrometry. This unusual bond has only previously been observed in thioester domains of complement and complement-like proteins where it is used to form covalent attachment to target molecules. The structure also reveals two intramolecular isopeptide bonds, one of these formed through a Lys/Asp residue pair, which are strategically positioned to confer protein stability. Removal of the internal thioester by allele-replacement mutagenesis in S. pyogenes severely compromises bacterial adhesion to model host cells. Although current paradigms of bacterial/host cell interaction envisage strong non-covalent interactions, the present study suggests cell adhesion could also involve covalent bonds.
Keywords: Cell Adhesion, Cell Surface Protein, Protein Chemistry, Protein Structure, X-ray Crystallography, Gram-positive Pili, Streptococcus Pyogenes, Host/Pathogen Interaction, Internal Thioester
Introduction
Adhesion to a host cell surface is often a key step in establishing a successful infection. Many specific adhesins are projected outwards from bacterial surfaces by extended polymeric protein filaments known as pili. These structures have received considerable attention due to their roles in enabling host colonization and potential as vaccine candidates (1–6). In Gram-positive bacteria, pili are assembled through intermolecular covalent linkages, formed by the action of sortases, which recognize specific sequence motifs on substrate proteins (7–10). An emerging paradigm for the structure of Gram-positive pili includes a minor pilus subunit displayed at the tip followed by a polymer of repeating subunits (composed of a protein frequently referred to as the “shaft” or “major” pilus subunit); this structure is then attached to the bacterial surface, frequently through a second minor pilus subunit that acts as a cell wall linker. In addition to intermolecular links between subunits, major pilus subunits also contain intramolecular isopeptide bonds (11–14) that confer proteolytic, thermodynamic and mechanical stability on these proteins (12, 13, 15, 16). Together, these bonds are thought to provide structural stability to these thin (1 molecule), extended (100s of molecules) structures and maintain pilus integrity during the early stages of interaction with host cells.
Streptococcus pyogenes (Group A Streptococcus) is an important human host-specific pathogen that can cause a wide variety of infections including potentially life-threatening diseases such as acute rheumatic fever, streptococcal toxic shock syndrome, and necrotizing fasciitis (17). Pili presented on the surface of the serotype M1 Group A Streptococcus strain SF370 (GAS)3 are required for efficient adhesion to model host cells and clinically relevant tissues (18–20, 51). These pili are composed of three distinct protein subunits: Spy0128, the major shaft subunit; Spy0130, which links the polymerized pilus to the cell-wall peptidoglycan layer; Spy0125, which has been localized at the tip of the pilus (as has its homologue in serotype M3 Group A Streptococcus strain AM3) (22, 51). Each of these proteins is encoded within a genetic locus, known as the FCT region, along with a specific sortase enzyme (pilin polymerase, Spy0129) that catalyzes the intermolecular linkages between pilus proteins. Recent advances have revealed the high resolution crystal structures of Spy0128 and a homologue of Spy0130 from Group A Streptococcus strain 90/306S (23); a crystal structure of the housekeeping sortase from GAS (that links the pilus to the cell wall) has also been reported (24). High resolution structural information on Spy0125 would generate a complete atomic-level description of all the components of a Gram-positive adhesive pilus.
Spy0125 is the pilus-presented adhesin in GAS, as deletion of the gene encoding this protein from the pathogen chromosome or preincubation of bacteria with Spy0125-specific anti-sera renders the bacteria essentially non-adherent (18, 51). Studies in other important Gram-positive pathogens, including Corynebacterium diphtheriae (25) and Streptococcus pneumoniae TIGR4 (26, 27), suggest similar roles for component pilus subunits despite there being considerable divergence in amino acid sequence between equivalent subunits across these pathogens. Very little is known about how Gram-positive pilus subunits mediate adhesion at the molecular level.
The region of Spy0125 responsible for adhering to host cells has been localized to a domain comprising residues Asn-286—Thr-723 (Spy0125 C-terminal region (CTR) (51)). Deletion of residues 53- 285 in GAS does not affect the ability of these bacteria to bind cultured host cells (51), and in some GAS serotypes this region has been lost from the genome (see supplemental Fig. 1), revealing it is dispensable for pilus assembly and Spy0125 function.
In this study the crystal structure of Spy0125-CTR is presented in two different crystal forms. The protein is formed from three domains. Two of these contain intramolecular isopeptide bonds, with one of these formed through a Lys/Asp pair resulting in the elimination of a water molecule. The third domain contains a highly unusual internal thioester bond formed between the side chains of a Cys and Gln residue; such thioesters are implicated in covalent linkages to surfaces (28). Removal of the thioester in GAS by allelic-replacement mutagenesis severely compromises pathogen binding to host cells, revealing it has a direct role in the pathogen/host interaction. Host/pathogen interactions that use strong non-covalent binding are well known; this work suggests a mechanism by which a bacterial adhesin has evolved to use a covalent bond to attach to a host cell surface.
EXPERIMENTAL PROCEDURES
Protein Production
The cloning of Spy0125 residues Asn-286—Thr-723 (Spy0125-CTR) has been described elsewhere (51). Spy0125-CTR was overexpressed in Escherichia coli BL21 (DE3) induced at A600 ∼ 0.4–0.6 with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were grown for a further 4 h before harvesting by centrifugation. For production of selenomethionine-labeled protein, E. coli B834 (DE3) cells were used with minimal media supplemented with l-selenomethionine. Cell pellets were resuspended in 50 mm HEPES, 150 mm NaCl, 10 mm imidazole, pH 7.5, supplemented with 5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride and lysed by sonication. Cell lysate was clarified by centrifugation, and the supernatant was applied to a Ni2+-immobilized metal ion affinity chromatography column (GE Healthcare). Proteins were eluted with an imidazole gradient. Fractions containing Spy0125-CTR were pooled and concentrated, then injected onto a Hi-Load 16/60 Superdex 200 gel filtration column (GE Healthcare) pre-equilibrated with 20 mm Tris-HCl, 150 mm NaCl, pH 7.5. Fractions containing purified Spy0125-CTR were pooled and concentrated to 10 mg/ml.
Crystallization and Data Collection
Initial crystals of native Spy0125-CTR were grown at 20 °C in a sitting drop plates setup using a robotic crystallization system, then optimized in 24-well plate hanging drop experiments. These latter experiments used 2 μl of protein (10 mg/ml) mixed with 2 μl of precipitant (precipitants used include 38–40% PEG 4000, 200 mm sodium acetate, and 100 mm trisodium citrate, pH 5.6–5.9, 34–36% PEG 5000-MME, 100 mm ammonium sulfate, 100 mm MES, pH 6.0–6.5). All native crystals obtained were of the B-form (see below). Selenomethionine-labeled protein crystallization used the conditions above and routinely gave rise to B-form crystals. In a small number of experiments, plate-like crystals of different morphology were obtained (subsequently named A-form crystals).
