Abstract
Rheb is a homolog of Ras GTPase that regulates cell growth, proliferation, and regeneration via mammalian target of rapamycin (mTOR). Because of the well established potential of activated Ras to promote survival, we sought to investigate the ability of Rheb signaling to phenocopy Ras. We found that overexpression of lipid-anchored Rheb enhanced the apoptotic effects induced by UV light, TNFα, or tunicamycin in an mTOR complex 1 (mTORC1)-dependent manner. Knocking down endogenous Rheb or applying rapamycin led to partial protection, identifying Rheb as a mediator of cell death. Ras and c-Raf kinase opposed the apoptotic effects induced by UV light or TNFα but did not prevent Rheb-mediated apoptosis. To gain structural insight into the signaling mechanisms, we determined the structure of Rheb-GDP by NMR. The complex adopts the typical canonical fold of RasGTPases and displays the characteristic GDP-dependent picosecond to nanosecond backbone dynamics of the switch I and switch II regions. NMR revealed Ras effector-like binding of activated Rheb to the c-Raf-Ras-binding domain (RBD), but the affinity was 1000-fold lower than the Ras/RBD interaction, suggesting a lack of functional interaction. shRNA-mediated knockdown of apoptosis signal-regulating kinase 1 (ASK-1) strongly reduced UV or TNFα-induced apoptosis and suppressed enhancement by Rheb overexpression. In conclusion, Rheb-mTOR activation not only promotes normal cell growth but also enhances apoptosis in response to diverse toxic stimuli via an ASK-1-mediated mechanism. Pharmacological regulation of the Rheb/mTORC1 pathway using rapamycin should take the presence of cellular stress into consideration, as this may have clinical implications.
Keywords: Apoptosis, ER Stress, Oxidative Stress, Raf, Ras, Rapamycin, Rheb, Chemical Shift Mapping, mTORC1, NMR Spectroscopy
Introduction
Ras homolog enriched in the brain (Rheb)6 is a small GTPase belonging to the Ras superfamily of guanine nucleotide-binding proteins and is related to Ras, Rap, and Ral (1). The function of Rheb has been studied in a variety of organisms, especially in Drosophila and mammalian cells. These results underscore the role of Rheb as a molecular switch in many cellular processes such as cell volume growth, cell cycle progression, neuronal axon regeneration, autophagy, nutritional deprivation, oxygen stress, and cellular energy status (2–4). The effects of Rheb are mediated via the mammalian target of rapamycin (mTOR), which exists in two different multiprotein complexes: the rapamycin-sensitive mTORC1, which is responsible for the modulation of protein translation, and TORC2, which mediates the spatial control of cell growth by regulating the actin cytoskeleton (5, 6). Known mTORC1 targets include ribosomal p70S6 kinase (S6K), the translational repressor 4E-BP1, and PRAS40 (7, 8).
Rheb activity is regulated by a dual mechanism. Insulin and other growth factors stimulate the GTP loading of Rheb via inhibition of tuberous sclerosis complex (TSC)1/TSC2, a tumor suppressor protein complex that acts as a Rheb GTPase-activating protein (GAP) (9, 10). In contrast to Ras, Rheb synthesis is up-regulated similar to immediate early genes after toxic insults or by growth factors, such as epithelial growth factor (EGF) or basic fibroblast growth factor (bFGF) (11). Because of the high concentration of GTP-bound Rheb under basal conditions, elevated levels of Rheb are sufficient to activate mTORC1 (5, 12).
Because of high sequence identity, we initially anticipated that Rheb would promote an H-Ras-like phenotype. Vast evidence exists for Ras activity acting as a protective agent both in non-neuronal (13) and neuronal systems (14–17). Furthermore, transgenic activation of neuronal Ras in the brain prevents degeneration in several lesion paradigms (17–19). Similar to Ras, Rheb signaling is also directly correlated with the promotion of survival. Correspondingly, attenuated mTOR signaling has been shown to induce apoptosis in cell lines (20, 21). In neurons, the Parkinson disease mimetic 6-hydroxy dopamine triggers neuronal death by suppressing the activation of mTOR (22). After axotomy of retinal ganglion cells, mTOR activity is suppressed and new protein synthesis impaired, contributing to degeneration. Reactivating mTOR by conditionally knocking out TSC1 leads to axon regeneration (4).
In contrast, in other cellular systems such as radiation-induced cell damage, up-regulated Rheb activity is associated with apoptosis (23). Furthermore, TSC deficiency results in severe insulin resistance (24) and triggers the unfolded protein response to regulate endoplasmic reticulum (ER) stress (25).
Taken together, this evidence shows that the balance of the mTOR pathway is essential to ensure the healthy state of the cell. However, whether enhanced Rheb activity will lead to cellular protection or increased vulnerability cannot be predicted.
Here, we investigated whether Rheb influences cell death induced by excitotoxic glutamate treatment in neurons. More specifically, we analyzed whether Rheb enhances or prevents the apoptosis of HeLa (cervical cancer) cells triggered by UV light, TNFα, and tunicamycin in an mTORC1-dependent manner.
Analyzing Ras-Rheb cross-talk, other studies have suggested that Rheb is involved in negative regulation of B-Raf and c-Raf-activity (26), suggesting that Rheb is a feedback inhibitor of Ras signaling that results in the antagonism of Ras (26–28). To investigate the interaction among Rheb, Ras, and c-Raf at the atomic level, we compared their signaling mechanisms and analyzed the perturbed chemical shifts upon the formation of the Rheb/c-Raf-Ras-binding domain (RBD) complex (29, 30). Finally, we correlated the solution structure and backbone dynamics of Rheb with its biochemical properties.
EXPERIMENTAL PROCEDURES
Chemicals, Antibodies, and Plasmids
All chemicals were purchased from Sigma unless stated otherwise. Inhibitors were obtained from Calbiochem and used at the following concentrations: rapamycin (20 nm), cycloheximide (2.5 μg/ml), TNFα (10 ng/ml), salubrinal (100 μm), and tunicamycin (1.5 μm). Antibodies were acquired from Cell Signaling with the exception of anti-FLAG-M2 and anti-HA (Sigma). Rat Rheb cDNA was a kind gift from A. Wittinghofer (Max-Planck-Institut, Dortmund, Germany) and subcloned into pcDNA3 fused to an N-terminal FLAG epitope. pASK-ΔN-c-Raf was a kind gift from U. Rapp (Medizinische Strahlenkunde und Zellforschung, Würzburg, Germany) and subcloned into pcDNA3-myc. In addition, pHA-ASK-1 was a gift from W. Min (Yale University, New Haven, CT). pBos-mRFP-histone 2B was applied as published previously (31), and the pSM2 scramble shRNA and pSM2-hRheb shRNA constructs were obtained from Open Biosystems.
Primary Neuron Cell Culture, Transfection, and Stimulation
Cortical cultures were prepared from the cortices of P0–P2 newborn NMRI mice according to a slightly modified version of the protocol established by Lessmann and Heumann (32). Primary cortical cultures were transfected using the calcium phosphate method described by Haubensak et al. (33). After 48 h, the cells were stimulated with 1 mm glutamate in balanced salt solution (130.0 mm NaCl, 5.4 mm KCl, 1.8 mm CaCl2, 5.5 mm glucose, 20.0 mm HEPES, pH 7.4) supplemented with 10 μm glycine for 1 h at 37 °C under conditions of 5% CO2. Twenty-four hours later, the cells were fixed with 4% paraformaldehyde and stained with bisbenzimide H3342 (Fluka). Using a fluorescence microscope (Olympus IX51, ×20), we determined apoptosis by counting transfected neurons showing fragmented or strongly condensed nuclei caused by DNA fragmentation.
Cell Culture, cDNA Transfection, and Inhibitor Stimulation
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 2 mm glutamine. Transient transfection was carried out for 48 h using Polyfect (Qiagen) according to the manufacturer's protocol or for 96 h in the case of shRNA transfection. Rapamycin was applied 2 h before stimulating the cells with TNFα and cycloheximide (for 4 h) or UV light (0.03 J/cm2 followed by 4 h incubation). Salubrinal was applied 2 h before UV irradiation and 30 min before tunicamycin stimulation.
