Abstract
Kv2.1 channels, which are expressed in brain, heart, pancreas, and other organs and tissues, are important targets for drug design. Flecainide and propafenone are known to block Kv2.1 channels more potently than other Kv channels. Here, we sought to explore structural determinants of this selectivity. We demonstrated that flecainide reduced the K+ currents through Kv2.1 channels expressed in Xenopus laevis oocytes in a voltage- and time-dependent manner. By systematically exchanging various segments of Kv2.1 with those from Kv1.2, we determined flecainide-sensing residues in the P-helix and inner helix S6. These residues are not exposed to the inner pore, a conventional binding region of open channel blockers. The flecainide-sensing residues also contribute to propafenone binding, suggesting overlapping receptors for the drugs. Indeed, propafenone and flecainide compete for binding in Kv2.1. We further used Monte Carlo-energy minimizations to map the receptors of the drugs. Flecainide docking in the Kv1.2-based homology model of Kv2.1 predicts the ligand ammonium group in the central cavity and the benzamide moiety in a niche between S6 and the P-helix. Propafenone also binds in the niche. Its carbonyl group accepts an H-bond from the P-helix, the amino group donates an H-bond to the P-loop turn, whereas the propyl group protrudes in the pore and blocks the access to the selectivity filter. Thus, besides the binding region in the central cavity, certain K+ channel ligands can expand in the subunit interface whose residues are less conserved between K+ channels and hence may be targets for design of highly desirable subtype-specific K+ channel drugs.
Keywords: Biophysics, Computer Modeling, Drug Action, Potassium Channels, Receptor Structure-Function, Site-directed Mutagenesis
Introduction
Potassium channels are crucial in physiology. Medically important drugs block the open pore, which is formed by four subunits. Each subunit consists of transmembrane α-helical segments S1–S6 and the membrane re-entering extracellular P-loop. The pore-forming domain is composed of four S5-P-S6 sequences. Each P-loop contains an extracellular S5-P linker, the pore helix (P-helix), the ascending limb with the signature sequence TVGYG, and the extracellular linker P-S6 (1).
Potassium channels are blocked by drugs of dramatically different chemical structures (2, 3). Pore-lining residues in S6 segments and P-loop turns affect binding of open channel blockers (4). In the experimentally determined (5–7) and predicted (8–11) structures of K+ channels with blockers, the latter occupy the central cavity.
In x-ray structures of K+ channels, four radial niches are seen between neighboring S6 segments and P-helices. Mutations in the niches affect binding of some K+ channel blockers (9, 12). The niche-lining residues are less conserved than the pore-lining residues and therefore may be targets for design of subtype-specific ligands of K+ channels (2).
Delayed rectifier Kv2.1 channels play important physiological roles in different organs and tissues including in the heart (13, 14), brain (15–17), pancreas (18–20), lung (21, 22), and placenta (23). Expression of Kv2.1 channels is altered under pathologic conditions like hyperthyroidism (24), infarction (25), and diabetes (26). This stimulates attempts to design Kv2.1-selective drugs.
An important step toward this goal is to understand how currently available Kv2.1-selective ligands block the channel. Structurally different antiarrhythmics propafenone (PROP)4 and flecainide (FLEC) block Kv2.1 more potently than other Kv channels (12, 27). Earlier systematic substitution of segments in PROP-sensitive channel Kv2.1 with those from PROP low-sensitive channel Kv1.2 allowed to identify five loci in the P-helix and S6, which are responsible for Kv2.1 selectivity to PROP (12). In the present study, we found that substitution of some of the PROP-sensing loci in Kv2.1 also decreased FLEC potency. To rationalize this observation, we created a Kv1.2-based homology model of Kv2.1 and used multiple Monte Carlo-energy minimizations (28) to search for energetically preferable binding modes. Computations predict that a large part of the FLEC molecule fits the niche between P-helix and S6, where trifluoromethyl groups bind to loci TITp46 (see Table 1 for residue labels) and Vi17, whereas the ammonium group exposes to the central cavity. A large part of the PROP molecule also binds in the niche, where it interacts with the TITp46 locus and approaches Vi17, whereas the aminopropyl group protrudes in the central cavity.
TABLE 1.
Our study, for the first time, identifies FLEC-sensing residues in Kv2.1, provides structural models of PROP and FLEC in Kv2.1, defines loci with direct and indirect effects on FLEC and PROP potencies, and highlights importance of the niches between P-helices and inner helices as targets for subtype-specific K+ channel blockers.
MATERIALS AND METHODS
In Vitro Mutagenesis and RNA Synthesis
The cDNAs for chimeras of rat Kv1.2 (GenBankTM accession no. X16003) and human Kv2.1 (GenBankTM accession no. X68302) were obtained using an overlap PCR and were cloned into the pGEM vector (29). The chimera details are given in the supplemental information. PCR-amplified DNA sequences were verified by using the BigDye terminator cycle sequencing kit (PerkinElmer Life Sciences). The sequence reactions were analyzed on an ABI 377 or Prism 310 automated sequencer (PerkinElmer Life Sciences). The cDNA encoding rat Kv1.2 and human Kv2.1 and their chimeras were transcribed to cRNA using a commercial kit (mMESSAGE mMACHINE, Ambion, Austin, TX) and T7 RNA polymerase. Denaturing agarose gel electrophoresis was used to check the quality of cRNA product of each reaction and to quantify the yield.
Preparation of Oocytes
South African clawed frogs (Xenopus laevis) were anesthetized in ethyl m-aminobenzoate (Sandoz, Basel, Switzerland), and small sections of the ovary were removed surgically. Oocytes were injected with 0.1 or 1.0 ng of cRNA in 50 nl of distilled water and were maintained under tissue culture conditions at 20 °C until used for experiments. The tissue culture solution was a modified Barth medium: 88 mmol/liter NaCl, 1 mmol/liter KCl, 1.5 mmol/liter CaCl2, 2.4 mmol/liter NaHCO3, 0.8 mmol/liter MgSO4, 5 mmol/liter HEPES, pH 7.4, which was supplemented with penicillin (100 international units/ml) and streptomycin (100 μg/ml).
