Abstract
Protein–protein interaction plays a major role in all biological processes. The currently available genetic methods such as the two-hybrid system and the protein recruitment system are relatively limited in their ability to identify interactions with integral membrane proteins. Here we describe the development of a reverse Ras recruitment system (reverse RRS), in which the bait used encodes a membrane protein. The bait is expressed in its natural environment, the membrane, whereas the protein partner (the prey) is fused to a cytoplasmic Ras mutant. Protein–protein interaction between the proteins encoded by the prey and the bait results in Ras membrane translocation and activation of a viability pathway in yeast. We devised the expression of the bait and prey proteins under the control of dual distinct inducible promoters, thus enabling a rapid selection of transformants in which growth is attributed solely to specific protein–protein interaction. The reverse RRS approach greatly extends the usefulness of the protein recruitment systems and the use of integral membrane proteins as baits. The system serves as an attractive approach to explore novel protein–protein interactions with high specificity and selectivity, where other methods fail.
INTRODUCTION
Protein–protein interaction plays a major role in all biological processes (1). Proteins act in large modular structures, also referred to as protein machines, to provide high specificity and selectivity (2). Following the completion of the genome projects of lower organisms with relatively small genome sizes and the rapid advance in the human genome project it is becoming evident that many putative proteins encoded by novel genes have no known function or sequence homology with proteins in the databank (3). For example, the budding yeast Saccharomyces cerevisiae genome project was completed in 1996 (4). However, one-third of the predicted open reading frames are still classified as proteins with unknown function (5). In a genome-wide approach to identify all possible protein–protein interactions of the yeast genome using the two-hybrid system, it is clearly evident that, of those protein pairs known to participate in protein–protein interaction (8%), proteins of membrane origin are under-represented (5). One possible explanation for this might be the fact that expression of these proteins in the yeast nucleus renders these proteins non-functional due to inappropriate folding (6,7). An alternative approach to study protein–protein interaction in the yeast cytoplasm is the recently developed Ras recruitment system (RRS; 8). This system is based on the translocation of a cytoplasmic Ras to the plasma membrane via protein–protein interaction. Ras membrane recruitment results in activation of a viability pathway in yeast. Although very powerful, RRS is limited in its ability to use membrane proteins as bait (8). This is due to the fact that fusion of a membrane protein to Ras will result in its membrane translocation independent of protein–protein interaction.
Here we describe a reverse RRS approach, specifically designed for the use of membrane proteins as bait. This system allows the identification of protein–protein interaction following library screening. Moreover, it greatly extends the usefulness of the protein recruitment systems for the analysis of nuclear, cytoplasmic and membrane proteins and may serve to explore new territories where other methods have failed.
MATERIALS AND METHODS
Plasmids
The rat pituitary cDNA library expression plasmid was constructed as follows. The rat pituitary cDNA was excised from the previously described rat pituitary library (9) by EcoRI–XhoI digestion and religation into pYes2 (Invitrogen) derived expression vector encoding for the oncogenic cytoplasmic Ras protein devoid of its membrane localization signal and stop codon. The library represents over 4 × 106 independent cloning events.
The methionine expression plasmid, p425-Met25, was previously described (10). The different Chp constructs were subcloned into p425-Met25 by HindIII–XhoI.
ChpAc.ΔN cDNA fragment was generated by PCR with appropriate oligonucleotides designed to contain EcoRI–XhoI sites (5′–3′, respectively) encoding for ChpAc starting at amino acid 33 (KCVLVGDG). The cDNA was fused initially to an myc-epitope tag and subsequently transferred to pMet expression plasmid by HindIII–XhoI digestion. The pMet–M–ChpAc encodes for full-length Chp fused to v-Src myristoylation signal. The Chp cDNA is inserted via EcoRI–XhoI into the YesMΔPolyA plasmid described by Aronheim et al. (11).