Apoptosis Assay
To visualize transfected cells and facilitate the quantification of apoptotic cells, a mRFP-histone 2B reporter construct was co-transfected at a molar ratio of 1:5 with the constructs of interest (31). Apoptotic mRFP-histone 2B-positive cells were counted under a fluorescence microscope (Olympus IX51, ×20) 4 h (TNFα + cycloheximide and UV irradiation) or 24 h (tunicamycin) after apoptotic stimulation to visualize mRFP-histone 2B. Apoptotic cells showed fragmented or strongly condensed nuclei caused by DNA fragmentation (supplemental Fig. S1).
Flow Cytometry
Cell volume was analyzed by flow cytometry using a CyFlow SL instrument (Partec). HeLa cells were trypsinized 72 h post-transfection and fixed with 4% paraformaldehyde. The distribution of forward scatter signals gated for transfected green fluorescent protein (GFP)-positive cells was recorded.
Protein Expression, Purification, and Western Blot
Rheb was expressed and purified as described previously (29). The RBD of c-Raf kinase was expressed and purified as described previously (34–36). Western blot analysis was also performed essentially as described previously (17).
NMR Spectroscopy
All spectra were recorded at 298 K on Bruker DRX600 and Bruker DRX750 spectrometers equipped with pulsed field gradients and triple resonance probe heads, processed with NMRPipe, and analyzed by NMRView (37, 38). The steady state heteronuclear NOE (nuclear Overhauser effect) experiments were recorded on a DRX 600 spectrometer at 298 K in an interleaved manner to reduce influence from possible instabilities in the experimental conditions (39). The NOE values were calculated from the ratio of the peak heights in the experiment with and without proton saturation (40–43).
Structure Calculation
Assignment, data handling, and calculations were performed using NMRView and XPLOR-NIH (38, 44, 45). The distance constraints were supplemented with torsion angle constraints derived from the HNHA experiment (46) and TALOS (47). Structure calculations were performed using standard protocols for simulated annealing constraint methods implemented in the program XPLOR-NIH and as published previously (44). Further details are given in supplemental Table S1 (Protein Data Bank code 2L0X).
RESULTS
Rheb Overexpression Enhances Apoptosis
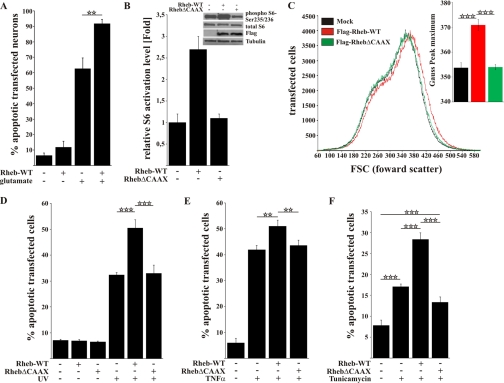
To investigate whether Rheb participates in survival or the regulation of cell death, we overexpressed FLAG-Rheb-WT in primary cortical neurons and subjected the cells to transient stimulation with excitotoxic concentrations of glutamate (1 mm, 1 h). Cell death was assessed 24 h later by nuclear staining and determining the percentage of fragmented or pyknotic nuclei within the population of transfected cells as indicated by co-expression of an EGFP-reporter plasmid. Although overexpression of FLAG-Rheb-WT did not induce cell death per se in primary cortical neurons, it did significantly enhance apoptosis by nearly 30% after excitotoxic stimulation (Fig. 1A; 62 versus 91%, mock versus FLAG-Rheb-WT, respectively).
FIGURE 1.
Rheb overexpression enhances the rate of cell death in a CAAX-box-dependent manner after toxic treatment. A, primary cortical neurons transfected with FLAG-Rheb-WT exhibited an increased rate of cell death 24 h after excitotoxic treatment with 1 mm glutamate compared with mock-transfected cells. B, in Rheb-WT- but not RhebΔCAAX-transfected HeLa cells, the mTOR pathway was strongly activated as shown by the activation level of the S6 protein on Western blot. C, the size of Rheb-WT-transfected HeLa cells was increased compared with RhebΔCAAX or mock-transfected HeLa cells. D, HeLa cells transfected with FLAG-Rheb-WT were more sensitive to UV light as measured by quantification of apoptotic cells 4 h after UV light treatment (0.03 J/cm2). Notably, the basal level of apoptotic cells did not change upon transfection with FLAG-Rheb-WT or FLAG-RhebΔCAAX. E, HeLa cells transfected with Rheb-WT had significantly higher rates of cell death after TNFα (10 ng/ml + 2.5 μg/ml cycloheximide) treatment compared with RhebΔCAAX or mock-transfected cells. F, ER stress-induced apoptosis in the presence of tunicamycin was increased in Rheb-WT-transfected cells but not in RhebΔCAAX-transfected cells. All bars represent mean ± S.E. of at least six independent experiments. **, p < 0.01; ***; p < 0.001 (determined by ANOVA followed by a Bonferroni post hoc test).
To test whether Rheb overexpression generally enhances apoptosis, we used a different, non-neuronal cell type. Indeed, Rheb overexpression significantly increased the sensitivity of HeLa cells to UV-induced apoptosis. However, the apoptosis-enhancing effect disappeared when RhebΔCAAX lacking the C-terminal farnesylation signal (27) was overexpressed (Fig. 1D andsupplemental Fig. S1). In contrast to Rheb-WT, RhebΔCAAX failed to activate mTORC1 activity, which was measured by ribosomal S6 protein phosphorylation and the cellular volume (Fig. 1, B and C). This result is in accordance with previous reports that Rheb-dependent mTORC1 activation depends on Rheb targeting to the membrane (48). Similarly, Rheb was observed to have CAAX-box-dependent apoptosis-enhancing effects when TNFα (Fig. 1E) or the ER stress inducer tunicamycin (Fig. 1F) was applied as apoptotic stimuli. Taken together, the data show that Rheb overexpression enhances the pro-apoptotic response of various mammalian cell types to different kinds of apoptotic stimuli in a CAAX-box-dependent manner.
Apoptosis Is Enhanced by Endogenous Rheb
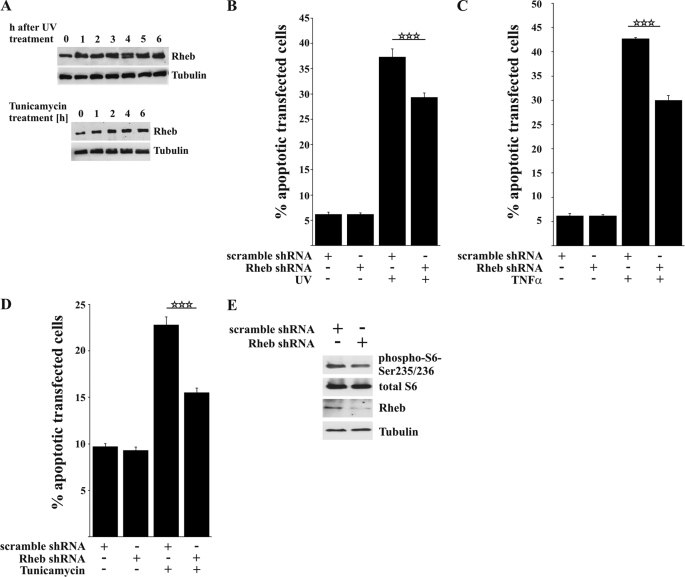
To assess whether endogenous levels of Rheb are required for the transmission of pro-apoptotic signals, we investigated the effects of a functional Rheb knockdown construct (Fig. 2E). Knocking down Rheb led to a significant decrease in apoptotic cells after stimulation with UV light, TNFα, and tunicamycin, indicating that endogenous Rheb functionally in participates the apoptotic response induced by these agents (Fig. 2, B–D). Notably, neither Rheb overexpression nor Rheb knockdown led to apoptosis in the absence of toxic stimuli, underscoring a cell stress-dependent functional switch by Rheb.