Electrophysiological Techniques
Oocytes with follicular tissues were investigated with the two-electrode voltage clamp technique. Microelectrodes were made from borosilicate glass and had resistances of 0.5–1 megohm for the current electrodes and 1–2 megohm for the potential electrodes when filled with 3 mol/liter KCl. The holding potential was −80 mV, and command potentials were applied up to a potential of +60 mV. The control bath fluid was a Ringer solution consisting of the following: 115 mmol/liter NaCl, 2 mmol/liter KCl, 1.8 mmol/liter CaCl2, 10 mmol/liter HEPES, pH 7.2. FLEC (acetate salt; up to 1000 μmol/liter, Meda Pharma), PROP (chloride salt; Sigma), and the R- and S-enantiomers of flecainide hydrochloride and propafenone (Dr. Margarete Fischer-Bosch, Institute for Clinical Pharmacology) was added to the bath solution and applied at least 30 s before eliciting currents. Solutions were applied with a concentration clamp technique (30), allowing an exchange of >90% of the extracellular solution within <10 ms. All experiments were performed at days 3 and 4 after injection of cRNA and were carried out at room temperature (22 ± 1 °C).
Data Acquisition and Analysis
The K+ currents obtained in two-electrode recordings were low pass-filtered at 1 kHz and transferred to a computer (pClamp program, Axon Instruments). The amplitudes of the total outward currents were corrected for leakage. Leakage currents and capacitive transients were subtracted online using a p/−4 pulse protocol.
The K+ current amplitudes were measured at the end of 500-ms depolarizing voltage steps. Conductance-voltage relations were obtained by normalizing the conductance data to the maximal value under control conditions and by fitting the data to the Boltzmann equation y = Gmax(1 + exp((V½ − V)/b)), where y is the normalized conductance, Gmax is the normalized maximal conductance, V½ is the potential of the half-maximal conductance, V is the voltage, and b is the slope factor. Concentration-response curves were determined by fitting the mean current values at different FLEC concentrations to the Langmuir equation y = (KD/c)n/(1 + (KD/c)n)), where y is the fraction of control current, KD is the half-blocking concentration, c is the FLEC concentration, and n is the Hill coefficient.
The voltage dependence of block was determined using the KD values, which were obtained from the fractional current f = IFLEC/ICTRL measured in the presence of FLEC (IFLEC) at a concentration [D] of 100 μmol/liter and under control conditions (ICTRL) at the end of the voltage step and KD = [D] × f/(1 − f). The fractional electrical distance (δ), i.e. the fraction of the transmembrane electrical field sensed by a single positive charge at the binding site, was determined by fitting the KD values with the equation KD = KD0 × exp(−zδFV/RT). KD0 represents the half-blocking concentration at the reference potential of 0 mV, V is the membrane potential, and z, F, R, and T have their usual meanings.
The measured values are given as mean ± S.E. Statistical significance was tested using a t test or a Mann-Whitney rank sum test. Values of p ≤ 0.01 were taken as statistically significant. Curve fitting and all statistical procedures were performed using the program SigmaPlot (Jandel Scientific, Erkrath, Germany).
Molecular Modeling
The homology model of Kv2.1 was based on a Kv1.2 x-ray structure, Protein Data Bank code 2A79 (31). Energetically optimal drug-channel complexes were searched by the Monte Carlo-energy minimization method (32) from multiple starting points using the ZMM program as described elsewhere (28). The starting positions and orientations of the drugs were seeded in the area covering all drug-sensing loci, except that in the P-S6 linker, which is not reachable from the inner pore. An AMBER force field (33) with a cut-off distance of 8 Å was used. The hydration energy was computed using the implicit solvent method (34). All ionizable residues were kept in a neutral form (35). Atomic charges of FLEC and PROP were calculated by AM1 method of MOPAC (36). Four potassium binding sites in the outer pore, T1–T4, were loaded as follows: sites T1 and T3 with K+ ions and sites T2 and T4 with explicit water molecules. Residues are designated (see Table 1) using a scheme universal for P-loop channels (37).
RESULTS
Block of Kv2.1 Currents by FLEC
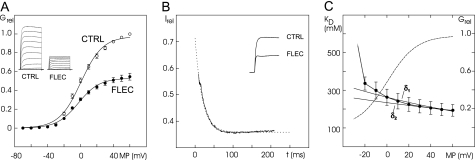
Kv2.1 channels expressed in Xenopus oocytes mediated outward currents at potentials positive to −30 mV as described (e.g. Ref. 12). FLEC (200 μmol/liter) decreased the outward currents over the entire potential range up to the most positive tested potential of +60 mV (Fig. 1A). At the most positive test pulses (+30 to +60 mV), a change in the activation kinetics became apparent with FLEC (Fig. 1A, FLEC; see small humps at the beginning of the current traces). Fitting the conductance values revealed a decrease of the maximal conductance (Gmax) by 46 ± 3% (Fig. 1A; n = 8), whereas conductance-voltage relations were not affected significantly (shift of potential of half-maximal conductance: −3.7 ± 0.6 mV; decrease of slope factor: −0.7 ± 0.2 mV). Thus, the effects of FLEC on Kv2.1 conductance are similar to those of PROP (12).
FIGURE 1.