Yeast manipulations
All yeast transfection and manipulations were carried out by standard procedures as described previously (8,9). Plasmid DNA was recovered from candidates as described previously (12). In order to efficiently repress the methionine (Met) promoter, we used 0.66 mM methionine in the medium (10).
Cell extract and western blot analysis
Culture (3 ml) was grown overnight at 24°C in different media. Cells were recovered by centrifugation and resuspended in 0.5 ml of H2O. Alkali lysis was performed at 4°C by addition of 85 µl of 1.85 M NaOH/7.4% β-mercaptoethanol for 10 min. Protein was recovered by the addition of 40 µl of 100% trichloroacetic acid followed by a 10 min incubation and a 10 min centrifugation. The precipitated extract was washed once with ice-cold acetone and air-dried. The pellet was recovered in SDS–protein sample buffer, loaded onto a 12.5% SDS–polyacrylamide gel and immobilized to a nitrocellulose filter. The filter was incubated in 10% low-fat milk in PBS and probed with anti-Myc monoclonal antibodies (9E10; Babco) followed by anti-mouse horseradish peroxidase (Transduction laboratories). Following extensive washing in PBS, a chemiluminescent reaction (Super signal; Pierce) was performed and the filter was exposed to autoradiography.
RESULTS
The reverse RRS approach
A method directed towards identification of protein–protein interactions involving membrane proteins was developed. The method, designated reverse RRS, is based on the fact that Ras localization to the plasma membrane is crucial for its function (8,13). Similar to RRS, expression of cytoplasmic Ras in yeast does not complement mutation in the Ras guanyl nucleotide exchange factor, CDC25-2 (14). However, membrane-bound mammalian Ras can efficiently complement CDC25-2 mutation (9,15). Ras membrane translocation can be achieved via protein–protein interaction (8) which can be readily monitored in a Cdc25-2 yeast strain by contributing to cell growth at the restrictive temperature (8).
A schematic diagram describing the basis of the method is shown in Figure 1. In principle, the expression of a cDNA encoding for a membrane protein with no additional sequences is expected to locate the protein to its natural environment, i.e. the membrane (Fig. 1A). In order to identify protein partners with a membrane protein of interest (‘the bait’), a cDNA encoding for a known protein partner for the bait (‘the prey’) or a cDNA library are fused to cytoplasmic Ras (Fig. 1B). The expression of either the bait or the prey alone is not expected to complement Cdc25-2 mutation. However, upon protein–protein interaction between the membrane bait and the prey protein (Ras-cDNA), Ras is expected to be localized to the plasma membrane resulting in efficient growth of Cdc25-2 cells at the restrictive temperature (Fig. 1C).
Figure 1.
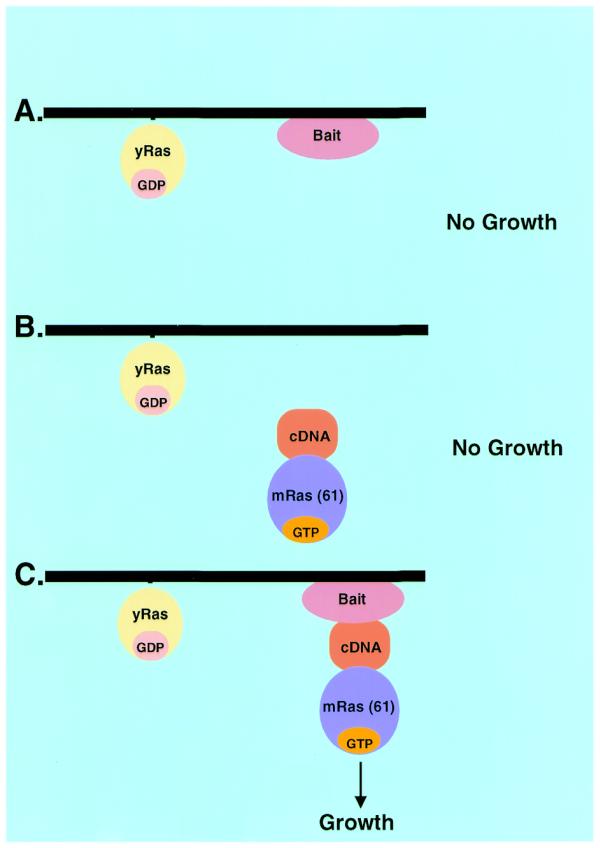
Schematic diagram describing the reverse RRS approach. The membrane protein of interest is expressed in a Cdc25-2 yeast strain with no additional sequences. The bait is located at the membrane thus preserving its functional conformation. (A) The bait protein is not expected to induce cell growth of Cdc25-2 yeast cells at the restrictive temperature. (B) A known protein partner for the bait or a cDNA library is expressed as a membrane fusion with the cytoplasmic oncogenic Ras protein. Unless the cDNA encodes for a membrane protein or is associated with a membrane component it is not expected to result in Ras translocation to the membrane and cell growth at the restrictive temperature. (C) Upon protein–protein interaction between the membrane bait and the protein prey fused to Ras, Ras is translocated to the plasma membrane which allows cell growth at the restrictive temperature.