Data used to solve the Spy0125-CTR structures were collected on BM14 at the European Synchrotron Radiation Facility, Grenoble, France from crystals grown using selenomethionine-labeled protein. Three-wavelength MAD experiments were collected from both A-form and B-form crystals; an additional redundant data set was also collected on the same A-form crystal. A dataset from native protein crystals was collected on station I03 at the Diamond Light Source, Oxford, UK.
Data Processing and Structure Solution, Refinement/Rebuilding, and Validation
All data were processed using iMOSFLM (29) and scaled with SCALA (30), as implemented in the CCP4 suite (31). Data collection statistics are given in Table 1. The AutoSol wizard in PHENIX (Version 1.4–111 (32)) was used to solve the structures from the MAD data. The native dataset, collected at Diamond Light Source, was solved by molecular replacement using PHASER and one chain from the final B-form model. Final models of all structures were produced through iterative rounds of refinement using REFMAC5 (33) and rebuilding with COOT (34). Structure validation used COOT and MOLPROBITY (35). Ramachandran plot analysis (using MOLPROBITY) reveals no residues flagged as outliers. The A-form crystal structure comprises residues Thr-291—Thr-603 in chain A (with the exception of residues Gly-316—Ala-324, which are disordered) and Thr-291—Asn-605 in chain B (residues Asn-318—Phe-322 are disordered in this chain). In the final stages of refinement the thioester and isopeptide links were generated, and Translation-Libration-Screw restraints (middle and top domains as separate groups) were used. The B-form crystal structure from the European Synchrotron Radiation Facility data comprises residues Thr-290—Pro-719 in chain A (with the exception of residues Asn-318—Ser-321 and Ile-373—Lys-376, which are disordered) and Pro-288—Val-720 in chain B (residues Asp-317—Asn-320 and Ile-373—Lys-376 are disordered in this chain). Translation-Libration-Screw restraints were used in the final stages of refinement with the middle, top, and bottom domains forming separate groups (the top domain of the B-chain was defined by two groups). Non-crystallographic symmetry restraints (tight on main chain, medium on side chain) were used throughout. The B-form crystal structure from the Diamond Light Source data comprises residues Thr-290—Pro-719 in the chain A (with the exception of residues Val-319—Ser-321, Ile-373—Lys-376, and Glu-598—His-604, which are disordered) and Pro-288—Pro-719 in chain B (residues Ile-373—Asp-374 are disordered in this chain). Translation-Libration-Screw and non-crystallographic symmetry restraints were used as above. All refinement statistics are given in Table 1. Structural superimpositions were performed with LSQMAN (36), and protein structure figures have been prepared with PyMOL.
TABLE 1.
X-ray data collection, refinement statistics, and model analysis
| Spy0125-CTR (Selenomethionine), A-form |
Spy0125-CTR (Selenomethionine), B-form |
Spy0125-CTR (native), B-form | ||||||
|---|---|---|---|---|---|---|---|---|
| Peak | Inflection | Remote | Peak 2 | Peak | Inflection | Remote | ||
| Data collection | ||||||||
| Space group | P1 | P1 | P1 | P1 | P212121 | P212121 | P212121 | P212121 |
| Unit cell parameters (Å, °) | a = 35.43, | a = 35.42, | a = 35.42, | a = 35.48, | a = 43.10, | a = 43.13, | a = 43.06, | a = 45.81, |
| b = 66.28, | b = 66.26, | b = 66.27, | b = 66.46, | b = 113.32, | b = 113.42, | b = 113.46, | b = 117.50, | |
| c = 101.75, | c = 101.69, | c = 101.63, | c = 101.80, | c = 175.36 | c = 175.49 | c = 175.64 | c = 177.75 | |
| α = 86.19, | α = 86.20, | α = 86.36, | α = 86.06, | |||||
| β = 90.04, | β = 90.06, | β = 90.12, | β = 90.06, | |||||
| γ = 75.02 | γ = 75.06 | γ = 75.09 | γ = 75.08 | |||||
| Wavelength (Å) | 0.978 | 0.979 | 0.975 | 0.978 | 0.978 | 0.979 | 0.975 | 0.970 |
| Resolution range (Å) | 38.47-2.10 (2.21-2.10) | 38.45-2.10 (2.21-2.10) | 38.50-2.10 (2.21-2.10) | 38.00-1.90 (2.00-1.90) | 56.66-2.90 (3.06-2.90) | 56.71-2.90 (3.06-2.90) | 56.73-2.85 (3.06-2.85) | 55.00-2.65 (2.79-2.65) |
| Rmerge (%) | 4.9 (11.5) | 3.9 (10.9) | 4.2 (12.2) | 7.4 (31.8) | 10.6 (35.6) | 10.6 (36.5) | 11.1 (41.7) | 10.7 (41.1) |
| I/σ(I) | 12.5 (7.2) | 16.4 (7.7) | 14.0 (6.9) | 15.4 (5.0) | 13.2 (5.2) | 13.3 (5.0) | 12.8 (4.5) | 18.6 (8.2) |
| Completeness (%) (all data, anomalous half data sets) | 97.0 (96.0), 96.0 (95.1) | 97.0 (96.0), 96.1 (95.2) | 97.0 (96.1), 96.1 (95.1) | 93.6 (74.9) | 100 (100), 100 (100) | 100 (100), 100 (100) | 100 (100), 100 (100) | 99.9 (100.0) |
| Redundancy (all data, anomalous half data sets) | 3.3 (3.3), 1.7 (1.7) | 3.3 (3.3), 1.6 (1.6) | 3.3 (3.3), 1.7 (1.7) | 7.0 (6.7) | 7.2 (7.3), 3.8 (3.8) | 7.2 (7.3), 3.8 (3.8) | 7.2 (7.3), 3.8 (3.8) | 16.7 (17.3) |
| No. of sites (SOLVE) | 6 | 6 | ||||||
| FOM (SOLVE/RESOLVE) | 0.33/0.67 | 0.25/0.67 | ||||||
| Refinement | ||||||||
| Resolution range (Å) | 38.00-1.90 (1.95-1.90) | 56.66-2.90 (2.98-2.90) | 55.00-2.65 (2.72-2.65) | |||||
| Rwork (%) | 20.6 (23.7) | 23.1 (30.9) | 22.8 (34.8) | |||||
| Rfree (%) | 23.2 (29.6) | 27.4 (41.0) | 26.8 (42.7) | |||||
| No. of non-hydrogen atoms (protein, waters) | 4896, 527 | 6721, 21 | 6723, 23 | |||||
| Root mean square bond (Å) | 0.015 | 0.005 | 0.015 | |||||
| Root mean square angles (°) | 1.41 | 0.91 | 1.42 | |||||
| ESU (based on ML) (Å) | 0.08 | 0.35 | 0.30 | |||||
Mass Spectrometry Analyses
For accurate intact mass determination, a sample of Spy0125-CTR with the His tag removed was prepared. The molecular mass was determined by electrospray mass spectrometry using an LTQ-FT mass spectrometer (ThermoElectron, Bremen, Germany). Mass spectrum plots and spectral deconvolution were generated using the QualBrowser program (ThermoElectron). Protein peak spectra were deconvoluted to determine the average molecular mass of the protein assuming “averagine” (0.2678% sulfur) composition.
