Abstract
To better understand influenza virus infection of pigs, we examined primary swine respiratory epithelial cells (SRECs, the primary target cells of influenza viruses in vivo), as a model system. Glycomic profiling of SRECs by mass spectrometry revealed a diverse range of glycans terminating in sialic acid or GalαGal. In terms of sialylation, α2–6 linkage was more abundant than α2–3, and NeuAc was more abundant than NeuGc. Virus binding and infection experiments were conducted to determine functionally important glycans for influenza virus infection, with a focus on recently emerged swine viruses. Infection of SRECs with swine and human viruses resulted in different infectivity levels. Glycan microarray analysis with a high infectivity “triple reassortant” virus ((A/Swine/MN/593/99 (H3N2)) that spread widely throughout the North American swine population and a lower infectivity human virus isolated from a single pig (A/Swine/ONT/00130/97 (H3N2)) showed that both viruses bound exclusively to glycans containing NeuAcα2–6, with strong binding to sialylated polylactosamine and sialylated N-glycans. Treatment with mannosamine precursors of sialic acid (to alter NeuAc/NeuGc abundances) and linkage-specific sialidases prior to infection indicated that the influenza viruses tested preferentially utilize NeuAcα2–6-sialylated glycans to infect SRECs. Our data indicate that NeuAcα2–6-terminated polylactosamine and sialylated N-glycans are important determinants for influenza viruses to infect SRECs. As NeuAcα2–6 polylactosamine glycans play major roles in human virus infection, the importance of these receptor components in virus infection of swine cells has implications for transmission of viruses between humans and pigs and for pigs as possible adaptation hosts of novel human influenza viruses.
Keywords: Glycoprotein, Glycosylation, Mass Spectrometry (MS), Receptor Structure-Function, Virus Entry, Influenza
Introduction
Influenza A viruses are significant pathogens of many species, including humans and pigs. Pigs have long been postulated as intermediate hosts in the generation of pandemic human influenza viruses because they are susceptible to human and avian viruses (1–6). Since the late 1990s, influenza viruses in North American pigs have evolved extensively, with the emergence of “triple reassortant” H3N2 (rH3N2) viruses that contain genes from human-, avian-, and swine-lineage viruses (7–9). Further reassortment with existing viruses led to the development of H1N1, H1N2, H3N1, and H2N3 reassortant viruses in North American pigs (10–13). Finally, at some point in past years, North American triple reassortant and Eurasian swine viruses underwent reassortment to create the human pandemic 2009 H1N1 influenza virus (14–16). These events document the natural occurrence of genetic reassortment among influenza viruses in pigs, demonstrate the importance of pigs in the epidemiology and evolution of influenza viruses for humans, and emphasize the need for a better understanding of influenza virus infection of pigs.
To determine what allowed the initial rH3N2 viruses to successfully emerge, spread, and be maintained in the swine population, we previously compared a representative rH3N2 virus (A/Swine/Minnesota/593/99 (Sw/MN)) with a human-lineage virus (A/Swine/Ontario/00130/97 (Sw/ONT)) isolated from a single pig during the same time period. Sw/MN was more infectious for pigs in vivo (17), and it infected a significantly higher number of cultured primary swine respiratory epithelial cells (SRECs,5 the target cells of influenza viruses in pigs) (18). Finally, using reverse genetics-generated Sw/MN × Sw/ONT reassortant and point mutant viruses, we demonstrated that the infectivity phenotypes in SRECs were strongly dependent upon three amino acids within the viral HA (hemagglutinin) gene (18).
Influenza viruses initiate infection via the virus HA protein binding to host cell sialic acids (Sia) typically linked α2–3 or α2–6 to galactose (Gal). Binding preference is widely believed to be a major determinant of influenza virus host range and species specificity, because avian viruses preferentially bind Siaα2–3Gal, whereas human and swine viruses bind Siaα2–6Gal (19–22). However, in addition to the Sia-Gal linkage, Sia species may influence virus infection and host range restriction (23). The main Sia species expressed in quail and chicken intestine (the site of influenza virus replication in these species) is N-acetylneuraminic acid (NeuAc) (24), whereas affinity for N-glycolylneuraminic acid (NeuGc)α2–3Gal is important for virus replication in the intestinal tract of mallard ducks (25). The human respiratory tract expresses solely NeuAc. (Humans lack expression of NeuGc because of a mutation in the CMAH gene required to convert NeuAc into NeuGc (26).) Both NeuAc and NeuGc are present in swine trachea (27), but the relative functional importance of NeuAc versus NeuGc for infection of pigs is not fully understood. Furthermore, recent data indicate that influenza virus/receptor interactions are more complex than the simple α2–3 versus α2–6 dichotomy would suggest (28). Finally, overall glycan topology is important for virus binding and transmission (29, 30). In light of all these findings, we sought to more fully understand the nature of glycans expressed on SRECs and to examine influenza virus infection of this novel swine cell system, with a focus on the reassortant viruses that have emerged and spread throughout the North American swine population over the past decade.