Effect of 200 μmol/liter FLEC on Kv2.1 K+ currents and conductance. A, open probability curves of mean conductance values from eight experiments. Inset, original recording of one experiment (maximal amplitude, 21.7 μA; voltage step duration, 500 ms). Shown are potential steps from −80 to +60 mV under control conditions (CTRL, open circles) and with FLEC (filled circles). The conductance values (Grel) were normalized to the respective maximal value under control conditions. B, change of fractional current (Irel), i.e. fraction of current with FLEC relative to current under control conditions (CTRL), with the duration (t) of voltage steps to +60 mV (except for the first 8 ms). The trace is the mean of seven experiments. Hatched line, fit of Irel with a monoexponential function. The fit is shown from 0 to 250 ms of the voltage step. Inset, original recording of currents at +60 mV (maximal amplitude, 25.7 μA; voltage step duration, 200 ms). C, voltage dependence of a fraction of block. The half-blocking concentrations (KD) at different potentials are given as mean ± S.E. of six experiments (filled circles, dotted line). The hatched line gives the open probability curve obtained by fitting the relative conductance values (Grel) current amplitude with a Boltzmann equation. The fit was performed with the measured KD0 of 263 μmol/l (δ1 = 0.17 ± 0.01) and with an unknown KD0 (δ2 = 0.10 ± 0.03, KD0 = 200 μmol/liter; n = 6, solid lines). The error bars indicate S.E. MP, membrane potential.
Time and Voltage Dependence of Block
Analyzing the time course of Kv2.1 block in the presence of 200 μmol/liter FLEC versus control currents showed block development within the first milliseconds of the test pulse (Fig. 1B). The decay of current could be fitted well with a monoexponential function with a time constant of 19 ± 2 ms at +60 mV (Fig. 1B). Extrapolating this fit of the block development to zero time at the test pulse beginning indicated an initial block of <28% (hatched line in Fig. 1B). This observation supports the view that FLEC mainly blocks the open Kv2.1 channel and that block efficacy increases with open probability.
The voltage dependence of block was calculated by the fraction of control current with FLEC in the potential range from −30 to +60 mV (Fig. 1C). The FLEC-induced block increased with positive going potentials as indicated by a steady decrease of the half-blocking concentrations (KD). The fractional electrical distance (δ), i.e. the fraction of the transmembrane electrical field sensed by the drug at the binding site referenced to the intracellular side of the channel, was calculated at potentials from +40 to +60 mV. In this potential range, the open probability was roughly maximal (change of maximal conductance < 9%; n = 8; Fig. 1C, hatched line), and the endogenous currents of the oocytes were small (<0.8 μA up to +80 mV in water-injected oocytes; n = 3; data not shown). With a pKa value of 9.3 for FLEC acetate and an intracellular pH value of 8.2 in oocytes (38), >90% of the intracellular FLEC should be positively charged. Calculating δ with the measured KD at the reference potential of 0 mV (KD0 = 263 μmol/liter) and with the estimated charge of FLEC (z = 0.9) yielded δ1 = 0.17 ± 0.01 (Fig. 1C; n = 6). At the potential of 0 mV, however, the open probability was ∼50% of the maximal value, suggesting that the theoretical KD for maximal open probability could be smaller than the computed one. The values were therefore refitted with the assumption of an unknown KD0. The fit revealed an electrical distance δ2 = 0.10 ± 0.03 and a KD0 of 200 ± 29 μmol/liter (Fig. 1C). Thus, the values of electrical distance are similar to those of PROP (δ1 = 0.16 and δ2 = 0.14, (12)) and of other substances found to block ion channels at intracellular parts of the channel molecule (δ = 0.16 for tetraethylammonium (39) and bupivacaine (40)).
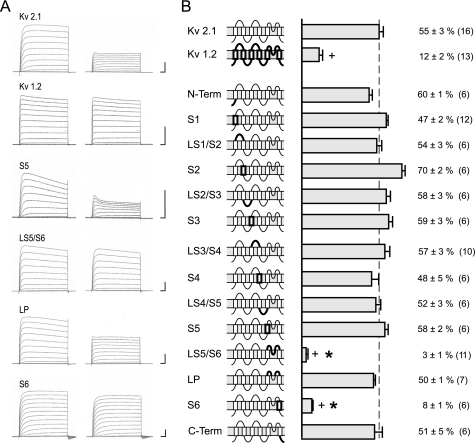
Effects of FLEC on Kv2.1/1.2 Chimeras
Kv1.2 channels were blocked less by FLEC than Kv2.1 channels. Thus, Kv1.2 currents showed a reduction of Gmax in the presence of 200 μmol/liter FLEC by 12 ± 2% in comparison to 55 ± 3% for Kv2.1 currents (Fig. 2). We took advantage of this different sensitivity and used a systematic set of Kv2.1/Kv1.2 chimeras to screen the channel domains for FLEC binding sites. We tested for insensitivity of the Kv2.1 channel to also discover a possible composite site of action.
FIGURE 2.
Effect of 200 μmol/l FLEC on K+ currents and maximal conductance of Kv1.2, Kv2.1, and Kv2.1 chimera. A, original current recordings of wild-type channels and chimera under control conditions (left) and with FLEC (right). Potentials steps from −80 up to +60 mV. Calibrations: 50 ms and 5 μA each. B, the schematic structures of the wild-type and mutant channels are shown on the left. The bars and numbers show the reduction (%) of maximal conductance as mean ± S.E. The numbers in brackets indicate the number of experiments. *, no statistically significant difference versus the Kv1.2 channel. +, statistically significant decrease versus the Kv2.1 channel. C-Term, C-terminal.