FIGURE 2.
Knocking down Rheb decreases cell death after toxic treatment. A, Western blot showing that endogenous Rheb expression increased after exposure to UV light or tunicamycin. B–D, HeLa cells were transfected with either a non-silencing scramble shRNA or human-specific Rheb shRNA and incubated for 96 h. The shRNA-mediated knockdown of endogenous Rheb significantly protected against UV light (B), TNFα (C), and tunicamycin-induced apoptosis (D). E, the efficiency of knocking down Rheb and inhibiting mTOR activation was tested by Western blot 96 h after transfection showing a moderate inhibition of S6 phosphorylation. All bars represent mean ± S.E. of at least six independent experiments. ***, p < 0.001 (determined using ANOVA followed by a Bonferroni post hoc test).
As dysregulated mTOR signaling stimulated the unfolded protein response and ER stress, we wanted to investigate whether the ER stress signaling pathway (49, 50) might be responsible for Rheb-enhanced apoptosis. As shown in supplemental Fig. S2A, Rheb was still able to enhance apoptosis in the presence of the ER stress-protective agent salubrinal (100 μm), despite clear inhibition of tunicamycin-induced apoptosis. Similarly, after apoptosis was induced with UV treatment, salubrinal was not able to prevent Rheb-enhanced apoptosis. This finding indicates that Rheb-mediated apoptosis occurs independent of the ER stress/unfolded protein response pathway (Fig. S2B).
Up-regulated and Basal Rheb Levels May Mediate Apoptosis
Because UV irradiation and tunicamycin (Fig. 2A) induced an up-regulation of Rheb protein in HeLa cells within 4 h, the time period of maximal cell death, we tested whether these stimuli would also activate the mTOR pathway. The irradiation of HeLa cells with UV light increased the phosphorylation level of the ribosomal S6 protein (Fig. 3C). However, we were unable to demonstrate further increases in S6 phosphorylation in the presence of Rheb-WT overexpression, possibly because of the saturating activation of mTOR after UV irradiation (Fig. 3C). Functional Rheb knockdown attenuated TNFα-stimulated apoptosis in the presence of cycloheximide, although the latter inhibited protein synthesis (Fig. 2C). Thus, up-regulation of endogenous Rheb mimicks transfection-induced overexpression, although the basal level of Rheb may be sufficient to mediate apoptosis.
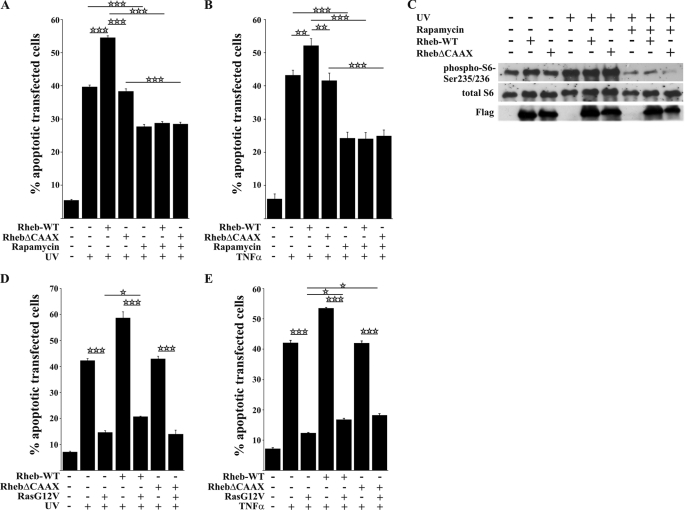
FIGURE 3.
Rheb-enhanced apoptosis after toxic treatment is mTOR-dependent. A and B, pretreatment (2 h) of transfected HeLa cells with rapamycin (20 nm) inhibited Rheb-enhanced apoptosis after UV light (A) or TNFα treatment (B). C, Western blot showing a strong induction of the mTOR pathway 4 h after stimulation with UV light in mock-transfected cells, which was comparable with the activation levels of Rheb-WT-transfected cells. Pretreatment with rapamycin blocked activation of the mTOR pathway. D and E, co-expression of permanently activated RasG12V almost completely protects HeLa cells against UV light as well as TNFα-induced cell death but cannot inhibit Rheb-enhanced cell death. All bars represent the mean ± S.E. of at least six independent experiments. * p < 0.05; **, p < 0.01; ***, p < 0.001 (determined using ANOVA followed by a Bonferroni post hoc test).
Rheb-enhanced Cell Death Is Sensitive to Rapamycin Treatment
The major TORC1 inhibitor rapamycin inhibits S6 phosphorylation to below basal levels (Fig. 3C) and reduces, but does not abolish, apoptosis after TNFα stimulation and UV irradiation, suggesting a pro-apoptotic effect of endogenous mTORC1 activity (Fig. 3, A and B). However, rapamycin treatment never completely blocked the cell death induced by toxic treatment in the absence or presence of Rheb-WT overexpression, indicating that other additional pro-apoptotic mechanisms may exist independent of the mTORC1 pathway.
Active H-RasG12V Opposes Apoptosis but Not the Specific Contribution of Rheb Overexpression
In accordance with a cytoprotective role of constitutively active Ras (18), H-RasG12V protected cells from apoptosis induced by adverse stimuli, whereas Rheb overexpression exacerbated the apoptotic response. Exposure to UV light or TNFα typically increased apoptotic numbers from 7–8 to 42–43%, and active RasG12V decreased it to 12–14% (Fig. 3, D and E). Rheb-enhanced apoptosis was also reduced, but only to ∼15–18%. Notably, in the presence of activated Ras, Rheb was still able to enhance apoptosis (compare bars 3 and 5 in Fig. 3, D and E). Considering the dissimilarity in signaling, despite a high degree of sequence similarity between Rheb and Ras, we sought to gain further insight into the mechanisms underlying the differential signaling pathways of these proteins using a structural approach.
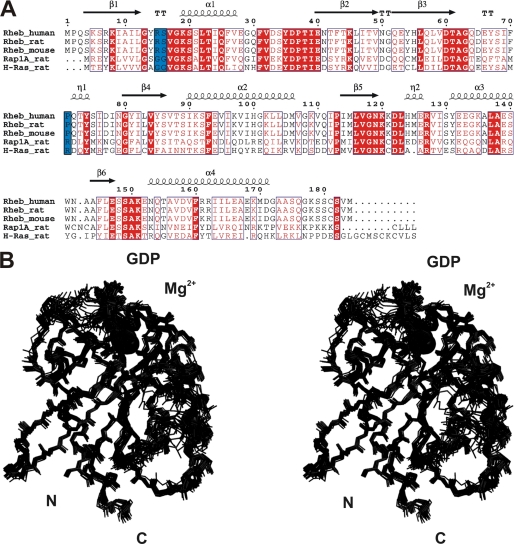
Overall Structure of Full-length Rheb in Solution
Based on the sequence alignment, Rheb is more closely related to Ras and Rap than other small G proteins (Fig. 4A). The switch I region extends from Asp-33 to Asn-41 and the switch II region from Gly-63 to Asn-79. In all small GTPases, these switch regions are involved in the recognition of and interaction with GAPs, guanine nucleotide exchange factors (GEFs), and effectors (51, 52). Small GTPases also carry a characteristic phosphate binding P-loop that, in Rheb, comprises residues Gly-13 to Ser-20. The final ensemble 〈SA〉 of 20 Rheb-GDP NMR structures is shown in stereo in Fig. 4B, and a summary of refinement statistics is given in supplemental Table S1. Overall, the structure is well defined, and the coordinate precision is rather high. The root mean square deviation between the ensemble of structures (〈SA〉) and the average structure of the ensemble (〈SA〉av) for the backbone atoms is ∼0.59 ± 0.11 Å for backbone atoms and 1.17 ± 0.17 Å for all heavy atoms. 99.5% of all backbone angles φ and ϕ are located in allowed regions of the Ramachandran plot (supplemental Table S1).
FIGURE 4.