Identification of intramolecular isopeptide bonds and characterization of the thioester linkage typically involved digestion of ∼5 μg of protein with either trypsin at 30 °C for ∼16 h or Asp-N and Glu-C at 37 °C for ∼16 h (for experiments with small molecule modifiers, ∼225 μg of Spy0125-CTR was incubated (2 h at 20 °C) with either 10 mm methylamine, 2 mm iodoacetamide, or 10 mm methylamine/2 mm iodoacetamide and precipitated with cold acetone before tryptic digest). Digests were stopped by the addition of TFA to a final concentration of 0.5%. Peptides were extracted from urea-containing samples using OmixR C18 tips (Varian, Walnut Creek, CA) and re-dissolved in 0.1% TFA. Peptides were analyzed by nano-LC-MS/MS on an LTQ-OrbitrapTM machine (Thermo Fisher Scientific Inc., Waltham, MA) with a nanoAcquity UPLCTM (Waters, Manchester, UK). Tandem mass spectra were extracted by BioWorks Version 3.3.1 (ThermoElectron). Peptide assignment and protein identification used Mascot (Matrix Science, London, UK; Version Mascot 2.2, in-house). The expected masses of isopeptide-containing fragments were calculated using on-line tools. Non-native sequences were detected by manual analysis of the raw data files using the Qualbrowser of Xcalibur (Thermo). Identification of the isopeptide bond linking residues Lys-297 and Asp-595 used a tryptic digest (Table 2). The 2+ and 3+ charged versions of this peptide were detected at the same retention time. An MS/MS spectrum was selected from each charge state and visually inspected for the presence of the expected fragment ions (supplemental Table 1 and Fig. 3A). Identification of the isopeptide bond linking residues Lys-610 and Asn-715 used an Asp-N/Glu-C digest (Table 2). The 3+, 4+, and 5+ charged versions of this peptide were detected at the same retention time. MS/MS spectra of the 3+- and 4+-charged versions were selected for visual inspection as described above (supplemental Table 2 and Fig. 3B). MAXQUANT (37) was used to quantify peptides in mass spectra.
TABLE 2.
Fragments generated by proteolytic digest to identify the isopeptide bonds in Spy0125-CTR and their observed/expected mass/charge ratios
| Isopeptide bond | Enzyme | Peptide 1a | Peptide 2a | Expected mass | Observed m/zb (expected m/z) | ||
|---|---|---|---|---|---|---|---|
| Da | |||||||
| Lys-297—Asp-595 | Trypsin | (K)YAIGDYSK | ME(D)K | 1546.734 | 774.3742+ (774.375) | 516.5853+ (516.586) | |
| Lys-610—Asn-715 | AspN/GluC | VIPVTHNLTLR(K)TVTGLAG | (N)NKEPVVPT | 2968.671 | 990.5653+ (990.565) | 743.1754+ (743.176) | 594.7415+ (594.742) |
a Residues in parentheses are part of the isopeptide bond.
b Charge state as indicated.
UV-Visible Spectroscopy and Fluorimetry
UV-visible spectroscopy was used to monitor solutions containing 0.2 mm 2,2′-dithiopyridine (DTP) in 50 mm HEPES, 150 mm NaCl, 6 m urea, pH 7.5, over time. Upon reaction with free thiols, DTP liberates 2-thiopyridine, which has an intense absorption band at 343 nm. Equivalent results are obtained using buffers without urea, but the reaction takes many hours to reach completion. Fluorescence spectra were obtained using a PerkinElmer Life Sciences LS55 fluorimeter at 25 °C using 5-nm excitation and emission slits.
Urea-induced unfolding of wild-type and C426A Spy0125-CTR was observed as the appearance of a peak at ∼365 nm (consistent with solvent exposure of a tryptophan residue, excitation at 255 nm) in 20 mm K2HPO4, pH 7.2 (protein at 2.5 μm), after incubation in urea solution (0–10 m) for 15 min. Excitation and emission spectra for both wild-type and C426A variant Spy0125-CTR proteins were found to be essentially identical (supplemental Fig. 5A). The emission spectra of both proteins was dominated by a peak at 307 nm (likely due to tyrosine fluorescence). All unfolding data were normalized to the fluorescence maximum at 307 nm (supplemental Fig. 5C), and the change in fluorescence at 360 nm was plotted (supplemental Fig. 5D).
Mutagenesis of the Thioester Linkage, Adhesion Assays, and Western Blots
The C426A point mutation in Spy0125 was generated by overlap extension PCR using primers FR1 (5′-TTACATACTGCAGGTTATGTATAATGGACATCCACAA-3′, forward) and RV1 (5′-TAGATCTGCATTAAATGCATAGACAACCTGTGAAC-3′, reverse) for fragment 1 and primers FR2 (5′-GTTCACAGGTTGTCTATGCATTTAATGCAGATCTA-3′, forward) and RV2 (5′-AAGTTCTCGAGTTTAAATTCGAGGTTTGTTTTATC-3′, reverse) for fragment 2; wild-type genomic DNA was used as the template. The products were mixed, annealed, and reamplified by PCR with primers FR1 and RV2. In addition to the mutation, the primers introduced a unique NsiI site (ATGCAT, underlined). For mutagenesis in GAS, the resulting PCR product was cloned between the PstI and XhoI sites of the vector pG+host 9. After transformation into S. pyogenes, the mutant allele was introduced into the chromosome by allele-replacement mutagenesis (38). Genomic DNA from S. pyogenes colonies was screened to identify bacteria containing the desired mutant by NsiI cleavage of PCR-amplified fragments that encompass the Spy0125 sequence. DNA from colonies positive for NsiI cleavage were then fully verified by sequencing. For production of recombinant C426A Spy0125-CTR protein, the appropriate coding sequence was amplified from mutagenized GAS genomic DNA by PCR and cloned into the pOPIN-F expression vector (39). The protein was expressed and purified as described for the wild-type protein after verifying the DNA sequence.