Development of double inducible promoters system
In order to be able to screen for novel proteins binding to the membrane bait, a cDNA expression library has to be constructed and fused to Ras (Fig. 1B). To do this, a rat pituitary cDNA library was constructed. The cDNA was inserted at the 3′-end of the cDNA encoding for oncogenic Ras mutant devoid of both its C-terminal CAAX box and termination codon. The expression of the library was designed under the control of the Gal1 promoter. As expected, ∼5% of the yeast transformants expressing the Ras-cDNA library exhibited efficient cell growth at the restrictive temperature, when grown under galactose-containing medium, independent of the expression of a specific protein bait (data not shown). Yeast cell growth is probably due to cDNAs encoding for membrane proteins or proteins that associate with yeast membrane components. This relatively high background activity required the development of an approach enabling efficient selection for yeast transformants that exhibit growth solely dependent on protein–protein interaction. To this end, we designed the expression of the membrane bait protein under the control of a distinct inducible promoter. The bait protein expression was designed under the control of the methionine promoter (10). This promoter is induced when cells are grown in a medium lacking methionine. In addition, the prey and bait expression plasmids provide the ability to complement the uracil and leucine auxotrophy, respectively. In order to be able to select for an event where the expression of the prey and the bait together are necessary for cell growth, it is absolutely necessary to be able to tightly control the expression of the bait plasmid independent of the expression of the prey expression plasmid. Therefore, we first tested the ability to regulate the level of expression of the Gal1 and Met promoters. We constructed plasmids encoding for an myc-epitope-tagged Ras protein designed under the control of alcohol dehydrogenase (ADH), Gal1 and Met promoters. The plasmids were used to transform Cdc25-2 yeast cells. Transformants were selected and grown in three different liquid media containing either glucose or galactose in the presence and absence of methionine. The extract derived from these transformants was subjected to SDS–PAGE and western blot analysis using anti-Myc antibodies (Fig. 2A). This analysis revealed that the expression level of the myc–Ras protein under the Gal1 and Met promoters are relatively similar when the cells are grown in galactose medium in the absence of methionine. However, the addition of Met to the medium completely abolished Ras expression derived from the methionine promoter. The Gal1 promoter was completely repressed when the cells were grown on glucose medium. This analysis strongly suggests that the expression of proteins under the control of the Gal1 and Met promoters could be tightly regulated. Once induced, the expression levels were comparable to the expression obtained from the constitutive ADH. These results prompted us to test whether these two inducible/repressible promoters could facilitate the selection for protein–protein interaction using the reverse RRS approach.
Figure 2.