In the first step, we replaced the Kv2.1 N and C termini, every segment (S1, S2, S3, S4, S5 and S6) and every linker (LS1/S2, LS2/S3, LS3/S4, LS4/S5, and LS5/S6 with and without the P-loop) by the respective parts of the Kv1.2 channel (Fig. 2). Original current recordings of the wild-type channels and the chimeras of the pore region under control conditions and with 200 μmol/liter FLEC (Fig. 2A) show that the current reduction by FLEC remains similar to Kv2.1 wild-type for the Kv2.1 chimera S5 and LP, whereas the current reduction by FLEC was strongly reduced in the chimera LS5/S6 and S6. The reduction of Gmax (Fig. 2B) in the chimeras substantially decreased reaching the Kv1.2 wild-type level only for the chimeras with the exchanged S5-S6 linker and the exchanged S6, whereas the exchange of the other extracellular linkers and segments did not cause a significant reduction of the block (Fig. 2B, asterisks indicate no statistical significant difference to Kv1.2 wild-type, n = 5–12). These results suggest a high affinity binding site for FLEC in the pore region. This is completely in line with the findings for PROP (12), but in contrast to PROP exchanges of the N and C termini and the intracellular linker LS4/S5 did not cause statistically significant current reductions by FLEC.
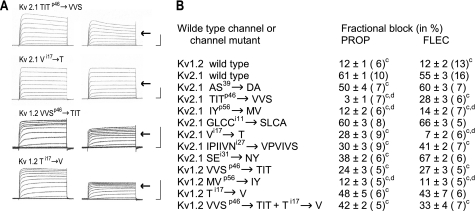
In the second step, Kv2.1 mutants with exchanged residues in the P-loop and S6 segment (Table 1) were tested for sensitivity to FLEC (Fig. 3). Original recordings of some mutants are shown in Fig. 3A. Within the P-region of Kv2.1, exchange TITp46 → VVS and especially exchange IYp56 → MV strongly reduced the sensitivity of the channel to FLEC, whereas the exchange ASp39 → DA had no statistically significant effect (Fig. 3B, FLEC). Thus, the amino acid exchanges that decreased the PROP potency (12, Fig. 3B, PROP) also decrease the FLEC potency, despite the fact that quantitative effects of some exchanges are different. Exchanges TITp46 → VVS and IYp56 → MV had the largest impact on the potencies of PROP and FLEC, respectively (Fig. 3B).
FIGURE 3.
Effect of 200 μmol/liter FLEC on K+ currents and conductance of mutants of Kv2.1 and Kv1.2. A, original current recordings of mutants most relevant for drug binding under control conditions (left) and with FLEC (right). Potentials steps from −80 up to +60 mV. 50 ms and 5 μA were used for each calibration. The arrows indicate the block of current for Kv2.1 wild-type channel. B, fractional block of maximal conductance by FLEC and PROP in % as mean ± S.E. Data for PROP are from Ref. 14. Numbers in brackets indicate the number of experiments. c, statistically significant difference from Kv2.1 block. d, no statistically significant difference from Kv1.2 block.
In the last step of this mutational approach, we replaced Kv2.1 residues in S6 (see Table 1) by those of Kv1.2 (Fig. 3). The exchange Gi8 → S and Ci11 → A in the outer part of S6 and SEi31 → NY in the inner part of S6 did not reduce the sensitivity to FLEC. In contrast, the exchanges IPIIVNi27 → VPVIVS and especially Vi17 → T significantly reduced the FLEC potency. The mutation of Vi17 reduced FLEC potency to that in the wild-type Kv1.2 (Fig. 3B, FLEC). Thus, as for PROP, effects of mutations were found in S6, but in contrast to PROP (Fig. 3B, PROP), these were more focused on the middle of S6.
Comparison of PROP and FLEC Sites of Action
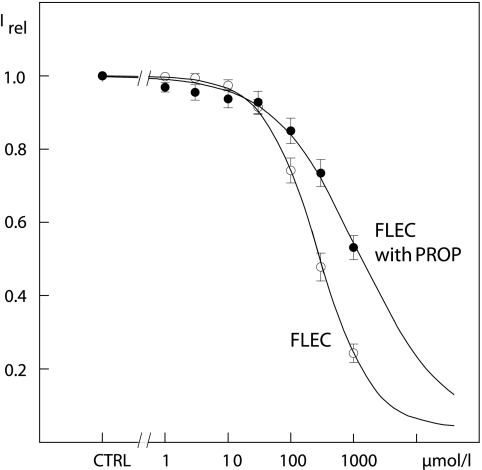
Because the effects of FLEC at Kv2.1 were similar to those described for PROP, we explored possible overlap of the receptors by measuring dose response relations of FLEC at +60 mV under control conditions and in the presence of 100 μmol/liter PROP shown to block around half of the maximum conductance (12). The block by PROP, however, also had a slow component with a time course >1 s (12). Therefore, even without application of FLEC, a small decrease of K+ currents was found with each voltage step in the presence of the PROP. To compensate for this effect, the PROP-induced reduction was estimated by fitting five voltage steps (preceding FLEC applications) to bi-exponential functions and by subtracting the extrapolated current decrease from the control current values. The experiments revealed a KD of 310 ± 50 μmol/liter FLEC for the reduction of Kv2.1 K+ currents at +60 mV (Fig. 4, n = 6). In the presence of PROP, KD increased to 1360 ± 230 μmol/liter FLEC (Fig. 4, n = 7). The differences were statistically significant with p = 0.002. Furthermore, the Hill coefficient, which indicates the number of FLEC molecules needed for block, decreased from 1.02 ± 0.05 under control conditions to 0.75 ± 0.13 in the presence of PROP. The increase of KD and the reduction of the Hill coefficient suggest that FLEC and PROP compete for the same or overlapping binding sites.
FIGURE 4.
Concentration-response curves for FLEC (open circles) and for FLEC in the background of 100 μmol/liter PROP (filled circles). The symbols represent currents relative to the maximal current under control conditions at +60 mV as mean ± S.E. of six and seven experiments. Irel, fraction of current with FLEC relative to current under control conditions (CTRL). The data were fitted to Langmuir equations.