Structure-based sequence alignment of Rheb from different species, as well as Ras and Rap1A from the rat, and stereoview of the backbone atoms (N, Cα, C′, and O) of the ensemble 〈〈SA〉〉 of 20 rRheb structures. A, invariant residues are highlighted in red-shaded boxes, and open red boxes indicate conserved residues. The secondary structural elements of Rheb are given above the sequence alignment. Residues in blue boxes are discussed in detail throughout the text. B, this superposition shows the lowest root mean square deviation values for the backbone atoms (N, Cα, and C′) of the regions with a regular secondary structure. The unstructured N-terminal residues Ser-1 to Lys-5 and C-terminal residues Ser-175 to Met-184 have been omitted for clarity.
The overall structure of Rheb adopts the typical canonical fold of small GTPases (53–55). Rheb has an αβ-fold that contains a β-sheet consisting of one antiparallel and five parallel β-strands (Fig. 5). The β-strands wrap around the C terminus of the α-helix, which is oriented perpendicular to the first α-helix. The convex side of the β-sheet is covered by three additional helices. The nucleotide binding site (GTP or GDP) is located at the C-terminal end of the β-sheet, similar to other nucleotide-binding, open β-sheet structures (54). This nucleotide binding site is formed by the switch I and switch II regions and the P-loop, as well as the N-terminal end of the first helix (53). Overall, the orientation of the bound GDP nucleotide is conserved in Rheb. In summary, β1 extends from Lys-5 to Leu-12, α1 from Lys-19 to Glu-28, β2 from Asn-41 to Val-49, β3 from Gln-52 to Asp-60, β4 from Gly-80 to Ser-86, α2 from Ile-90 to Val-107, β5 from Ile-114 to Asn-119, α3 from Tyr-131 to Trp-141, β6 from Ala-144 to Glu-147, and α4 from Asn-153 to Ile-170 (Fig. 4A). The resonances of the N-terminal residues Ser-1 to Lys-5 and the C-terminal residues Ser-175 to Met-184 could not be observed in the spectra, presumably because of chemical exchange. The topology of the fold is identical to that found for Ras and the crystallographic study of Rheb (53–55). However, important differences were found for the (flexible) switch I and switch II structural elements. Furthermore, the C-terminal helix in the NMR structure extends to residue Ile-170 (Fig. 5).
FIGURE 5.
Ribbon drawing of a representative member of the ensemble structures of rRheb. The N and C termini are indicated. The unstructured N-terminal residues Ser-1 to Lys-5 and C-terminal residues Ser-175 to Met-184 have been omitted for clarity. This figure was generated using PyMol (83, 84).
Conformational Dynamics of the Switch I and Switch II Regions of Rheb
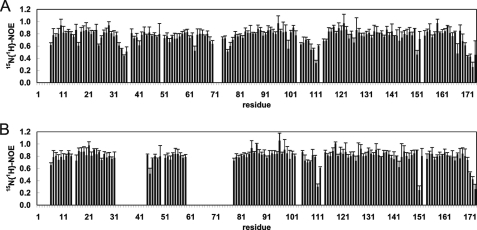
The heteronuclear NOE data in Fig. 6 clearly show that the switch I and, to a lesser extent, the switch II region of GDP-bound Rheb is flexible in solution due to genuine mobility rather than a lack of experimental restraints. In addition, the turn including Pro-113 has values less than 0.6 for the heteronuclear NOE, which is a clear indication of increased flexibility on the pico- to nanosecond time scale and also observed for the C-terminal residues Asp-171 to Ala-174. Obviously, these regions undergo motions faster than the overall tumbling time of the protein. In the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of Rheb bound to Gpp(NH)p (the nonhydrolyzable analog of GTP), the resonance signals for the switch I (residues Val-32 to Phe-43) and II (residues Asp-60 to Ile-78) regions were broadened beyond detection (Fig. 6B), presumably due to a chemical exchange at an intermediate rate on the NMR time scale, in marked contrast to Rheb-GDP. Interestingly, this difference between the GDP- and Gpp(NH)p-bound forms of Rheb could not directly be inferred from the crystal structure (55). Due to absent resonances in Rheb-Gpp(NH)p, no quantitative differences could be extracted for the switch I and switch II regions.
FIGURE 6.
Steady state heteronuclear NOE for the backbone amides of rRheb in its GDP- (A) and Gpp(NH)p-bound (B) states. Residues for which no results are shown correspond to either prolines or residues for which relaxation data could not be extracted. The switch I region extends from residue Asp-33 to Asn-41 and the switch II region from residue Gly-63 to Asn-79. For details, refer to the text under “Experimental Procedures” and “Results.”
Rheb Interacts with c-Raf-RBD with Low Affinity in a Ras-like Manner
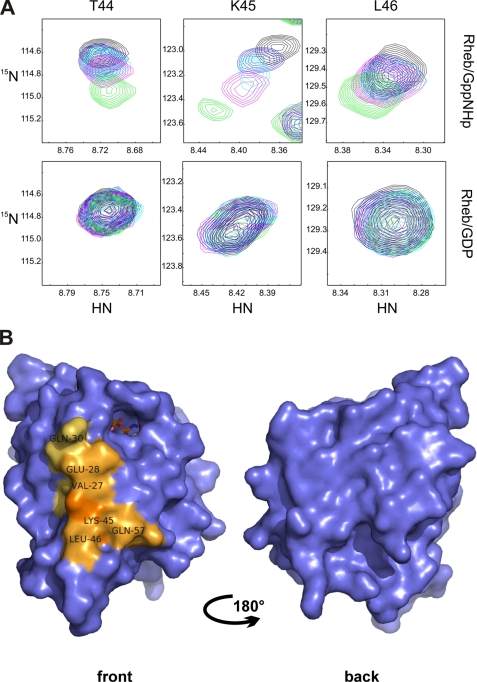
The functional and physical interactions between Rheb and Raf proteins were suggested previously (26, 28). We examined whether the RBD of c-Raf might possibly bind to Rheb because of similarities between Ras and Rheb in the switch I region. We used [U-15N]Rheb bound to either GDP or Gpp(NH)p to show that the RBD of c-Raf selectively binds to Rheb-Gpp(NH)p in multidimensional NMR spectroscopy (Fig. 7A). We mapped the binding site of c-Raf-RBD on the Rheb surface (Fig. 7B), which resembled the effector-type interaction between Ras and c-Raf kinase. The binding site includes Rheb residues Glu-28, Gln-30, Thr-44, Lys-45, Leu-46, and Gln-57. The c-Raf-RBD selectively bound to Rheb-Gpp(NH)p, as no significant chemical shift perturbation was observed for Rheb-GDP even in the presence of a 6-fold molar excess of c-Raf-RBD. Rheb has a lower affinity for c-Raf kinase compared with Ras (56). Although the affinity of the latter interaction is 120 nm (34, 57), the affinity of Rheb for c-Raf kinase is ∼200 ± 80 μm according to our NMR titration experiments. The novel GTP-Ras effector mNORE1 is capable of inducing apoptosis; therefore, we questioned whether it may bind to Rheb, thereby mediating cell death (58). In contrast to c-Raf-RDB, the RBD of mNORE1 did not show any affinity for activated or inactivated Rheb (data not shown).
FIGURE 7.