For immunoblotting, cell-wall proteins were prepared as described (51). Samples were run on gradient (4–15% w/v) SDS-PAGE gels and blotted onto a nitrocellulose membrane. The membrane was then probed with specific anti-Spy0125 sera followed by a peroxidase conjugated anti-rabbit IgG. Adhesion assays were performed as described previously (51). Rhodamine-labeled phalloidin was used to stain HaCaT cells and visualization of GAS cells used a FITC (fluorescein)-labeled polyclonal antibody to Group A Streptococcus (Acris Antibodies). Samples were mounted and analyzed by confocal laser microscopy. For each image used, adhesion was quantified by determining the fluorescence from a single bacterium then dividing the total measured fluorescence by this value to give the total number of bacteria binding. Three independent adhesion experiments with n = 21 repeats (n = 3, n = 12, and n = 6 for the three experiments) and n = 51 repeats (n = 12, n = 24 and n = 15 for the three experiments) for the wild-type and C426A variant strain, respectively. Binding of the Δspy0125 variant strain is essentially zero (18, 51). Within each independent experiment average values for bacteria binding and their S.D. were scaled such that wild-type binding = 100%. Data from the three experiments were then combined to give the final analysis. Statistical significance was determined using Student's t tests (two-tailed, unequal variance between samples within each experiment), with p values of 0.013, 1.46 × 10−6, and 3.35 × 10−5 for each experiment respectively (comparing wild-type to the C426A variant).
RESULTS
Spy0125-CTR (residues Asn-286—Thr-723) was cloned and expressed in E. coli. The purified protein has been crystallized in two forms (referred to as A-form and B-form), and the structure was determined to 1.9 and 2.65 Å resolution, respectively (a second B-form crystal structure was determined to 2.9 Å, Table 1). Each crystal form contains two molecules in the asymmetric unit, with the A-form crystal structure spanning residues Thr-291 to Asn-605 and the B-form structures spanning residues Pro-288 to Val-720 (full details of model compositions are provided under “Experimental Procedures”).
Overall Structure of Spy0125-CTR
Spy0125-CTR adopts a three domain structure, described here as middle, top, and bottom (Fig. 1), and is very different to the recently determined RrgA pilus adhesin from S. pneumoniae TIGR4 (27). The middle domain comprises residues 291–372 and 590–597, the top domain comprises residues 390–583, and the bottom domain comprises residues 603–719; intervening residues form links between the domains. The middle and bottom domains adopt β-sandwich folds. Searches for similarity to domains of known structure (40) reveal most significant hits to the N2 domain of GBS52, a minor pilin from Streptococcus agalactiae (41) (root mean square deviation (r.m.s.d.) = 1.55 Å for 72 equivalent Cα atoms) and the C-terminal domain of Spy0128, the major pilus subunit in GAS (13) (r.m.s.d. = 1.27 Å for 96 equivalent Cα atoms) for the middle and bottom domains, respectively. The structural similarity between the C-terminal (bottom) domains of Spy0125 and Spy0128 is likely important for recognition by the pilin polymerase that catalyzes pilus biogenesis in GAS. Searches with the top domain do not reveal significant similarity to any known protein structure, and this domain appears to represent an indel event within the middle domain (it forms an apparent insertion toward the N-terminal end of strand βG of the GBS52 N2 structure but otherwise does not disrupt the fold).
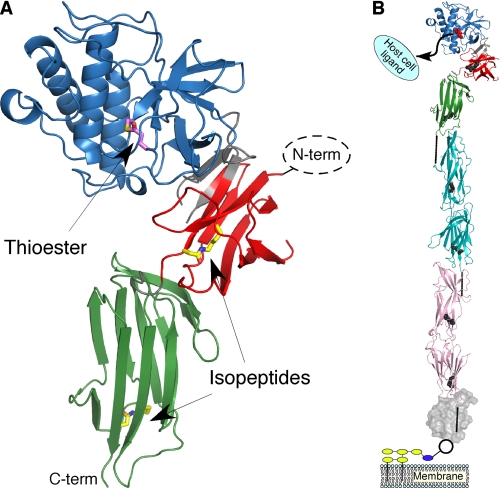
FIGURE 1.
The structure of Spy0125-CTR and its location in the GAS pilus. A, shown is a ribbon representation of the middle, top, and bottom domain colored red, blue, and green, respectively; the termini are also labeled. Residues composing the thioester bond (Cys-426 and Gln-575) are shown with their carbon atoms in magenta. Residues composing the intramolecular isopeptide bonds (Lys-297 and Asp-595 in the middle domain, Lys-610 and Asn-715 in the bottom domain) are shown with their carbon atoms in yellow. Regions connecting the domains are shown in gray. The missing Spy0125-NTR (not expressed in the construct) would be positioned to the right of the middle domain. B, a structural model of an intact GAS pilus is shown. Spy0125 is colored as above except the thioester and isopeptide residues are shown in red and gray. Two molecules of Spy0128 are shown (representing the polymerized pilus, cyan and pink with intramolecular isopeptide bonds colored as above). Spy0130 is presented as a gray molecular surface. The location of the intermolecular isopeptide bonds are shown as solid lines. Yellow and blue ovals represent cell wall and cell wall precursors, respectively.
The relative orientation and intimate contacts between the middle and top domains are conserved in the molecules of the two Spy0125-CTR crystal forms presented here. An overlay of residues 295–600 (spanning the top/middle domains) of the two molecules in the B-form crystal reveals a shift in the position of the bottom domain comprising an ∼14 Å translation and a small rotation (the linker region between the two domains (residues 598–602) re-orients to enable this translation). It has not been possible to definitively position a model for the bottom domain in the A-form crystal. Residual electron density and available space in the lattice (supplemental text and Fig. 2A) suggests this domain is present in the crystal but does not adopt a fixed position. The structures in both crystal forms, therefore, reveal an inherent flexibility in the linker region between the bottom domain of Spy0125-CTR and the rest of the protein. Additional description of the structures is given in the supplemental text and Fig. 2.
The Middle and Bottom Domains of Spy0125-CTR Contain Intramolecular Isopeptide Bonds
Spy0128, SpaA and BcpA, the major pilus subunits of GAS, C. diphtheriae and Bacillus cereus, respectively, all contain intramolecular isopeptide bonds (12–14). Such bonds are also revealed in the structure of Spy0125-CTR; one in the bottom domain between residues Lys-610 and Asn-715 (Fig. 2A) and a second between residues Lys-297 and Asp-595 in the middle domain (Fig. 2B). Whereas intramolecular isopeptide bond formation between Lys and Asn residues results in the release of an NH3 unit, the equivalent bond between the side chains of Lys and Asp eliminates a water molecule. The presence of both these isopeptide bonds in Spy0125-CTR has been confirmed using mass spectrometry (Table 2, supplemental Tables 1 and 2 and Figs. 3A and 3B).
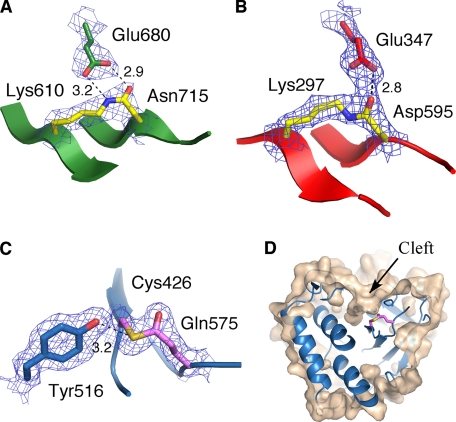
FIGURE 2.