We further tested FLEC enantiomers (200 μmol/liter each) and PROP enantiomers (100 μmol/liter each) and found that the differences of the channel-blocking potencies of enantiomers are statistically insignificant. In particular, Kv2.1 currents showed a reduction of Gmax by 39 ± 6% (n = 6) with (R)-FLEC and by 43 ± 6% (n = 6) with (S)-FLEC. The reduction of Gmax was 56 ± 4% (n = 7) with (R)-PROP and 63 ± 3% (n = 7) for (S)-PROP (data not shown).
Mapping the FLEC Receptor
The current study shows that FLEC is an open channel blocker that enters the open pore from the cytoplasm. Locus IYp56 at the P-S6 linker is unlikely to directly interact with FLEC. Indeed, mutations at the linker are known to affect slow inactivation kinetics, e.g. (9). Other loci would be reachable by FLEC from the inner pore (Fig. 5A). An unbiased multi-Monte-Carlo energy minimization search predicted low energy complexes with the FLEC ammonium group at the focus of P-helices (Fig. 5, A–C). In this respect, the complex resembles co-crystals of KcsA with tetrabutylammonium (5, 7). Bis-(trifluoroethoxy)-phenyl moiety filled a trifurcating niche between the P-helix and two S6 segments (Fig. 5B). Group m-CF3 occupied a cavity in the niche, which is lined by Vi17, Ii18, and two flanking leucine residues from the S5 motif LILFL. Another cavity was occupied by group o-CF3, which approached the methyl group of Tp46 (Fig. 5C). These interactions are consistent with the lipophilic properties of group CF3 (41). Residues Mp47, Tp48, Tp49, Vi15, and Ii18, which are common between Kv1.2 and Kv2.1, also stabilize FLEC binding. In particular, Tp49 donated an H-bond to the FLEC ether oxygen (Fig. 5C). Thus, multi-Monte-Carlo energy minimization docking predicted a complex, in which two of the four FLEC-sensing loci directly interact with the ligand and all functional groups of FLEC contribute to the ligand binding.
FIGURE 5.
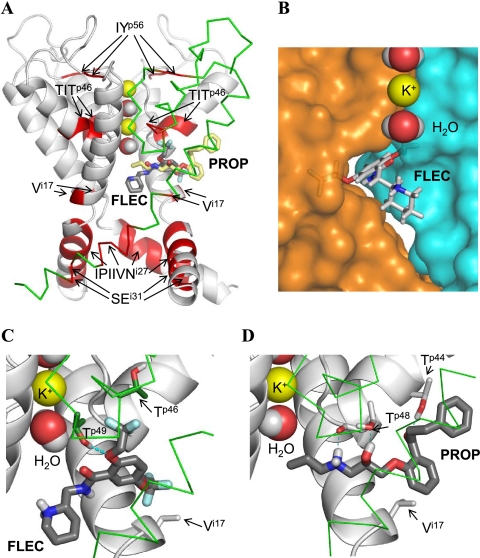
PROP and FLEC in Kv2.1. A, overall view of the pore domain. Three subunits of the channel are shown with gray ribbons (α-helices) and gray rods. For clarity, one subunit is shown by green Cα tracing and S5 helices are not shown. K+ ions and water molecules in the selectivity filter region are space-filled. Ligands (sticks with red oxygens, blue nitrogens, dark gray carbons in FLEC, and yellow carbons in PROP) are superimposed to indicate overlapping binding sites. Channel segments whose substitutions substantially affect action of FLEC and/or PROP (Table 1 and Fig. 3) are highlighted by red. Note that segment TITp46 and residue Vi17 are close to the ligands, whereas other segments are far from the ligands (see text for further explanations). B, close-up view at FLEC in Kv2.1. The piperidine ring is in the central cavity, and the aromatic ring is in the niche between the neighboring subunits, whose surfaces are colored blue and orange. A potassium ion and two water molecules in the selectivity filter are space-filled. C and D, close-up view at FLEC and PROP, respectively, in the channel. The ligands are shown by thick sticks with dark gray carbons, red oxygens, blue nitrogens, white polar hydrogens, and cyan fluorine atoms. Channel residues, whose direct interactions with the ligands are proposed in this study, are shown by thin sticks, and their carbons atoms are colored as respective backbones. Note that FLEC interacts with the proposed residues, which are located in the neighboring subunits (gray carbons of Vi17 versus green carbons of Tp46 and Tp49), whereas PROP interacts with Tp44, Tp48, and Vi17 in the same subunit (gray carbons).
Mapping the PROP Receptor
High flexibility of PROP, which contains 12 rotatable bonds, decreases chances to find the global minimum in an unbiased search. Exchange TITp46 → VVS largely decreased the PROP potency (Fig. 3B) suggesting that Tp44 likely forms an H-bond with PROP. Therefore, we used distance constrains to near different polar groups of PROP and Tp44 and Monte Carlo-minimized corresponding complexes with and then without the constraints. The search yielded a low energy complex in which the carbonyl group of PROP accepted an H-bond from Tp44 (Fig. 5D), and the hydroxy group donated an H-bond to the side chain of Tp48, whereas the ammonium group donated an H-bond to the backbone carbonyl of Tp48. The latter is at the P-loop turn and does not accept an H-bond from the P-helix backbone and hence is a particularly attractive site for the ligand H-bond donor. A disubstituted phenyl ring bound between Ii18 and Ii12 in neighboring subunits and approached Vi17 as close as 5 Å. Another phenyl ring of the ligand bound between methyl group of Tp48 and Gi8 in the neighboring subunit.