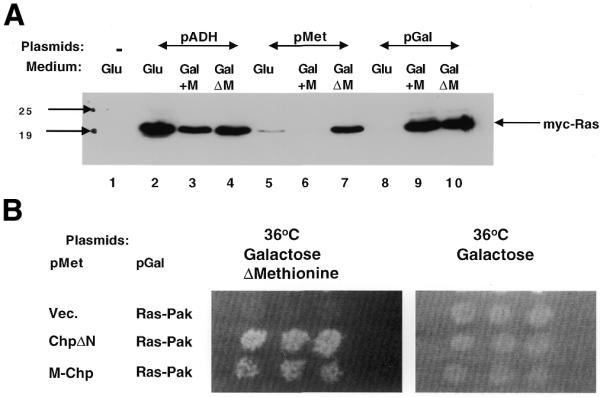
The regulation of the Gal1 and Met promoters. (A) Western blot analysis with anti-Myc antibodies. The regulation of the ADH (lanes 1–4), Met (lanes 5–7) and Gal1 (lanes 8–10) promoters was examined in three distinct media. The expression of myc-epitope Ras protein (myc–Ras; lanes 2–10) or Ras (lane 1) was detected with anti-Myc antibodies. Whole cell extract was derived from yeast transformants expressing the myc–Ras protein grown on either glucose (Glu)- or galactose (Gal)-containing medium in the presence or absence of methionine (GalΔM). The migration of myc–Ras and protein size markers are indicated. (B) Methionine-dependent growth of transformants expressing protein interacting pairs. Cdc25-2 transformants expressing the indicated protein pairs under the indicated promoter were grown on glucose medium at 24°C. The plate was replica plated onto galactose-containing plates in which methionine was either included or excluded from the medium. The plates were incubated at 36°C. Transformants expressing protein pairs under the control of Met and Gal1 promoters, respectively, are able to grow only on plates lacking methionine.
To test the feasibility of this approach we first tested the interaction between known interacting protein pairs designed under the control of Gal1 and Met promoters. We used the novel small GTPase, recently isolated in our laboratory, designated Chp (11), fused to v-Src myristoylation signal (M–Chp) and its protein partner Pak65 fused to Ras (Ras–Pak) designed under the control of Met and Gal1 promoters, respectively. Transformants expressing both proteins exhibited efficient growth only when grown on galactose-containing medium in the absence of methionine (Fig. 2B). Similar results were obtained when Ras–Pak and M–Chp were expressed under the control of Met and Gal1 promoters, respectively (data not shown). The ability to detect interaction between Ras–Pak and Chp using the RRS in yeast is dependent on membrane localization of Chp through the v-Src myristoylation signals (not shown). This is due to the fact that Chp, unlike all Rho-GTPases, does not contain the typical C-terminal consensus CAAX box (16). Surprisingly, we found that transformants expressing the Chp protein lacking the N-terminal domain (ChpAc.ΔN) and the Ras–Pak protein, displayed efficient growth at the restrictive temperature (Fig. 2B). This interaction no longer required the v-Src myristoylation signals, suggesting that, upon truncation of the N-terminal of Chp, the protein translocates to the plasma membrane.
Screening for ChpAc.ΔN interacting proteins
The fact that ChpΔN was able to rescue Cdc25-2 cells when expressed with Ras–Pak and the observation that both the Met and Gal1 promoters are tightly regulated, prompted us to use ChpAc.ΔN as a model system for membrane bait to screen a rat pituitary cDNA expression library fused to Ras, using the reverse RRS approach. A flow chart for the screen is depicted in Figure 3. In principle, about 200 000 transformants (5% of the complexity of the library) were plated on glucose plates containing the appropriate growth selection compounds and subsequently replica plated on to galactose-containing plates either in the absence or presence of methionine. Thirty yeast colonies that exhibited efficient growth on the galactose plate lacking methionine (as compared to the growth obtained on the plate containing methionine) were selected and exposed to a secondary growth-dependent test on galactose-containing plates in the presence or absence of methionine. Two out of the 30 colonies selected exhibited reproducible growth on galactose medium in the absence of methionine but not on galactose medium containing methionine (Fig. 4). Plasmid DNA extracted from these clones was reintroduced with the specific bait (ChpAc.ΔN), Met empty expression plasmid and myristoylated full-length ChpAc. (M–ChpAc.) (Fig. 5). Both clones #1 and #2 exhibit efficient growth in the presence of the original ChpAc.ΔN bait. Clone #1 shows no interaction with the full-length Chp in the activated form but interacts with the myristoylated dominant negative full-length Chp (data not shown). Clone #2 interacts weakly with myristoylated activated full-length Chp (Fig. 5) but very efficiently with the dominant negative Chp (data not shown). Collectively, these results indicate that the growth of the transformants isolated was dependent on the expression of both the bait- and the prey-encoding proteins.