Intramolecular isopeptide and thioester bonds in the structure of Spy0125-CTR. A, shown is the isopeptide bond in the bottom domain between Lys-610 and Asn-715; Glu-680 is also shown. B, shown is the isopeptide bond in the middle domain between Lys-297 and Asp-595; Glu-347 is also shown. C, shown is the internal thioester link between Cys-426 and Gln-575; also shown is residue Tyr-516, which forms a hydrogen bond to the sulfur atom. D, the thioester bond is buried in a cleft on the Spy0125-CTR surface. The cut-away molecular surface of the protein is shown in wheat (calculated with a 1.2 Å probe) with the top domain ribbon in blue. Panels A–C show the appropriate final σ-weighted 2 Fobs − Fcalc (calculated with model phases, ϕcalc) electron density map contoured at 1.2 σ and hydrogen bonding patterns (with distances in Å).
The Top Domain of Spy0125-CTR Contains an Internal Thioester Bond
In addition to the intramolecular isopeptide bonds, connectivity between the side chains of Cys-426 and Gln-575 was also observed in the electron density maps. This continuous electron density links the Sγ atom of the Cys to the Cδ atom of the Gln (Fig. 2C). The presence of this bond in Spy0125-CTR was confirmed using mass spectrometry and UV-visible spectroscopy. The experimentally determined intact mass of Spy0125-CTR was 49,288.5 Da, which compares to the theoretical mass of 49,342.0 Da calculated from the sequence. This 53.5 Da difference can be interpreted as a loss of three H2O and/or NH3 units, 2 of which are accounted for in formation of the intramolecular isopeptide bonds (1 H2O and 1 NH3 unit). The third is consistent with the loss of an NH3 unit from formation of a thioester between Cys-426 and Gln-575.
In contrast to the intramolecular isopeptide bonds, a peptide mass consistent with the presence of an intact thioester linkage between Cys-426 and Gln-575 is not present in the LC-MS spectrum of proteolytic digests of Spy0125-CTR (using trypsin, trypsin/AspN, AspN/GluC, or chymotrypsin). Perhaps this is not surprising given the different chemistries of these bonds and their relative susceptibility to nucleophilic attack. Inspection of tryptic digest LC-MS/MS data for the individual peptides that compose this link reveals a peptide containing position 575 is routinely observable, but significantly, a Glu side chain was recovered more frequently at this position than Gln (an increase of ∼1 Da in mass to give a peptide of 2283.15 Da compared with the expected 2282.17 Da for Gln-575, supplemental Table 3). On average (>four runs), the Glu-575 peptide is ∼7-fold enriched in the spectrum compared with the Gln-575 peptide (supplemental Fig. 4A, quantified using MAXQUANT (37)). This conversion is consistent with hydrolysis of the thioester during sample preparation but also with deamidation of the Gln-575 side chain (it should be noted that no other residue displayed any evidence of deamidation in the LC-MS/MS data). To confirm the presence of the thioester in Spy0125-CTR, the protein was incubated with the small molecule nucleophile methylamine, which would be predicted to attack the internal thioester (producing methylated Gln-575 and a free thiol group on Cys-426 (28)) and iodoacetamide (which would alkylate Cys-426 in its free thiol form). As expected, incubation with methylamine produced a previously unobserved tryptic peptide of mass 2296.18 Da, consistent with methylation of Gln-575 (supplemental Table 3); no other Gln or Asn residue displayed such a modification under the assay conditions used (and in a C426A variant, no modification was observed, confirming the reactivity of Spy0125-CTR toward methylamine is dependent on the thioester). Incubation with iodoacetamide alone produces a minimal amount of alkylated peptide containing Cys-426 (mass = 1700.78 Da); this presumably represents a small quantity of free Cys-426 thiol in the protein, which is also observed by UV-visible spectroscopy (see below). However, no differences in the wild-type peptide-containing position 575 were detected. To observe significant levels of the peptide-containing Cys-426 required Spy0125-CTR to be either reduced and alkylated (with DTT and iodoacetamide) during denaturation (before tryptic digest) or for the digested peptides to be reduced (the free thiol peptide is observed) or reduced and alkylated (the carbamidomethylated peptide is observed); see supplemental Table 3. Significantly, preincubation of Spy0125-CTR with methylamine and iodoacetamide resulted in an ∼30-fold increase in the alkylated Cys peptide in the mass spectrum (supplemental Table 3 and Fig. 4B, quantified with MAXQUANT). Searches of the raw LC-MS/MS data from non-reduced samples (except the methylamine and iodoacetamide incubation) revealed a mass consistent with a dimer of the Cys-426-containing peptide linked by a disulfide (mass = 3285.51 Da); this dimer must be an artifact of sample preparation.
In addition to the LC-MS/MS analysis, UV-visible spectroscopy shows that Cys-426 is only susceptible to modification by the thiol-specific reagent DTP in the presence of methylamine (Fig. 3A). Reaction of DTP with thiols liberates 2-thiopyridine, which has an intense absorption band at 343 nm. As Spy0125-CTR only has a single cysteine residue (that is involved in the thioester linkage), reaction of Spy0125-CTR with DTP should only be possible when the thioester is broken; for example, by reaction with methylamine. Furthermore, UV-visible data allows an estimation of the fraction of cysteine thiols in Spy0125-CTR that are addressable after the addition of methylamine. Changes in absorbance spectra were monitored over time from 270 to 390 nm (Fig. 3B). The addition of protein to buffer gave an initial A280 reading from which a protein concentration of 1.42 μm was calculated (extinction coefficient of 35535 m−1cm−1 at 280 nm (42)). The change in absorbance at 343 nm upon the addition of 10 mm methylamine (Fig. 3C) reveals 1.14 μm 2-thiopyridine is liberated in the reaction (using an extinction coefficient of 8080 m−1cm−1 at 343 nm for 2-thiopyridine), and there must, therefore, be an equal amount of newly exposed thiols. This reveals that ∼80% of thiols are only addressable by DTP after exposure to methylamine. The remaining ∼20% most likely represents protein in which the thioester has not formed properly; this is consistent with the small increase in absorbance at 343 nm upon the addition of protein to DTP solution in the absence of methylamine.