Chemical shift differences between rRheb bound to GDP or Gpp(NH)p upon titration with the RBD of c-Raf kinase. A, the upper panel shows representative regions of the 1H-15N HSQC spectra for Thr-44, Lys-45, and Leu-46 of rRheb-GppNHP titrated with c-Raf-RBD (molar ratio of rRheb/c-Raf-RBD ranging from 1:0 (shown in black) to 1:3 (shown in green)). The lower panel shows the corresponding regions of the 1H-15N-HSQC spectra of Rheb-GDP titrated with c-Raf-RBD (molar ration of rRheb/c-Raf-RBD ranging from 1:0 (shown in black) to 1:6 (shown in green)). B, significant chemical shift pertubation for Rheb bound to Gpp(NH)p upon titration with the RBD of c-Raf kinase projected onto the accessible surface of rRheb. The differences are represented in light to dark orange depending on the magnitude of the observed weighted chemical shift differences. For details, refer to the text under “Experimental Procedures” and “Results.” This figure was generated using PyMol (83, 84).
c-Raf Opposes Rheb-enhanced Apoptosis
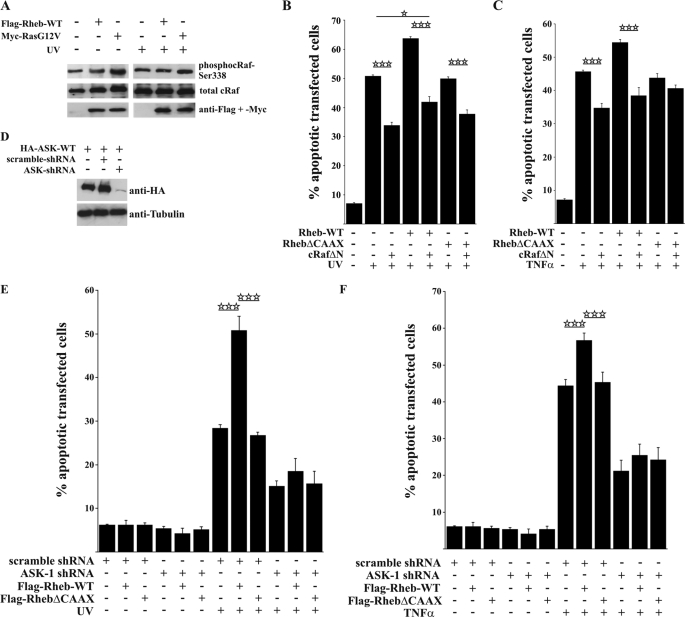
Having established that Rheb binds specifically to c-Raf-RBD with low affinity in a Ras-like pattern, we tested whether this weak interaction might have functional implications in cells by measuring c-Raf activity using a phospho-specific c-Raf antibody against the activating phosphorylation site, Ser-338 (27). H-RasG12V increased phosphorylation of c-Raf (Fig. 8A). However, we were unable to detect major changes in the phosphorylation level of c-Raf in the presence of Rheb-WT, which is consistent with the notion that, because of the low binding affinity in the micromolar range, direct regulation of the c-Raf kinase might not be possible, despite the interaction specificity.
FIGURE 8.
Active c-Raf does not inhibit Rheb-enhanced apoptosis. A, Western blot showing that, in transfected HeLa cells, Rheb expression does not influence the activation level of endogenous c-Raf as indicated by no differences at the Ser-338 phosphorylation site of c-Raf. B and C, co-expression of a constitutively active c-Raf mutant significantly protects against UV light and TNFα-induced apoptosis but is not able to completely inhibit Rheb-enhanced apoptosis. D, representative Western blot illustrating the efficiency of ASK-1 knockdown. E and F, HeLa cells were transfected with scramble or human ASK-1 shRNA plus FLAG-Rheb-WT or FLAG-RhebΔCAAX and incubated for 96 h. ASK-1 knockdown led to a general decrease in cell death and inhibited Rheb-enhanced apoptosis after UV irradiation as well as TNFα stimulation, as shown by a lack of significant differences in cell death. All bars represent mean ± S.E. of at least six independent experiments. *, p < 0.05; ***, p < 0.001 (determined using ANOVA followed by a Bonferroni post hoc test).
To further characterize the role of c-Raf, we used a constitutively active c-RafΔN mutant (59) that resulted in a protective effect against UV light and TNFα-induced apoptosis (Fig. 8, B and C). Although active c-RafΔN opposed apoptosis in all cases tested, Rheb was still able to enhance apoptosis, indicating that c-Raf and Rheb signal through independent mechanisms (compare bars 3 and 5 in Fig. 8B).
The effects by c-RafΔN could be due to either the lack of an N-terminal RBD region, preventing the interaction with Ras, or an inhibitory action of the RBD on Raf kinase activity. To test whether the kinase activity is necessary for the anti-apoptotic c-RafΔN activity, we co-expressed an inactive c-Raf mutant, c-RafΔN-K375M, with FLAG-Rheb-WT and FLAG-RhebΔCAAX and treated HeLa cells with UV light and TNFα. Interestingly, inactive c-RafΔN-K375M was still able to inhibit Rheb-enhanced apoptosis to nearly the same extent as c-RafΔN (supplemental Fig. S3).
What Is the Mechanism Behind the Apoptosis-enhancing Effect of Rheb?
Our data did not show a role for Ras, Raf kinase, or ER stress in Rheb-enhanced apoptosis, and there was no associated change in the activity of Akt kinase (data not shown). Apoptosis signal-regulating kinase 1 (ASK-1) plays an essential role in apoptosis (60, 61). Upon shRNA-mediated knockdown of ASK-1 (Fig. 8D), we observed a decrease in apoptosis from 50.9 ± 3.2% to 18.5 ± 3.1% for UV treatment and from 56.7 ± 1.9% to 25.4 ± 2.9% for TNFα in the presence of Rheb (Fig. 8, E and F). Notably, in the presence of shRNA knockdown of ASK-1, apoptosis was not significantly enhanced by Rheb overexpression (compare bars 10 and 11 in Fig. 8, E and F). Taken together, the data suggest that Rheb-enhanced apoptosis is mediated by ASK-1 activity. Next, we inhibited the ASK-1 downstream targets p38 and c-Jun kinase and found that this opposed apoptosis (supplemental Fig. S4). Whether additional ASK-1 effectors are involved in Rheb-enhanced apoptosis remains to be determined.
DISCUSSION
Here, we have described a switch in the physiological function of Rheb from a growth-promoting to a cell death-mediating GTPase. Overexpression of membrane lipid-anchored Rheb stimulates S6 phosphorylation and increases cell volume without inducing any detectable levels of apoptosis. After acute apoptotic stress, however, Rheb leads to a rapamycin-sensitive enhancement of apoptosis.
Cell Stress-dependent Switch to a Death-mediating GTPase
Considering the various reports demonstrating survival and regeneration via the activated mTOR pathway (4), we were surprised to find that Rheb facilitated, rather than attenuated, excitotoxic glutamate-induced cell death. As the mechanisms of excitotoxic cell death are still being debated (62), we decided to further study the Rheb-mediated apoptotic mechanism in a more defined non-neuronal cell system by applying three different types of apoptosis inducers that initiate ER and/or mitochondrial signaling pathways: UV stress (the release of toxic oxygen radicals), DNA fragmentation, and activation of p38 and c-Jun kinase. TNFα and cycloheximide cause cell death via a defined signaling cascade and the glycosylation inhibitor tunicamycin via a well described ER-unfolding response program (63). In all cases, Rheb-enhanced apoptosis was diminished when a Rheb construct lacking the C-terminal lipidation anchor was overexpressed, which suggests that intracellular membrane localization is essential for its function. Recently, Rheb was shown to stimulate exclusively the mTORC1 pathway in vitro, leading to enhanced binding and phosphorylation of 4E-BP1, but mTORC2 signaling was not affected. Together with the observed sensitivity to rapamycin, we concluded that Rheb-enhanced apoptosis is mediated via the activation of mTORC1. In addition, we demonstrate that shRNA suppression of endogenously expressed Rheb attenuates the apoptotic effects (Fig. 2), clearly identifying Rheb as an apoptosis-mediating protein. However, further research is necessary to determine where Rheb-dependent and Rheb-independent pro-apoptotic pathways converge.
Rheb Activation Per Se Does Not Induce Apoptosis
Here, we show that overexpression of Rheb alone is not sufficient to trigger apoptosis. Numerous reports have shown that high Rheb GTP loading stimulated through TSC suppression and enhanced mTOR activity is an important aspect of the regulation of various processes of growth and survival (64) or neuronal differentiation (65). Furthermore, inappropriate activation of the Rheb/mTOR/S6 cassette causes cell survival deficiencies (66). Bnip3 decreases Rheb-GTP levels, thereby mediating the hypoxia-induced inhibition of mTOR (67), and PRAS40 inhibits mTORC1 kinase-mediated phosphorylation of the elf-4E-binding protein, thereby promoting apoptosis. Similarly, in the nervous system, axon regeneration and polarity are promoted by TSC deficiency (4), and overexpression of Rheb induces the formation of multiple axons in hippocampal neurons (68). Taken together, this evidence indicates that the regulation of Rheb GTP loading is vital for normal cell physiology. However, we have shown here that various toxic stimuli, such as excitotoxic glutamate treatment, UV irradiation, TNFα/cycloheximide, and tunicamycin, cause a dramatic switch in Rheb function from promoting growth to being an apoptosis effector protein.