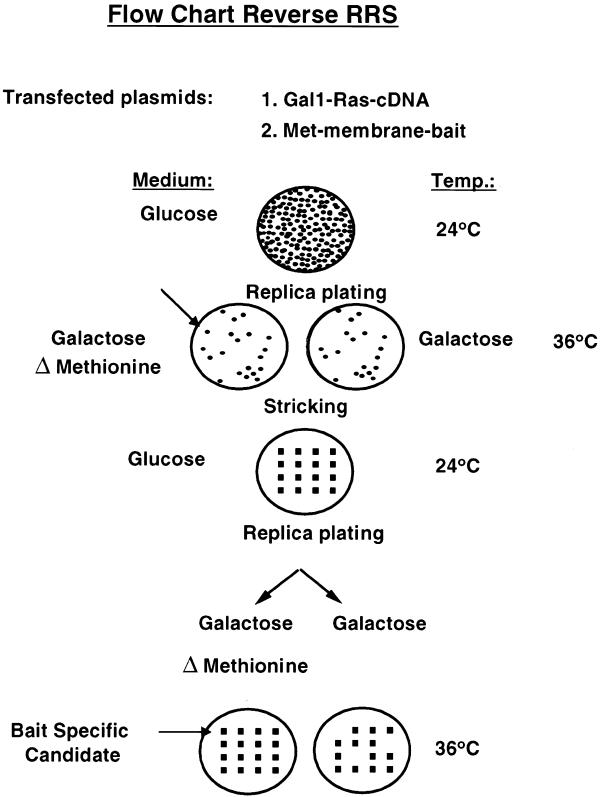
Figure 3.
Flow chart for the reverse RRS. Cdc25-2 cells are cotransfected with the membrane bait and a cDNA library fused to Ras. The expression of the bait and Ras prey proteins are designed under the control of the Met and Gal1 promoters, respectively. Transformants are selected on glucose minimal plates lacking leucine and uracil at 24°C. After 5–7 days, plates are replica plated onto galactose-containing plates either containing or lacking methionine and incubated at 36°C. Transformants that exhibit efficient growth on the plates lacking methionine and no growth on the plate containing methionine are selected on a glucose plate and grown at 24°C. The selected transformants are retested for their methionine-dependent growth. Those transformants that pass this secondary methionine-dependent test are considered candidates and are further pursued. DNA is extracted from candidates and the library plasmid is identified by a restriction digest. The DNA is used to retransform Cdc25-2 cells with either the specific bait or non-specific bait.
Figure 4.