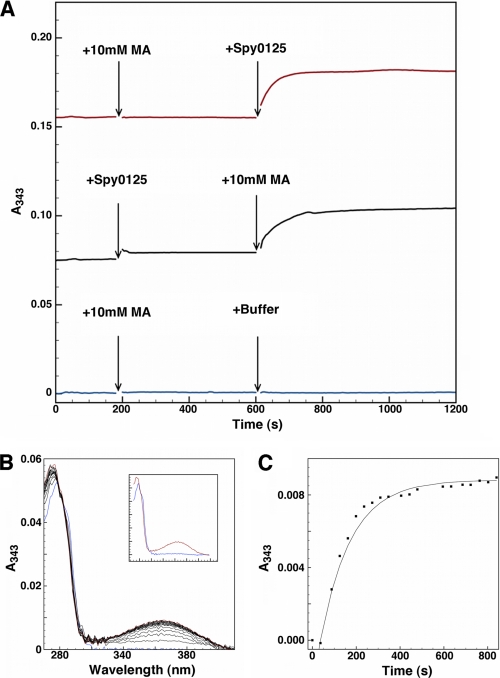
FIGURE 3.
UV-visible spectroscopy is shown. A, the addition of Spy0125-CTR after 10 mm methylamine (red line) or 10 mm methylamine (MA) after protein solution (black line) to 0.2 mm DTP in 50 mm HEPES, 150 mm NaCl, 6 m urea, pH 7.5, results in an increase in absorbance at 343 nm (control, addition of buffer, is shown as a blue line); spectra were taken every 5 s. B, absorbance change over time is shown after the addition of 10 mm methylamine to a solution containing 1.42 μm Spy0125-CTR and 0.2 mm DTP (zeroed against stable buffer + DTP base line). Blue, protein spectrum before methylamine addition; red, final spectrum taken after >800 s. Spectra were taken every 45 s. Inset, the same data show only the first and last spectrum. C, shown is the fit of the increase in absorbance at 343 nm over time (data from panel B) to a first-order exponential.
The Spy0125-CTR C426A Variant Remains Resistant to Chemical Denaturation
Using intrinsic protein fluorescence (Spy0125-CTR contains a single tryptophan and 23 tyrosine residues) as a probe for protein folding reveals that Spy0125-CTR is very resistant to chemical denaturation; a reversible folding/unfolding transition is only observed at a urea concentration of ∼8 m (supplemental Fig. 5). To observe whether the thioester significantly contributes to protein stability, this assay was repeated with the Spy0125-CTR C426A variant protein, which prevents the thioester forming. This mutated protein is also highly resistant to chemical denaturation (supplemental Fig. 5, unfolding transition was observed at a urea concentration ∼6 m). As the single tryptophan residue within the top domain, the fluorescence emission will be sensitive to the conformational state of this domain.
Removal of the Thioester Bond from GAS Pili Severely Compromises Adhesion to Host Cells
To test the importance of the thioester in pilus-mediated adhesion to host cells, a C426A mutation was introduced into GAS by allelic-replacement mutagenesis, replacing the native copy in the genome. Cell-wall fractions prepared from bacterial cultures confirmed that this variant protein is expressed, secreted, and incorporated into pili (Fig. 4A). Assays testing the ability of this variant GAS strain to adhere to HaCaT cells (a human keratinocyte cell line used as a model for GAS adhesion) revealed that bacteria presenting the C426A mutant protein display substantially reduced levels of binding to host cells (∼75% reduction) in comparison to wild type (Fig. 4B).
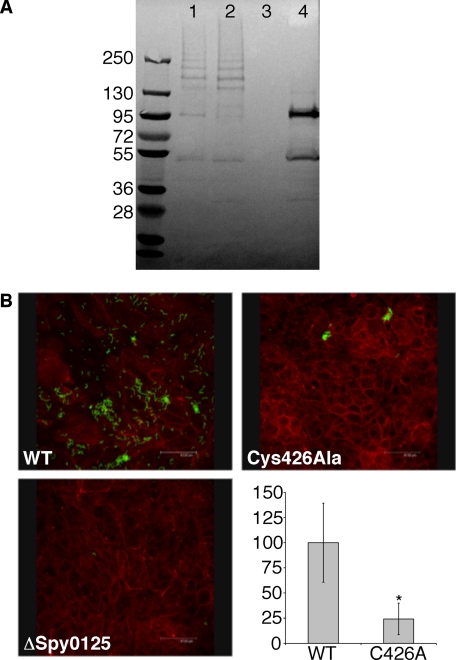
FIGURE 4.
The C426A variant of Spy0125 is compromised in its ability to adhere to host cells. A, an immunoblot shows the incorporation of variant Spy0125 into pili in the cell wall of GAS (using anti-Spy0125 sera). Lane 1, wild-type bacteria; lane 2, Spy0125 C426A variant; lane 3, Δspy0125; lane 4, Δspy0128. The equivalent laddering in the wild-type and mutant shows both strains incorporate Spy0125 into their pili. Lane 4 shows Spy0125 does not form a high molecular mass ladder in the absence of Spy0128; the band at ∼50 kDa likely represents mature Spy0125-CTR retained in the cell wall. B, GAS (green, visualized with a fluorescent anti-GAS antibody) binding to HaCaT cells (stained red with rhodamine-labeled phalloidin). Top left, wild-type bacteria; top right, Spy0125 C426A variant; bottom left, Δspy0125; bottom right, level of adhesion by the C426A mutant strain (C426A) compared with wild type. The asterisk indicates significant differences from the wild-type strain, with p values < 0.013.
DISCUSSION
The role of Gram-positive pili in cell adhesion and the mechanisms by which they are assembled has been the focus of intense research since their initial discovery (10). In M1 Group A Streptococcus strain SF370, the minor pilus subunit, Spy0125, is localized to the pilus tip where it acts as an adhesin to mediate host/pathogen interactions. Spy0125 is positioned away from the surface of the bacteria through a polymer of Spy0128 (the major pilus subunit), and this is presumed to promote receptor binding. The Spy0128 polymer is formed through intermolecular isopeptide bonds (catalyzed by a sortase) between the side chain of Lys-161 and the C terminus of the next Spy0128 molecule (13). This same Lys side chain forms the covalent link with the C terminus of Spy0125, defining the tip of the pilus (in this case only the C terminus of Spy0128 can react with a second Spy0128 molecule (51)). At the base of the pilus, the C terminus of Spy0128 is covalently linked to the side chain of Lys-120 in Spy0130 (23). The entire macromolecular assembly is then incorporated into the cell wall.
Recently, the role of the intramolecular isopeptide bonds in imparting resistance to mechanical stress in Spy0128 has been demonstrated (16). Two intramolecular isopeptide bonds are present in the Spy0125-CTR structure, which as in all proteins where they have been identified to date, are strategically positioned within the β-sandwich folds adjacent to either domain boundaries or the C-terminal sorting signal (Fig. 1). The Spy0125-CTR structure reveals an intramolecular isopeptide bond formed through the side chains of Lys-297 and Asp-595. To date, all well characterized intramolecular isopeptide bonds are formed thorough Lys and Asn side chains. A Lys/Asp intramolecular isopeptide bond has recently been reported in the structure of another S. pyogenes protein, fibronectin-binding protein (Fbab-B, PDB: 2X5P), as part of a structural proteomics effort (43).