Rheb-enhanced Apoptosis Is Not Specifically Affected by H-Ras and c-Raf Kinase
Although, H-Ras strongly opposed UV- or TNFα-induced apoptosis, significant enhancement by Rheb was still measured in the presence of H-Ras (Fig. 3, D and E). Similar Rheb-induced enhancement was found in the presence of active or inactive c-Raf kinase (Fig. 8B and supplemental Fig. S3), indicating some dramatic dissimilarity in signaling between Ras and Rheb after apoptotic challenge. To investigate whether these differential signaling mechanisms derive from changes at the structural level, we used a protein NMR-based approach.
The Switch I and Switch II Regions of Rheb Possess Unique Structural Features, Explaining Differences in Ras/Rap Signaling
The amino acid composition of switch I in Rheb-GDP differs significantly from the conformation of switch I in Ras and Rap (55). The difference predominantly includes the N-terminal portion of switch I, including Asn-41 and Thr-44. In the NMR structure, switch I and, in part, switch II of Rheb-GDP are generally less precise because of fluctuations on the pico- to nanosecond time scale, as observed in the heteronuclear NOE experiment. This experiment clearly shows that the switch I and, to a lesser extent, switch II loops are ill-defined in the solution state because of genuine mobility rather than a lack of experimental restraints; this has also been observed for the structure of Ras in solution (54). Nonetheless, Ser-34 of Rheb, Glu-31 in Ras, and Lys-31 in Rap have been shown to be crucial in determining the specificity for their effectors (69). Therefore, Ser-34 in Rheb might select for different effectors than those of Ras and Rap. Although the overall sequence similarity found for switch I is rather high, the eliminated charge (Arg in Ras versus Thr-44 in Rheb) and the shorter side chain of this amino acid will also most likely have an impact on the function of Rheb.
In the NMR structure, the ensemble of conformations of the flexible switch II region also deviates from the switch II conformations observed for Ras in solution (54). Switch II itself contains only a short helix-like stretch from Pro-71 to Ser-75, and it packs against the long α-helix composed of residues Ser-89 to Gly-108, which is parallel to the central β-sheet of Rheb (Fig. 5). This interaction is almost exclusively based on hydrophobic residues. This unique switch II conformation puts Gln-64 in a position that prevents its side chain from interacting with the bound nucleotide. The 1H-15N HSQC spectrum of activated Rheb bound to Gpp(NH)p, a nonhydrolyzable analog of GTP, suggests that switch I adopts multiple conformations in solution that interconvert on an intermediate time scale, as the corresponding resonances are broadened beyond detection. This finding supports the idea that switch I might adopt different conformations in active and inactive Rheb. Therefore, the conformation of switch I observed in solution for Rheb-GDP might deviate substantially from some structures of the ensemble of conformations expected for the Gpp(NH)p-bound state because of the broadened resonances in the 1H-15N HSQC spectrum (Fig. 6B). Some of these conformations might be catalytically important for the hydrolysis of GTP bound to Rheb. Further elucidation is needed, by studying the interaction between Rheb and its genuine GAP.
The major difference among small GTPases occurs in the switch II region. In the crystal structures of Rheb, switch II remains virtually unchanged in the GDP- and GTP-bound states, and only residues Pro-71 to Ser-75 constitute a helix-like structure that is much shorter than the helix found in Ras (51, 55). In contrast to the crystal structure, switch II as well as switch I of Rheb-Gpp(NH)p presumably adopts multiple conformations in solution, fluctuating on the micro- to millisecond time scale, as their resonances are broadened beyond detection in the 1H-15N HSQC spectrum. This observation suggests that switch I and switch II are capable of adopting several (two or more) different stable conformations, which are in slow interconversion, in contrast to Rheb-GDP. Interestingly, this difference has also been observed for Ras, and it has been suggested that this conformational flexibility might be functionally relevant for the GTP/GDP cycle, as well as for determining the binding selectivity of GAPs and GEFs (51, 54, 56, 70). Obviously, the interaction between Rheb and its GAP and/or GEF is also characterized by conformational selectivity.
The peculiar difference between the switch II conformations in Rheb and Ras is probably related to the different amino acid sequences. The Phe-70/Pro-71/Gln-72 motif in Rheb and R15G and S16G in its P-loop are not present in Ras (Fig. 4A). The conformational space of switch II in Rheb is restricted because of the limitations of the φ angle for Pro-71. Therefore, the different amino acid composition of the P-loop might be linked structurally to the conformation of the switch II region of Rheb and could suggest a different mechanism of (intrinsic) hydrolysis.
Nucleotide-dependent Low Affinity Interaction between Rheb and c-Raf-RBD
The observed low affinity between Rheb and c-Raf-RBD is most likely caused by two important changes in the primary structure of Rheb. Unlike Ras, Rheb carries an Asn instead of an Asp at position 41 and a Thr instead of an Arg at position 44. Both residues play an important role in the Ras/Raf interaction, contributing to affinity and determining the selectivity of the complex through the complementary surface charges of Ras and c-Raf kinase (71, 72). This interaction obviously cannot be achieved in the Rheb-Raf complex, thus providing a molecular explanation for the observed low affinity, which is based mainly on the interaction between hydrophobic residues located at the interface. Despite the very moderate affinity between activated Rheb-Gpp(NH)p and c-Raf kinase, Rheb does not bind unselectively to any RBD; for example, we could not detect NORE1-RBD binding to Rheb-Gpp(NH)p under the experimental conditions used for the titration of Rheb-Gpp(NH)p with c-Raf kinase RBD. Thus, we exclude the possibility here that Rheb-GTP-enhanced apoptosis is mediated by binding to NORE1, a known apoptosis effector for Ras (73).
The Mechanism Underlying Rheb-enhanced Apoptosis
Our data are compatible with recent reports suggesting that stimulation of the mTOR pathway in TSC-deficient neuronal or non-neuronal cells increases the ER stress response program, thereby lowering the threshold for apoptosis (25, 63, 74). However, using the ER stress-protective agent salubrinal, we were unable to suppress Rheb-enhanced apoptosis, suggesting that ER stress is an inducer but not a mediator of the pro-apoptotic function of Rheb.
Consistent with the low affinity between c-Raf and Rheb-Gpp(NH)p, this interaction is not associated with a regulation of c-Raf kinase after the overexpression of Rheb in HeLa cells. Furthermore, a c-Raf-mediated anti-apoptotic effect occurs in the absence of the N-terminal c-Raf domain that encompasses Raf-RBD. We cannot determine whether the previously described down-regulation of c-Raf activity by Rheb in HEK cells is due to cell type-specific signaling or different culturing conditions (26).
While this manuscript was under review, Ma et al. (75) reported that Rheb may exert a protective effect against certain apoptotic stimuli such as etoposide, and this finding was confirmed by us (data not shown). Thus, the cellular readout of Rheb signaling may rely on the specific type of apoptotic stimulus used.
ASK-1 Knockdown Prevents Rheb-enhanced Apoptosis
Similar to Rheb, ASK-1 overexpression may promote differentiation, but depending on the cellular context it can also induce apoptosis and plays a major role in stress responses (60, 76, 77). Accordingly, we found that knocking down ASK-1 not only strongly reduced UV or TNFα-induced apoptosis, but the specific enhancement by Rheb overexpression was abolished, similar to rapamycin treatment (Fig. 8). We could not decide whether the remaining 10–15% of apoptosis was due to an inefficient knockdown or an additional unknown pro-apoptotic signaling pathway.