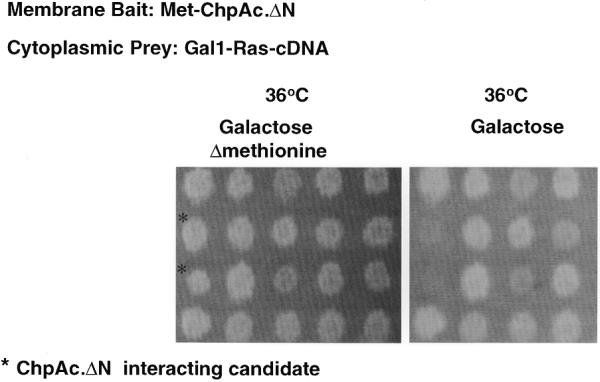
Secondary methionine dependency test for the ChpAc.ΔN interacting candidates. Cdc25-2 cells were cotransfected with Met expression plasmid encoding for myc–ChpAc.ΔN protein bait and Yes expression plasmid encoding for oncogenic cytoplasmic Ras protein fused to rat pituitary cDNA library. 200 000 transformants were plated on 20 glucose plates lacking leucine and uracil and incubated at 24°C. Following the first replica plating ∼5% of the transformants exhibited efficient growth on galactose-containing medium at the restrictive temperature. Only 30 clones showed preferential growth on the galactose-containing plate lacking methionine. Subsequently, those transformants (only 20 are shown) were subjected to a secondary methionine test which resulted in the identification of two clones (indicated by *) that did not grow on the galactose plate containing methionine (right panel), but grew efficiently in the absence of methionine (left panel).
Figure 5.
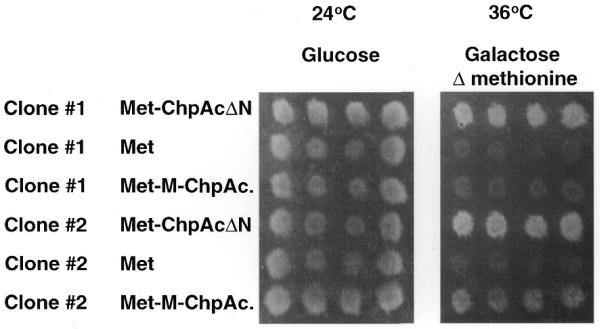
Specificity test for the library plasmids extracted from candidate clones #1 and #2. DNA plasmids isolated from candidate clones #1 and #2 were subjected to restriction digest and the library plasmid was identified. The plasmid was used to cotransfect Cdc25-2 cells with the Met expression vector encoding the original bait [i.e. ChpAc. deleted in its N-terminal domain (ChpAc.ΔN)], the Met expression vector (Met) and the Met expression vector encoding for full-length myristoylated Chp activated (Met–M–ChpAc.). Transformants were selected and used to replica plate onto appropriate plates containing galactose and lacking methionine incubated at 36°C.
DISCUSSION
In this paper we describe the development of a novel approach to the study of protein–protein interaction: the reverse RRS approach. This approach is specifically designed for membrane receptors, ion channels and transporters that span the membrane several times. Expression of these proteins in their natural membrane environment preserves their unique three-dimensional binding surfaces. This system is shown to efficiently identify novel protein–protein interactions, which exhibit specific interaction in yeast with the original membrane bait used. Using the ChpAc.ΔN as bait we have identified two distinct cDNAs encoding for proteins that exhibit specific interaction. Clone #1 encodes for placenta growth factor (PlGF accession number L40030). The direct link between a growth factor and Chp is not obviously explained; however, it is well known that in a genetic screen the expression of proteins in a compartment where they are usually excluded may result in the identification of false interaction. On the other hand, the sequence of clone #2 revealed the isolation of an uncharacterized protein (mouse EST accession number AA789582). Although the biological importance of these interactions is yet to be determined, the reverse RRS screening protocol was shown to efficiently identify plasmids encoding for proteins that reproduced the phenotypic growth in a bait-specific manner. Similarly, the first screening protocol for the two-hybrid system resulted in the isolation of proteins interacting with the leucine zipper of c-Jun with no obvious functional entity (17).
Although the two-hybrid system has been used to identify protein interaction with the cytoplasmic tail of different receptors such as the tumor necrosis factor (18,19) and the insulin receptor (20–22) these receptors contain a single transmembrane domain. Similarly, protein interaction with small GTPases which are membrane associated were also identified, e.g. those from the Rho-GTPases (23,24).