Models in Fig. 5 show Kv2.1 with (R)-PROP and (S)-FLEC. We also docked (S)-PROP and (R)-FLEC. For each pair of enantiomers, the difference of ligand-binding energies calculated using solvent exposure- and distance-dependent dielectric function (42) is less than 2 kcal/mol. Because our computational methodology does not take into consideration the librational entropy and some other contributions to the free energy, the predicted binding energies are not expected to be precise enough to correlate them with statistically insignificant differences in the potency of enantiomers. However, superpositions of the channel-bound enantiomers (supplemental Fig. S1) show that the same residues of the channel interact with the same moieties of enantiomers, whereas positions, orientations, and conformations of the enantiomers are similar. For example, the ammonium groups of the FLEC enantiomers are at the focus of P-helices, close to the inner pore cavity center, whereas methylene groups of the piperidine ring fill the cavity below its center (supplemental Fig. S1A). The ammonium groups of the PROP enantiomers donate H-bonds to the backbone carbonyl of Tp48, whereas hydroxy groups of Tp44 and Tp48 are engaged in H-bonding with the carbonyl and hydroxy groups, respectively (supplemental Fig. S1B).
Testing the Modeling Predictions
As a test for the proposed ligand-binding modes in Kv2.1, we have transferred into Kv1.2 the residues, which have been found in our model to be critical for the FLEC and PROP binding in Kv2.1 (Fig. 3). Original tracings of the Kv2.1 mutants and the corresponding Kv1.2 mutants under control conditions and with FLEC are shown in Fig. 3A. In complete agreement with the model, the current decrease by FLEC in Kv2.1 wild-type (arrows in Fig. 3A), which was strongly reduced by mutations TITp46 → VVS and especially Vi17 → T, was partly restores in Kv1.2 mutant VVSp46 → TIT and almost completely restored in the Ti17 → V mutant (Fig. 3A). As far as Gmax is concerned, replacement of the Kv1.2 motif VVSp46 with motif TIT increased ∼2-fold potency of both FLEC and PROP (Fig. 3B). A 4-fold increase of Kv1.2 sensitivity to both blockers was observed upon the point mutation Ti17 → V. Furthermore, the FLEC effect on the Kv1.2 Ti17 → V mutant showed no statistically significant difference to the FLEC effect in the Kv2.1 wildtype. We further transferred to Kv1.2 two components of the FLEC and PROP receptors in Kv2.1 in combination, TITp46 and Vi17 (Fig. 3B). The resulting chimera was ∼3-fold more sensitive to FLEC and PROP than the wildtype Kv1.2. Thus, for unknown reasons the combined substitutions were not more effective than the single ones.
Finally, we transferred to Kv1.2 residues IYp56, which do not interact with PROP and FLEC in our model, but had shown strong effects in the electrophysiological experiments. In line with our conclusions that FLEC and PROP do not bind at the extracellular mouth of the channel, the mutation did not change sensitivity of Kv1.2 to FLEC and PROP (Fig. 3B).
DISCUSSION
Voltage-gated K+ channels in general and Kv2.1 channels in particular are important drug targets. Although x-ray structures of several K+ channels in the open and closed states are available, structure-based design of subtype-specific K+ channel drugs remains a challenging problem. One of the reasons is incomplete knowledge of the drugs binding sites and binding modes. Here, we sought to explore structural determinants of selectivity for structurally different antiarrhythmics, PROP and FLEC, toward Kv2.1 versus other Kv channels. These drugs exhibit just a moderate Kv2.1-blocking potency, but this potency is higher than on other Kv channels (27).
Our electrophysiological data suggest that FLEC decreases the Kv2.1 currents by an open channel block from the cytoplasm. Thus, only a slight block of the current at the beginning of the voltage pulse was found, and the block increased with time. The current increase with FLEC was faster than under control conditions. Furthermore, the block increased with positive going potentials suggesting its correlation with the open probability. The fractional electrical distance (δ) of FLEC was in the range 0.10–0.17, indicating that FLEC acts at the intracellular side of the channel. Several substances known to block the inner pore of K+ channel have similar range of δ values. These are internally applied tetraethylammonium with δ = 0.16 (39), quinidine with δ = 0.19 (43), bupivacaine with δ = 0.16 (44), and PROP with δ = 0.14–0.16 (12). All these data support our conclusion that the mechanism of Kv2.1 block by FLEC is similar to that by PROP (12).
Numerous studies indicate that blockers of the open P-loop channels bind in the inner cavity to the pore-facing residues in positions i15, i18, and i22 (37). Residues Tp49 at the top of the cavity also contribute to binding of Kv blockers (45). Our present and previous (12) studies identified new loci in Kv2.1, whose exchange with corresponding residues in Kv1.2 diminished the blocking potency of FLEC and PROP. Three of the loci do not face the inner pore and two of them occur deep in the niches. Despite PROP and FLEC have dramatically different chemical structures, our results suggest that both drugs can bind in the niche and expose their hydrophobic ammonium groups to the inner pore to block the current (Fig. 5).
The ammonium group is considered traditionally as a fingerprint of hydrophobic cations like tetrabutylammonium. The ammonium nitrogen of FLEC occurs at the focus of P-helices (Fig. 5A). This position is slightly below the level where the nitrogen atom of tetrabutylammonium is seen in co-crystals of KcsA with tetrabutylammonium (5, 7). The NH2+ group of PROP, which forms an H-bond with Tp48, is ∼4 Å away from the pore axis. At this distance, it would still repel permeating K+ ions electrostatically. In addition, the hydrophobic methyl group of PROP riches the pore axis below the selectivity filter, approximately at the level T5, where a K+ ion binds in the central cavity of KcsA (Fig. 5, A and D). At this position, the methyl group would block the ion permeation by interfering with the hydration shell of a K+ ion approaching the selectivity filter.