Implications in Pathology and the Normal Organism
In the normal organism, some toxic stress conditions are likely to convert Rheb to a death effector protein. We hypothesized that Rheb is a death effector in the following physiological or pathophysiological situations: (i) during brain ischemia under excitotoxic glutamate conditions, causing a massive calcium overload and neuronal cell death (78); (ii) during ER stress produced by a toxic accumulation of proteins (79); (iii) during the growth of human malignant glioma cells, in which rapamycin protects from hypoxia-induced cell death (80); (iv) in a mouse model of Alzheimer disease, in which rapamycin abolishes cognitive deficits and reduces amyloid-β levels (81); and (v) during normal aging because feeding mice rapamycin late in life extends their life span (82). In these physiological or pathophysiological situations, the mTOR pathway could switch from a growth-controlling pathway to one mediating apoptosis.
We concluded that the application of cell stress stimuli drives Rheb in rapamycin and ASK-1-sensitive pro-apoptotic functions, underscoring the awareness that drugs targeting mTOR may have opposite effects depending on the presence or absence of specific types of cellular stress. This knowledge may have therapeutic implications for the application of rapamycin in clinical settings.
Supplementary Material
Acknowledgments
We are grateful to Sabine Laerbusch, Gregor Barchan, Martin Gartmann, and Hans-Jochen Hauswald for expert technical help. The 750-MHz spectra were recorded at the SON NMR Large Scale Facility in Utrecht, which is funded by European Union Project “EU-NMR-European Network of Research Infrastructures for Providing Access & Technological Advancement in Bio-NMR” (FP-2005-RII3, Contract 026145). We are grateful also to Prof. Dr. von Kiedrowski for providing generous access to the DRX600 spectrometer.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 642, A6.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
The atomic coordinates (code 2L0X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Rheb
- Ras homolog enriched in brain
- mTOR
- mammalian target of rapamycin
- TSC
- tuberous sclerosis complex
- GAP
- GTPase-activating protein
- RBD
- Ras-binding domain
- ER
- endoplasmic reticulum
- GEF
- guanine nucleotide exchange factor
- Gpp(NH)p
- guanosine 5′-(β,γ-imido)triphosphatey
- ANOVA
- analysis of variance
- HSQC
- heteronuclear single-quantum coherence
- ASK
- apoptosis signal-regulating kinase 1.
REFERENCES
- 1.Reuther G. W., Der C. J. (2000) Curr. Opin. Cell Biol. 12, 157–165 [DOI] [PubMed] [Google Scholar]
- 2.Aspuria P. J., Tamanoi F. (2004) Cell. Signal. 16, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 3.Manning B. D., Cantley L. C. (2003) Trends Biochem. Sci. 28, 573–576 [DOI] [PubMed] [Google Scholar]
- 4.Park K. K., Liu K., Hu Y., Smith P. D., Wang C., Cai B., Xu B., Connolly L., Kramvis I., Sahin M., He Z. (2008) Science 322, 963–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. (2003) Mol. Cell 11, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 6.Hall M. N. (2008) Transplant Proc. 40, S5-S8 [DOI] [PubMed] [Google Scholar]
- 7.Sato T., Nakashima A., Guo L., Tamanoi F. (2009) J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell. Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 9.Li Y., Corradetti M. N., Inoki K., Guan K. L. (2004) Trends Biochem. Sci. 29, 32–38 [DOI] [PubMed] [Google Scholar]
- 10.Inoki K., Li Y., Xu T., Guan K. L. (2003) Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagata K., Sanders L. K., Kaufmann W. E., Yee W., Barnes C. A., Nathans D., Worley P. F. (1994) J. Biol. Chem. 269, 16333–16339 [PubMed] [Google Scholar]
- 12.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 13.Raaijmakers J. H., Bos J. L. (2009) J. Biol. Chem. 284, 10995–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borasio G. D., John J., Wittinghofer A., Barde Y. A., Sendtner M., Heumann R. (1989) Neuron 2, 1087–1096 [DOI] [PubMed] [Google Scholar]
- 15.Borasio G. D., Barde Y. A., Sendtner M., Heumann R., John J., Wittinghofer A. (1989) Biol. Chem. Hoppe-Seyler 370, 615 [Google Scholar]
- 16.Bonni A., Brunet A., West A. E., Datta S. R., Takasu M. A., Greenberg M. E. (1999) Science 286, 1358–1362 [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarty K., Serchov T., Mann S. A., Dietzel I. D., Heumann R. (2007) Eur. J. Neurosci. 25, 1971–1981 [DOI] [PubMed] [Google Scholar]
- 18.Heumann R., Goemans C., Bartsch D., Lingenhöhl K., Waldmeier P. C., Hengerer B., Allegrini P. R., Schellander K., Wagner E. F., Arendt T., Kamdem R. H., Obst-Pernberg K., Narz F., Wahle P., Berns H. (2000) J. Cell Biol. 151, 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen H. H., Briem T., Dzietko M., Sifringer M., Voss A., Rzeski W., Zdzisinska B., Thor F., Heumann R., Stepulak A., Bittigau P., Ikonomidou C. (2004) Neurobiol. Dis. 16, 440–453 [DOI] [PubMed] [Google Scholar]
- 20.Teachey D. T., Obzut D. A., Cooperman J., Fang J., Carroll M., Choi J. K., Houghton P. J., Brown V. I., Grupp S. A. (2006) Blood 107, 1149–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn M., Li W., Yu C., Rahmani M., Dent P., Grant S. (2005) Mol. Cancer Ther. 4, 457–470 [DOI] [PubMed] [Google Scholar]
- 22.Malagelada C., Jin Z. H., Greene L. A. (2008) J. Neurosci. 28, 14363–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kita K., Wu Y. P., Sugaya S., Moriya T., Nomura J., Takahashi S., Yamamori H., Nakajima N., Suzuki N. (2000) Biochem. Biophys. Res. Commun. 274, 859–864 [DOI] [PubMed] [Google Scholar]
- 24.Shah O. J., Hunter T. (2006) Mol. Cell. Biol. 26, 6425–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozcan U., Ozcan L., Yilmaz E., Düvel K., Sahin M., Manning B. D., Hotamisligil G. S. (2008) Mol. Cell 29, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karbowniczek M., Robertson G. P., Henske E. P. (2006) J. Biol. Chem. 281, 25447–25456 [DOI] [PubMed] [Google Scholar]
- 27.Clark G. J., Kinch M. S., Rogers-Graham K., Sebti S. M., Hamilton A. D., Der C. J. (1997) J. Biol. Chem. 272, 10608–10615 [DOI] [PubMed] [Google Scholar]
- 28.Im E., von Lintig F. C., Chen J., Zhuang S., Qui W., Chowdhury S., Worley P. F., Boss G. R., Pilz R. B. (2002) Oncogene 21, 6356–6365 [DOI] [PubMed] [Google Scholar]
- 29.Berghaus C., Schwarten M., Heumann R., Stoll R. (2007) Biomol. NMR Assign. 1, 45–47 [DOI] [PubMed] [Google Scholar]
- 30.Schwarten M., Berghaus C., Heumann R., Stoll R. (2007) Biomol. NMR Assign. 1, 105–108 [DOI] [PubMed] [Google Scholar]
- 31.Goemans C. G., Boya P., Skirrow C. J., Tolkovsky A. M. (2008) Cell Death Differ. 15, 545–554 [DOI] [PubMed] [Google Scholar]
- 32.Lessmann V., Heumann R. (1997) Brain Res. 763, 111–122 [DOI] [PubMed] [Google Scholar]