Several methods were developed for the identification of protein–protein interactions outside the nucleus (25,26). However, their applicability for integral membrane proteins and the screening feasibility are yet to be shown. In addition, a recent article describes the development of a novel approach to monitor integral membrane protein–protein interactions in yeast (27). The method is based on disruption of a G-protein signaling cascade monitored by inhibition of a β-galactosidase reporter-based assay. Although the interaction between known proteins is shown, a screening protocol has to be developed. One obvious problem that may arise is the ability to monitor a decrease in reporter activity and selection of positive clones while screening a library.
The use of membrane proteins as bait when expressed in their natural environment, the membrane, using the reverse RRS approach would provide an attractive method to explore novel proteins associating with multiple membrane targets. For example, the hetero-oligomerization between different members of G-protein-coupled receptors (GPCRs) was recently demonstrated for the neurotransmitters somatostatin and dopamine receptors (28). This interaction results in their enhanced functional activity. It will be interesting to explore the protein interaction with such oligomers in the presence of their corresponding ligands.
The existence of yeast homologs for mammalian membrane proteins may result in complication with the reverse RRS approach due to interaction of the encoded prey with the yeast homologs independent of the interaction with the expressed mammalian counterpart. However, the ease in manipulation of the yeast genome by gene knockout can overcome this problem. Moreover, the existence of yeast mutants in different pathways can be used in complementation assays, with the objective to test the functionality of the protein prior to screening. This may provide evidence that the membrane protein indeed fully preserves its activity and potentially its protein binding surfaces. In addition, GPCRs, for example, can be used to screen for protein binding either in the presence or absence of their specific ligands or agonists.
Moreover, in many instances where a prey protein is identified following a regular RRS library screen, it is often useful to screen for additional proteins interacting with the prey protein. However, based on the experience with both the two-hybrid system and the RRS, it is well known that protein–protein interactions initially identified in one orientation with respect to the bait and prey fusions do not necessarily function in the reverse orientation (5). Using reverse RRS one can simply use the prey protein as bait and perform screening with the reverse library using the reverse RRS approach, thus preserving the bait in its binding conformation as initially identified.
In summary, we have described a novel approach to the study of protein–protein interactions specifically designed for membrane proteins. The system can be used to identify interactions with known proteins as well as identification of novel proteins following a library screening approach. Ultimately, this system would enable exploring novel territories where other genetic systems have failed.
Acknowledgments
ACKNOWLEDGEMENTS
We wish thank to Ms Aviva Cohen for excellent technical assistance and Dr P. Frankel for critical reading of the manuscript. This research was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities–Charles H. Revson Foundation.
References
- 1.Mendelsohn A.R. and Brent,R. (1999) Protein interaction methods – toward an endgame. Science, 284, 1948–1950. [DOI] [PubMed] [Google Scholar]
- 2.Blackstock W.P. and Weir,M.P. (1999) Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol., 17, 121–127. [DOI] [PubMed] [Google Scholar]
- 3.Dunham I., Shimizu,N., Roe,B.A., Chissoe,S., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. [DOI] [PubMed] [Google Scholar]
- 4.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 563–567. [DOI] [PubMed] [Google Scholar]
- 5.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 6.Fields S. and Song,O.K. (1989) A novel genetic system to detect protein-protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 7.Colas P. and Brent,R. (1998) The impact of two-hybrid and related methods on biotechnology. Trends Biotechnol., 16, 355–363. [DOI] [PubMed] [Google Scholar]