The Lys-297/Asp-595 isopeptide bond (which adopts a trans configuration) occupies the same position in the middle domain of Spy0125 as the Lys/Asn isopeptide bond in the structurally homologous N2 domain of GBS52, suggesting Asn/Asp residues can be functionally redundant in forming such bonds. The Lys-610/Asn-715 isopeptide bond in Spy0125-CTR (which adopts a cis configuration) occupies the same space as that found in the structurally similar domain of Spy0128. As in other proteins, a conserved Glu residue and cluster of hydrophobic residues associate with each isopeptide bond and are presumed to be important for their formation. In Spy0125, Glu-347 forms a hydrogen bond to the O moiety of the Lys-297/Asp-595 isopeptide bond, and residues Leu-307, Ala-310, Leu-312, Pro-351, Tyr-354, Ala-357, and Met-593 form the hydrophobic cluster. Glu-680 forms a bi-dentate interaction with the O and NH moieties of the Lys-610/Asn-715 isopeptide bond; residues Phe-623, Phe-625, Tyr-686, Val-688, and Phe-713 form the hydrophobic cluster. Presumably these isopeptide bonds also impart resistance to mechanical stress in Spy0125, although this has yet to be tested experimentally.
The most striking feature of the Spy0125-CTR structure is the presence of an internal thioester bond. To date, internal thioesters have only been identified in complement components C3, C4 (28), α2-macroglobulin (44), and complement-like proteins from the innate immune system of insects (45). These bonds appear to form spontaneously, with protein folding providing the structural environment necessary for thioester formation (46). The activity of these proteins is tightly regulated, but after activation, which results in a conformational change (47, 48), the thioester becomes exposed and is available to react with nucleophilic groups via the acyl moiety (for review, see Ref. 28). This results in covalent attachment to adjacent target surfaces. For complement proteins, covalent binding to activating surfaces (including the surface of pathogens) via this route is critical for their activity in the innate immune response. The identity of the residue at position 1106 (human C4 numbering) influences the mechanism of acylation of the glutamyl moiety, dictating a preference for amino- or hydroxyl-based nucleophiles (49). Also, formation of the internal thioester in these proteins creates a thiolactone ring comprised of four residues in the sequence Cys-Gly-Glu-Gln (for review, see Ref. 28); this ring structure supports autolysis of the protein between the Glu and the Gln under denaturing conditions (28, 50). Cys-426 and Gln-575, which form the thioester bond in Spy0125-CTR, are positioned on adjacent β-strands in the structure and are separated in sequence (149 residues); therefore, a thiolactone ring is not formed, and autolysis has not been observed. The thioester bond is located within a shallow cleft on the surface of the top domain and is partially solvent-exposed (Fig. 3D). The most prominent features of the cleft walls are contributions from main-chain carbonyl groups of residues Asn-428, Ala-429, and Leu-431 and the side chains of Ser-433, Tyr-412, and Tyr-516. The OH group of Tyr-516 forms a hydrogen bond to the Sγ atom of the thioester (Fig. 2C).
Although at this stage it cannot be completely ruled out, a role for the highly unusual internal thioester bond in maintaining protein integrity/stability in Spy0125, akin to the intramolecular isopeptide bonds, seems unlikely as it is susceptible to nucleophilic attack. Also, the thioester is not strategically positioned close to a domain boundary, which all stabilizing intramolecular isopeptide bonds identified to date have been. Furthermore, both wild-type Spy0125-CTR and the C426A variant are resistant to chemical denaturation, suggesting that the thioester is not important for the overall stability of the protein. If the primary role of this bond was structural stabilization, then its removal would be expected to have a more dramatic effect on protein folding. A more attractive hypothesis which has direct parallels with thioester-containing complement proteins, is that upon host binding the thioester reacts with a cell surface ligand, covalently linking the pathogen to the host. Removal of the thioester from Spy0125 in intact pili displayed on the surface of GAS (using site-directed mutagenesis to create a C426A variant in the bacterial genome) has revealed the importance of this bond; the resulting bacteria are severely compromised in their ability to adhere to host cells (∼75% reduction). The residual binding observed may derive from the same interface used by the wild-type protein to adhere to the host cell surface, but the absence of the thioester significantly limits the ability of the bacteria to be retained at this site. This is important as it suggests Spy0125 uses molecular recognition to present the thioester to a suitable target, and this may have a regulatory role (akin to conformational changes in complement proteins after activation). Although current paradigms of bacterial adhesin/host cell receptor binding envisage strong non-covalent interactions, these results suggest a mechanism by which a pathogen could use a thioester bond in an adhesin molecule to covalently attach to a host cell surface. The divergent environment of the thioester site in Spy0125, when compared with complement proteins, suggests this activity has evolved independently. Spy0125-mediated adhesion may be one of the first steps in the host/pathogen interaction of GAS and would localize other virulence factors to the host cell surface, promoting infection.
In this model of host cell interaction, the pathogen is linked to its target by a covalent polymer comprising a thioester-mediated bond (could be either an amide or ester bond; see below) linking the pilus tip to a host cell receptor and intermolecular isopeptide (amide) bonds linking (a) Spy0125 to a major pilus subunit, (b) the major subunit to itself, and (c) the major subunit to Spy0130. Finally, the basal subunit anchors the pilus to the peptidylglycan layer via another amide bond. This entire structure is also stabilized by intramolecular isopeptide bonds positioned at domain/molecule boundaries.
A significant challenge now is to identify the receptor(s) on the host cell to which the Spy0125 adhesin binds and conclusively demonstrate whether a thioester-mediated covalent bond is formed at this site. From the Spy0125-CTR structure alone it is not possible to predict the mechanism by which the thioester bond could interact with host cells; whether it is directly attacked by an amino-based nucleophile (as occurs in C4A) or whether an equivalent to His-1106 (as found in C3 and C4B) exists to promote reactivity toward hydroxyl groups. Spy0125 has close homologues in a number of S. pyogenes serotypes (see supplemental Fig. 1), but other serotypes encode proteins that share sequence similarity with the rlr pilus adhesin (RrgA in S. pneumoniae TIGR4). RrgA contains an integrin-1-like domain with a MIDAS (metal ion-dependent adhesion site) motif (27), which is known to mediate protein-protein interactions (21). It is interesting that different Gram-positive pathogens and indeed different serotypes within a species have evolved very distinct pilus-presented adhesins that may be important for defining host tissue/cell range and therefore impact upon disease.
Supplementary Material
Acknowledgment
We thank the staff of the European Synchrotron Radiation Facility and Diamond Light Source for access to data collection facilities.
This work has been funded by the Royal Society (to M. J. B.) and Medical Research Council (United Kingdom) Grant G0400849 (to M. A. K.) and Studentship G0600309 (to J. A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. 1–5, and Tables 1–4.