Although FLEC and PROP share ligand-sensing loci, quantitative effects of the exchange of loci on drug potency are different. Thus, exchange TITp46 → VVS decreased the PROP potency 30-fold, but the FLEC potency only 2-fold (Fig. 3B). This is consistent with our models where PROP accepts a strong H-bond from Tp44 that is lost in the mutant, whereas FLEC accepts a weak H-bond from Tp46 that would retain in the mutant. Diminished hydrophobicity of Sp46 versus Tp46 can account for the lower potency of FLEC in Kv1.2. Another mutation, Vi17 → T, diminished potency of FLEC almost 8-fold, but potency of PROP only 2-fold. In view of our models, the hydrophobic Vi17 in Kv2.1 provides a favorable environment for the lipophilic CF3 group of FLEC, but this environment is lost in mutant Vi17 → T. PROP potency was less sensitive to the mutation because the disubstituted phenyl ring approached Vi17 but did not make direct contacts with it. Replacing locus IPIIVNi27 decreased FLEC potency by 13% but decreased PROP potency by 30% (Fig. 3B). Although Ii22 lines the access path for both drugs to the pore, in our models, it interacts with PROP but not FLEC.
Exchange IYp56 → MV in the extracellular side of P-loop had a similar effect on the potency of both drugs. This locus is far from the drugs in our models (Fig. 5A), implying that the mutation allosterically affects the potency of the drugs. Indeed, mutations in the P-S6 linker are known to affect kinetics of C-type inactivation in Kv channels (46). The similar effect of the IYp56 → MV exchange on the potency of both drugs (Fig. 3B) also support an allosteric mechanism behind the observed effect of this exchange on the drugs potency.
Thus, mutational, electrophysiological, and modeling data of our study indicate a common binding site for FLEC and PROP in the niche between P-helix and the inner helix S6. From this niche, the drugs expose their ammonium groups in the inner pore to block the current. The interface between pore-forming subunits was proposed as the binding site of some ligands of Ca2+ channels (47, 48). Our present study provides evidence that some open K+ channel blockers can also bind mainly in the interface of the pore-forming subunits. This interface may be a new target for design of subtype-specific K+ channel drugs, demand for which is high (2).
Supplementary Material
Acknowledgments
We thank Denis B. Tikhonov for helpful discussions, Olaf Pongs (Center for Molecular Neurobiology, Hamburg) for the construction of the chimeras, Sabine Rothmann (MEDA Pharma) for flecainide acetate, and Dr. Michel Eichelbaum (Dr. Margarete Fischer-Bosch, Institute for Clinical Pharmacology) for the enantiomers of flecainide and propafenone. Flecainide acetate was kindly provided by 3M Health Care Limited. Computations were performed using the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET).
This work was supported by grants from the Hertie Foundation (to M. M.) and from the Natural Sciences and Engineering Research Council of Canada (to B. S. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
- PROP
- propafenone
- FLEC
- flecainide.
REFERENCES
- 1.Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 2.Wulff H., Zhorov B. S. (2008) Chemical Reviews 108, 1744–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaczorowski G. J., Garcia M. L. (1999) Curr. Opin. Chem. Biol. 3, 448–458 [DOI] [PubMed] [Google Scholar]
- 4.Sanguinetti M. C., Tristani-Firouzi M. (2006) Nature 440, 463–469 [DOI] [PubMed] [Google Scholar]
- 5.Zhou M., Morais-Cabral J. H., Mann S., MacKinnon R. (2001) Nature 411, 657–661 [DOI] [PubMed] [Google Scholar]
- 6.Faraldo-Gómez J. D., Kutluay E., Jogini V., Zhao Y., Heginbotham L., Roux B. (2007) J. Mol. Biol. 365, 649–662 [DOI] [PubMed] [Google Scholar]
- 7.Lenaeus M. J., Vamvouka M., Focia P. J., Gross A. (2005) Nat. Struct. Mol. Biol. 12, 454–459 [DOI] [PubMed] [Google Scholar]
- 8.Bruhova I., Zhorov B. S. (2007) BMC Struct. Biol. 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreker T., Grissmer S. (2005) Mol. Pharmacol. 68, 966–973 [DOI] [PubMed] [Google Scholar]
- 10.Rossokhin A., Teodorescu G., Grissmer S., Zhorov B. S. (2006) Mol. Pharmacol. 69, 1356–1365 [DOI] [PubMed] [Google Scholar]
- 11.Andér M., Luzhkov V. B., Aqvist J. (2008) Biophys. J. 94, 820–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madeja M., Leicher T., Friederich P., Punke M. A., Haverkamp W., Musshoff U., Breithardt G., Speckmann E. J. (2003) Mol. Pharmacol. 63, 547–556 [DOI] [PubMed] [Google Scholar]
- 13.Xu H., Barry D. M., Li H., Brunet S., Guo W., Nerbonne J. M. (1999) Circ. Res. 85, 623–633 [DOI] [PubMed] [Google Scholar]
- 14.Schultz J. H., Volk T., Ehmke H. (2001) Circ. Res. 88, 483–490 [DOI] [PubMed] [Google Scholar]