- 8.Broder Y.C., Katz,S. and Aronheim,A. (1998) The Ras recruitment system, a novel approach to the study of protein-protein interactions. Curr. Biol., 8, 1121–1124. [DOI] [PubMed] [Google Scholar]
- 9.Aronheim A., Zandi,E., Hennemann,H., Elledge,S. and Karin,M. (1997) Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol., 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronheim A., Broder,Y.C., Cohen,A., Fritsch,A., Belisle,B. and Abo,A. (1998) Chp, a homologue of the GTPase Cdc42, activates the JNK pathway and is implicated in reorganizing the actin cytoskeleton. Curr. Biol., 8, 1125–1128. [DOI] [PubMed] [Google Scholar]
- 12.Robzyk K. and Kassir,Y. (1992) A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res., 20, 3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock J.F., Magee,A.I., Childs,J. and Marshall,C.J. (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell, 57, 1167–1177. [DOI] [PubMed] [Google Scholar]
- 14.Petitjean A., Higler,F. and Tatchell,K. (1990) Comparison of thermosensitive alleles of the CDC25 gene involved in the cAMP metabolism of Saccharomyces cerevisiae. Genetics, 124, 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronheim A. (1997) Improved efficiency Sos recruitment system: expression of the mammalian GAP reduces isolation of Ras GTPase false positives. Nucleic Acids Res., 25, 3373–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall C. (1996) Ras effectors. Curr. Opin. Cell Biol., 8, 197–204. [DOI] [PubMed] [Google Scholar]
- 17.Chevray P.M. and Nathans,D. (1992) Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl Acad. Sci. USA, 89, 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinnaiyan A.M., O’Rourke,K., Tewari,M. and Dixit,V.M. (1995) FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell, 81, 505–512. [DOI] [PubMed] [Google Scholar]
- 19.Wallach D., Boldin,M.P., Kovalenko,A.V., Malinin,N.L., Mett,I.L. and Camonis,J.H. (1998) The yeast two-hybrid screening technique and its use in the study of protein-protein interactions in apoptosis. Curr. Opin. Immunol., 10, 131–136. [DOI] [PubMed] [Google Scholar]
- 20.Furlanetto R.W., Dey,B.R., Lopaczynski,W. and Nissley,S.P. (1997) 14-3-3 proteins interact with the insulin-like growth factor receptor but not the insulin receptor. Biochem. J., 327, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey B.R., Furlanetto,R.W. and Nissley,S.P. (1998) Cloning of human p55 gamma, a regulatory subunit of phosphatidylinositol 3-kinase, by a yeast two-hybrid library screen with the insulin-like growth factor-I receptor. Gene, 209, 175–183. [DOI] [PubMed] [Google Scholar]
- 22.Xu P., Jacobs,A.R. and Taylor,S.I. (1999) Interaction of insulin receptor substrate 3 with insulin receptor, insulin receptor-related receptor, insulin-like growth factor-1 receptor, and downstream signaling proteins. J. Biol. Chem., 274, 15262–15270. [DOI] [PubMed] [Google Scholar]
- 23.Van Aelst L., Barr,M., Marcus,S., Polverino,A. and Wigler,M. (1993) Complex formation between Ras and Raf and other protein kinases. Proc. Natl Acad. Sci. USA, 90, 6213–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joberty G., Perlungher,R.R. and Macara,I.G. (1999) The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol. Cell. Biol., 19, 6585–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnsson N. and Varshavski,A. (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl Acad. Sci. USA, 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozawa T., Nogami,S., Sato,M., Ohya,Y. and Umezawa,Y. (2000) A fluorescent indicator for detecting protein-protein interactions in vivo based on protein splicing. Anal. Chem., 72, 5151–5157. [DOI] [PubMed] [Google Scholar]
- 27.Ehrhard K.N., Jacoby,J.J., Fu,X.-Y., Jahn,R. and Dohlman,H.G. (2000) Use of G-protein fusions to monitor integral membrane protein-protein interactions in yeast. Nat. Biotechnol., 18, 1075–1079. [DOI] [PubMed] [Google Scholar]
- 28.Rocheville M., Lange,D.C., Kumar,U., Patel,S.C., Patel,R.C. and Patel,Y.C. (2000) Receptors for dopamine and somatostatin: formation of hetero-oligomers with enhanced functional activity. Science, 288, 154–157. [DOI] [PubMed] [Google Scholar]