The atomic coordinates and structure factors (codes 2xi9, 2xic, and 2xid) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GAS
- serotype M1 Group A Streptococcus strain SF370
- CTR
- C-terminal region
- DTP
- 2,2′-dithiopyridine.
REFERENCES
- 1.Mandlik A., Swierczynski A., Das A., Ton-That H. (2008) Trends Microbiol. 16, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proft T., Baker E. N. (2009) Cell Mol. Life Sci. 66, 613–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ton-That H., Schneewind O. (2004) Trends Microbiol. 12, 228–234 [DOI] [PubMed] [Google Scholar]
- 4.Scott J. R., Zähner D. (2006) Mol. Microbiol. 62, 320–330 [DOI] [PubMed] [Google Scholar]
- 5.Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006) Nat. Rev. Microbiol. 4, 509–519 [DOI] [PubMed] [Google Scholar]
- 6.Waksman G., Hultgren S. J. (2009) Nat. Rev. Microbiol. 7, 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzik J. M., Oh S. Y., Schneewind O. (2009) J. Biol. Chem. 284, 12989–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaminathan A., Mandlik A., Swierczynski A., Gaspar A., Das A., Ton-That H. (2007) Mol. Microbiol. 66, 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ton-That H., Marraffini L. A., Schneewind O. (2004) Mol. Microbiol. 53, 251–261 [DOI] [PubMed] [Google Scholar]
- 10.Ton-That H., Schneewind O. (2003) Mol. Microbiol. 50, 1429–1438 [DOI] [PubMed] [Google Scholar]
- 11.Budzik J. M., Marraffini L. A., Souda P., Whitelegge J. P., Faull K. F., Schneewind O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budzik J. M., Poor C. B., Faull K. F., Whitelegge J. P., He C., Schneewind O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19992–19997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. (2007) Science 318, 1625–1628 [DOI] [PubMed] [Google Scholar]
- 14.Kang H. J., Paterson N. G., Gaspar A. H., Ton-That H., Baker E. N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16967–16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H. J., Baker E. N. (2009) J. Biol. Chem. 284, 20729–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alegre-Cebollada J., Badilla C. L., Fernández J. M. (2010) J. Biol. Chem. 285, 11235–11242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham M. W. (2000) Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbot E. L., Smith W. D., Siou G. P., Chiriboga C., Smith R. J., Wilson J. A., Hirst B. H., Kehoe M. A. (2007) Cell Microbiol. 9, 1822–1833 [DOI] [PubMed] [Google Scholar]
- 19.Manetti A. G., Zingaretti C., Falugi F., Capo S., Bombaci M., Bagnoli F., Gambellini G., Bensi G., Mora M., Edwards A. M., Musser J. M., Graviss E. A., Telford J. L., Grandi G., Margarit I. (2007) Mol. Microbiol. 64, 968–983 [DOI] [PubMed] [Google Scholar]
- 20.Mora M., Bensi G., Capo S., Falugi F., Zingaretti C., Manetti A. G., Maggi T., Taddei A. R., Grandi G., Telford J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 22.Quigley B. R., Zähner D., Hatkoff M., Thanassi D. G., Scott J. R. (2009) Mol. Microbiol. 72, 1379–1394 [DOI] [PubMed] [Google Scholar]
- 23.Linke C., Young P. G., Kang H. J., Bunker R. D., Middleditch M. J., Caradoc-Davies T. T., Proft T., Baker E. N. (2010) J. Biol. Chem. 285, 20381–20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Race P. R., Bentley M. L., Melvin J. A., Crow A., Hughes R. K., Smith W. D., Sessions R. B., Kehoe M. A., McCafferty D. G., Banfield M. J. (2009) J. Biol. Chem. 284, 6924–6933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandlik A., Das A., Ton-That H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilleringmann M., Ringler P., Müller S. A., De Angelis G., Rappuoli R., Ferlenghi I., Engel A. (2009) EMBO J. 28, 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izoré T., Contreras-Martel C., El Mortaji L., Manzano C., Terrasse R., Vernet T., Di Guilmi A. M., Dessen A. (2010) Structure 18, 106–115 [DOI] [PubMed] [Google Scholar]
- 28.Law S. K., Dodds A. W. (1997) Protein Sci. 6, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie A. G. (2006) Acta Crystallogr. D. Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 30.Evans P. (2006) Acta Crystallogr. D. Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, n. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 32.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleywegt G. J., Zou J. Y., Kjeldgaard M., Jones T. A. (2001) International Tables for Crystallography, Vol. F. Crystallography of Biological Macromolecules, Kluwer Academic Publishers, Dordrecht [Google Scholar]
- 37.Cox J., Mann M. (2008) Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 38.Fontaine M. C., Lee J. J., Kehoe M. A. (2003) Infect. Immun. 71, 3857–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berrow N. S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D. I., Owens R. J. (2007) Nucleic Acids Res. 35, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnan V., Gaspar A. H., Ye N., Mandlik A., Ton-That H., Narayana S. V. (2007) Structure 15, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oke M., Carter L. G., Johnson K. A., Liu H., McMahon S. A., Yan X., Kerou M., Weikart N. D., Kadi N., Sheikh M. A., Schmelz S., Dorward M., Zawadzki M., Cozens C., Falconer H., Powers H., Overton I. M., van Niekerk C. A., Peng X., Patel P., Garrett R. A., Prangishvili D., Botting C. H., Coote P. J., Dryden D. T., Barton G. J., Schwarz-Linek U., Challis G. L., Taylor G. L., White M. F., Naismith J. H. (2010) J. Struct. Funct. Genomics 11, 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu C. T., Pizzo S. V. (1994) Lab. Invest. 71, 792–812 [PubMed] [Google Scholar]
- 45.Cherry S., Silverman N. (2006) Nat. Immunol. 7, 911–917 [DOI] [PubMed] [Google Scholar]
- 46.Pangburn M. K. (1992) FEBS Lett. 308, 280–282 [DOI] [PubMed] [Google Scholar]
- 47.Janssen B. J., Christodoulidou A., McCarthy A., Lambris J. D., Gros P. (2006) Nature 444, 213–216 [DOI] [PubMed] [Google Scholar]
- 48.Janssen B. J., Huizinga E. G., Raaijmakers H. C., Roos A., Daha M. R., Nilsson-Ekdahl K., Nilsson B., Gros P. (2005) Nature 437, 505–511 [DOI] [PubMed] [Google Scholar]
- 49.Dodds A. W., Ren X. D., Willis A. C., Law S. K. (1996) Nature 379, 177–179 [DOI] [PubMed] [Google Scholar]
- 50.Sim R. B., Sim E. (1981) Biochem. J. 193, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith W. D., Pointon J. A., Abbot E., Kang H. J., Baker E. N., Hirst B. H., Wilson J. A., Banfield M. J., Kehoe M. A. (2010) J. Bacteriol. 192, 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.