- 15.Wible B., Murawsky M. K., Crumb W. J., Jr., Rampe D. (1997) Brain Res. 761, 42–50 [DOI] [PubMed] [Google Scholar]
- 16.Misonou H., Mohapatra D. P., Park E. W., Leung V., Zhen D., Misonou K., Anderson A. E., Trimmer J. S. (2004) Nat. Neurosci. 7, 711–718 [DOI] [PubMed] [Google Scholar]
- 17.Chung J. J., Li M. (2005) FEBS J. 272, 3743–3755 [DOI] [PubMed] [Google Scholar]
- 18.Roe M. W., Worley J. F., 3rd, Mittal A. A., Kuznetsov A., DasGupta S., Mertz R. J., Witherspoon S. M., 3rd, Blair N., Lancaster M. E., McIntyre M. S., Shehee W. R., Dukes I. D., Philipson L. H. (1996) J. Biol. Chem. 271, 32241–32246 [DOI] [PubMed] [Google Scholar]
- 19.MacDonald P. E., Sewing S., Wang J., Joseph J. W., Smukler S. R., Sakellaropoulos G., Wang J., Saleh M. C., Chan C. B., Tsushima R. G., Salapatek A. M., Wheeler M. B. (2002) J. Biol. Chem. 277, 44938–44945 [DOI] [PubMed] [Google Scholar]
- 20.Jacobson D. A., Kuznetsov A., Lopez J. P., Kash S., Ammälä C. E., Philipson L. H. (2007) Cell Metab. 6, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel A. J., Lazdunski M., Honoré E. (1997) EMBO J. 16, 6615–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archer S. L., Wu X. C., Thébaud B., Nsair A., Bonnet S., Tyrrell B., McMurtry M. S., Hashimoto K., Harry G., Michelakis E. D. (2004) Circ. Res. 95, 308–318 [DOI] [PubMed] [Google Scholar]
- 23.Wareing M., Bai X., Seghier F., Turner C. M., Greenwood S. L., Baker P. N., Taggart M. J., Fyfe G. K. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R437–446 [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama A., Kambe F., Kamiya K., Seo H., Toyama J. (1998) Cardiovasc. Res. 40, 343–351 [DOI] [PubMed] [Google Scholar]
- 25.Huang B., Qin D., El-Sherif N. (2000) J. Cardiovasc. Electrophysiol. 11, 1252–1261 [DOI] [PubMed] [Google Scholar]
- 26.Qin D., Huang B., Deng L., El-Adawi H., Ganguly K., Sowers J. R., El-Sherif N. (2001) Biochem. Biophys. Res. Commun. 283, 549–553 [DOI] [PubMed] [Google Scholar]
- 27.Rolf S., Haverkamp W., Borggrefe M., Musshoff U., Eckardt L., Mergenthaler J., Snyders D. J., Pongs O., Speckmann E. J., Breithardt G., Madeja M. (2000) Naunyn Schmiedebergs Arch. Pharmacol. 362, 22–31 [DOI] [PubMed] [Google Scholar]
- 28.Tikhonov D. B., Zhorov B. S. (2007) Biophys. J. 93, 1557–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liman E. R., Tytgat J., Hess P. (1992) Neuron 9, 861–871 [DOI] [PubMed] [Google Scholar]
- 30.Madeja M., Musshoff U., Speckmann E. J. (1991) J. Neurosci. Methods 38, 267–269 [DOI] [PubMed] [Google Scholar]
- 31.Long S. B., Campbell E. B., Mackinnon R. (2005) Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 32.Li Z., Scheraga H. A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6611–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiner S. J., Kollman P. A., Case D. A., Singh U. C., Chio C., Alagona G., Profeta S., Weiner P. K. (1984) J. Am. Chem. Soc. 106, 765–784 [Google Scholar]
- 34.Lazaridis T., Karplus M. (1999) Proteins 35, 133–152 [DOI] [PubMed] [Google Scholar]
- 35.Momany F. A., McGuire R. F., Burgess A. W., Scheraga H. A. (1975) J. Phys. Chem. 79, 2361–2381 [Google Scholar]
- 36.Dewar M. J. S., Zoebisch E. G., Healy E. F., Stewart J. J. P. (1985) J. Am. Chem. Soc. 107, 3902–3909 [Google Scholar]
- 37.Zhorov B. S., Tikhonov D. B. (2004) J. Neurochem. 88, 782–799 [DOI] [PubMed] [Google Scholar]
- 38.Kauder O., Madeja M., Speckmann E.-J., Lehmenkuhler A. (1991) Pfleugers Arch. Eur. J. Physiol. 418, R77 [Google Scholar]
- 39.Choi K. L., Mossman C., Aubé J., Yellen G. (1993) Neuron 10, 533–541 [DOI] [PubMed] [Google Scholar]
- 40.Valenzuela C., Delpón E., Tamkun M. M., Tamargo J., Snyders D. J. (1995) Biophys. J. 69, 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yale H. L. (1959) J. Med. Pharm. Chem. 1, 121–133 [DOI] [PubMed] [Google Scholar]
- 42.Garden D. P., Zhorov B. S. (2010) J. Comput. Aided Mol. Des. 24, 91–105 [DOI] [PubMed] [Google Scholar]
- 43.Snyders J., Knoth K. M., Roberds S. L., Tamkun M. M. (1992) Mol. Pharmacol. 41, 322–330 [PubMed] [Google Scholar]
- 44.Valenzuela C., Snyders D. J., Bennett P. B., Tamargo J., Hondeghem L. M. (1995) Circulation 92, 3014–3024 [DOI] [PubMed] [Google Scholar]
- 45.Decher N., Pirard B., Bundis F., Peukert S., Baringhaus K. H., Busch A. E., Steinmeyer K., Sanguinetti M. C. (2004) J. Biol. Chem. 279, 394–400 [DOI] [PubMed] [Google Scholar]
- 46.Schmitz A., Sankaranarayanan A., Azam P., Schmidt-Lassen K., Homerick D., Hänsel W., Wulff H. (2005) Mol. Pharmacol. 68, 1254–1270 [DOI] [PubMed] [Google Scholar]
- 47.Hockerman G. H., Peterson B. Z., Johnson B. D., Catterall W. A. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 361–396 [DOI] [PubMed] [Google Scholar]
- 48.Tikhonov D. B., Zhorov B. S. (2008) J. Biol. Chem. 283, 17594–17604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.