- 33.Haubensak W., Narz F., Heumann R., Lessmann V. (1998) J. Cell Sci. 111, 1483–1493 [DOI] [PubMed] [Google Scholar]
- 34.Herrmann C., Martin G. A., Wittinghofer A. (1995) J. Biol. Chem. 270, 2901–2905 [DOI] [PubMed] [Google Scholar]
- 35.Wohlgemuth S., Kiel C., Krämer A., Serrano L., Wittinghofer F., Herrmann C. (2005) J. Mol. Biol. 348, 741–758 [DOI] [PubMed] [Google Scholar]
- 36.Rösch P., Wittinghofer A., Tucker J., Sczakiel G., Leberman R., Schlichting I. (1986) Biochem. Biophys. Res. Commun. 135, 549–555 [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 38.Johnson R. D., Bluemler P., Rafey R., Brodbeck D. (1994) Abstr. Pap. Am. Chem. Soc. 207, 138 [Google Scholar]
- 39.Stoll R., Renner C., Buettner R., Voelter W., Bosserhoff A. K., Holak T. A. (2003) Protein Sci. 12, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoll R., Renner C., Ambrosius D., Golob M., Voelter W., Buettner R., Bosserhoff A. K., Holak T. A. (2000) J. Biomol. NMR 17, 87–88 [DOI] [PubMed] [Google Scholar]
- 41.Stoll R., Lee B. M., Debler E. W., Laity J. H., Wilson I. A., Dyson H. J., Wright P. E. (2007) J. Mol. Biol. 372, 1227–1245 [DOI] [PubMed] [Google Scholar]
- 42.Rehmann H., Brüning M., Berghaus C., Schwarten M., Köhler K., Stocker H., Stoll R., Zwartkruis F. J., Wittinghofer A. (2008) FEBS Lett. 582, 3005–3010 [DOI] [PubMed] [Google Scholar]
- 43.Nowaczyk M., Berghaus C., Stoll R., Rögner M. (2004) Phys. Chem. Chem. Phys. 6, 4878–4881 [Google Scholar]
- 44.Schwieters C. D., Kuszewski J. J., Clore G. M. (2006) Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 46.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 47.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 48.Buerger C., DeVries B., Stambolic V. (2006) Biochem. Biophys. Res. Commun. 344, 869–880 [DOI] [PubMed] [Google Scholar]
- 49.Heath-Engel H. M., Chang N. C., Shore G. C. (2008) Oncogene 27, 6419–6433 [DOI] [PubMed] [Google Scholar]
- 50.Breckenridge D. G., Germain M., Mathai J. P., Nguyen M., Shore G. C. (2003) Oncogene 22, 8608–8618 [DOI] [PubMed] [Google Scholar]
- 51.Vetter I. R., Wittinghofer A. (2001) Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 52.Herrmann C. (2003) Curr. Opin. Struct. Biol. 13, 122–129 [DOI] [PubMed] [Google Scholar]
- 53.Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 54.Kraulis P. J., Domaille P. J., Campbell-Burk S. L., Van Aken T., Laue E. D. (1994) Biochemistry 33, 3515–3531 [DOI] [PubMed] [Google Scholar]
- 55.Yu Y., Li S., Xu X., Li Y., Guan K., Arnold E., Ding J. (2005) J. Biol. Chem. 280, 17093–17100 [DOI] [PubMed] [Google Scholar]
- 56.Ito Y., Yamasaki K., Iwahara J., Terada T., Kamiya A., Shirouzu M., Muto Y., Kawai G., Yokoyama S., Laue E. D., Wälchli M., Shibata T., Nishimura S., Miyazawa T. (1997) Biochemistry 36, 9109–9119 [DOI] [PubMed] [Google Scholar]
- 57.Kiel C., Filchtinski D., Spoerner M., Schreiber G., Kalbitzer H. R., Herrmann C. (2009) J. Biol. Chem. 284, 31893–31902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harjes E., Harjes S., Wohlgemuth S., Müller K. H., Krieger E., Herrmann C., Bayer P. (2006) Structure 14, 881–888 [DOI] [PubMed] [Google Scholar]
- 59.Le Mellay V., Troppmair J., Benz R., Rapp U. R. (2002) BMC Cell Biol. 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsukawa J., Matsuzawa A., Takeda K., Ichijo H. (2004) J. Biochem. 136, 261–265 [DOI] [PubMed] [Google Scholar]
- 61.Hayakawa T., Matsuzawa A., Noguchi T., Takeda K., Ichijo H. (2006) Microbes Infect. 8, 1098–1107 [DOI] [PubMed] [Google Scholar]
- 62.Semenova M. M., Mäki-Hokkonen A. M., Cao J., Komarovski V., Forsberg K. M., Koistinaho M., Coffey E. T., Courtney M. J. (2007) Nat. Neurosci. 10, 436–443 [DOI] [PubMed] [Google Scholar]
- 63.Di Nardo A., Kramvis I., Cho N., Sadowski A., Meikle L., Kwiatkowski D. J., Sahin M. (2009) J. Neurosci. 29, 5926–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 65.Tavazoie S. F., Alvarez V. A., Ridenour D. A., Kwiatkowski D. J., Sabatini B. L. (2005) Nat. Neurosci. 8, 1727–1734 [DOI] [PubMed] [Google Scholar]
- 66.Shah O. J., Wang Z., Hunter T. (2004) Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 67.Li Y., Wang Y., Kim E., Beemiller P., Wang C. Y., Swanson J., You M., Guan K. L. (2007) J. Biol. Chem. 282, 35803–35813 [DOI] [PubMed] [Google Scholar]
- 68.Li Y. H., Werner H., Püschel A. W. (2008) J. Biol. Chem. 283, 33784–33792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wittinghofer A., Nassar N. (1996) Trends Biochem. Sci. 21, 488–491 [DOI] [PubMed] [Google Scholar]
- 70.Spoerner M., Herrmann C., Vetter I. R., Kalbitzer H. R., Wittinghofer A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98, 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nassar M., Horn G., Herrmann C., Scherer A., McCormick F., Wittinghofer A. (1995) Nature 375, 554–560 [DOI] [PubMed] [Google Scholar]
- 72.Nassar N., Horn G., Herrmann C., Block C., Janknecht R., Wittinghofer A. (1996) Nat. Struct. Biol. 3, 723–729 [DOI] [PubMed] [Google Scholar]
- 73.Stieglitz B., Bee C., Schwarz D., Yildiz O., Moshnikova A., Khokhlatchev A., Herrmann C. (2008) EMBO J. 27, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghosh S., Tergaonkar V., Rothlin C. V., Correa R. G., Bottero V., Bist P., Verma I. M., Hunter T. (2006) Cancer Cell 10, 215–226 [DOI] [PubMed] [Google Scholar]
- 75.Ma D., Bai X., Zou H., Lai Y., Jiang Y. (2010) J. Biol. Chem. 285, 8621–8627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takeda K., Matsuzawa A., Nishitoh H., Ichijo H. (2003) Cell Struct. Funct. 28, 23–29 [DOI] [PubMed] [Google Scholar]
- 77.Bunkoczi G., Salah E., Filippakopoulos P., Fedorov O., Müller S., Sobott F., Parker S. A., Zhang H., Min W., Turk B. E., Knapp S. (2007) Structure 15, 1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bano D., Nicotera P. (2007) Stroke 38, 674–676 [DOI] [PubMed] [Google Scholar]
- 79.Scheper W., Hoozemans J. J. (2009) Curr. Med. Chem. 16, 615–626 [DOI] [PubMed] [Google Scholar]
- 80.Ronellenfitsch M. W., Brucker D. P., Burger M. C., Wolking S., Tritschler F., Rieger J., Wick W., Weller M., Steinbach J. P. (2009) Brain 132, 1509–1522 [DOI] [PubMed] [Google Scholar]
- 81.Spilman P., Podlutskaya N., Hart M. J., Debnath J., Gorostiza O., Bredesen D., Richardson A., Strong R., Galvan V. (2010) PLoS One 5, e9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harrison D. E., Strong R., Sharp Z. D., Nelson J. F., Astle C. M., Flurkey K., Nadon N. L., Wilkinson J. E., Frenkel K., Carter C. S., Pahor M., Javors M. A., Fernandez E., Miller R. A. (2009) Nature 460, 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delano W. L., Lam J. W. (2005) Abstr. Pap. Am. Chem. Soc. 230, U1371–U1372(abstr.) [Google Scholar]
- 84.Delano W. L. (2004) Abstr. Pap. Am. Chem. Soc. 228, U313–U314(abstr.) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.