Lectin binding studies show that SRECs express both α2–3- and α2–6-linked Sia (18) as seen in pigs (5), although recent data indicate that α2–6-linked are more abundant in the upper respiratory tract of pigs (32, 33) and have previously been shown to be a useful system that parallels the characteristics of influenza virus infection of pigs (18, 31). To define the natural glycan profile of cultivated SRECs, the total N- and O-linked glycans expressed on SRECs were determined using ultrasensitive mass spectrometric (MS) analysis. Our results demonstrate that the cells contain a wide variety of complex bi-, tri-, and tetra-antennary N-glycans terminating in Sia or GalαGal. Sialidase treatments demonstrated the presence of both α2–3- and α2–6-sialylated glycans, and NeuAc is much more abundant than NeuGc. Branched polylactosamine glycans are also expressed. Glycan microarray binding indicated that both viruses bound exclusively to NeuAcα2–6 glycans and bound strongly to sialylated polylactosamine and sialylated N-glycans. Infection studies in SRECs with modulated sialic acids demonstrated that the viruses examined preferentially utilize NeuAc over NeuGc and α2–6- over α2–3-sialylated glycans to infect SRECs.
To our knowledge, this is the first detailed description of glycans expressed by swine respiratory epithelial cells. The data described here provide insight into the characteristics of specific glycans that influenza viruses utilize to infect primary swine cells and indicate a possible basis for transmission of influenza viruses between humans and pigs.
MATERIALS AND METHODS
Cells and Viruses
Madin Darby canine kidney (MDCK) cells were grown in minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin/amphotericin (Invitrogen). SRECs were isolated and grown as published previously (18). Briefly, aseptically collected distal swine tracheal specimens were rinsed with phosphate-buffered saline (PBS) and placed in a Pronase/DNase tissue-dissociation solution for 72 h at 4 °C. Cells were then incubated at 37 °C in uncoated cell culture dishes for 2 to 6 h, and nonadherent (epithelial) cells were collected and seeded into type VI collagen (Sigma)-coated cell culture flasks (adherent fibroblasts were discarded). SRECs were grown in serum-free, hormone-supplemented bronchial epithelial cell growth media (BEGM®, Lonza, Walkersville, MD) and maintained at 37 °C in a 5% CO2 atmosphere up to passage five. The collection and use of SRECs was approved by the Animal Care and Use Committee of the School of Veterinary Medicine, University of Wisconsin-Madison. The following viruses were used in this study: A/Brazil/1137/99 (H1N1) and A/Brazil/02/99 (H3N2) human isolates and A/Swine/MN/593/99 (triple reassortant H3N2) (34), A/Swine/ONT/00130/97 (human-lineage swine H3N2 isolate) (34), A/Swine/NC/44173/00 (classical swine H1N1), and A/Swine/IND/9K035/99 (triple reassortant H1N2) (35) swine isolates. Viruses were grown in MDCK cells, and virus titrations were performed in MDCK cells, and 50% tissue culture infectious dose (TCID50) values were calculated using the method of Reed and Muench (36). To ensure highly accurate measurements of infectious particles, viruses were titered in three independent experiments uniquely using one-fifth log dilutions in quadruplicate, and infected cells were identified by immunocytochemical staining for viral nucleoprotein expression as described previously (17).
Harvesting SRECs for Glycan Analysis
At passage three, confluent SRECs in 12-well culture plates were washed twice with PBS and incubated with Enzyme-free Cell Dissociation Buffer (Invitrogen) for 2 h. Cells were gently scraped off the plates, washed three times with PBS, flash-frozen in liquid nitrogen, and stored at −80 °C until further analysis.
Matrix-assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) Glycomic Analysis of SRECs
The SRECs (typically 10 million cells) were subjected to sonication, reduction, carboxymethylation, and trypsin digestion as described previously (37). The N-glycans were released by digestion with peptide N-glycosidase F (EC 3.5.1.52; Roche Applied Science) in 50 mm ammonium bicarbonate, pH 8.5, for 24 h at 37 °C and purified by reverse-phase C18 Sep-Pak (Waters) chromatography. The purified N-glycans were subsequently permethylated or subjected to enzymatic digestions (see below) before permethylation. O-Glycans were released by reductive elimination in 400 μl of potassium borohydride (54 mg/ml in 0.1 m KOH) at 45 °C for 16 h. The reaction was terminated by slow addition of glacial acetic acid, followed by Dowex 5W-X8(H+) 50–100 mesh (Sigma) chromatography and borate removal.
Sialidase Digestion
The underivatized N-glycans were dissolved in 150 μl of 50 mm sodium acetate buffer, pH 5.5, and incubated at 37 °C with Sialidase S (Streptococcus pneumoniae, Glyco, 170 milliunits) or sialidase A (Arthrobacter ureafaciens, Glyco, 170 milliunits) for 24 h. The digested samples were lyophilized, permethylated and purified on a C18 Sep-Pak (Waters).
Enzymatic Digestion of Glycans Prior to MALDI-TOF and Gas Chromatography/Mass Spectrometry (GC-MS)
Endo-β-galactosidase Digestion
The underivatized N-glycans were dissolved in 100 μl of 50 mm ammonium acetate buffer, pH 5.5, and incubated at 37 °C with 0.05 units of endo-β-galactosidase (Escherichia freundii, AMS Biotechnology). After 24 h, a second 0.05 unit of enzyme was added and incubated for an additional 24 h. The sample was lyophilized, permethylated, and purified on a C18 Sep-Pak cartridge (Waters).
α-Galactosidase Digestion
A portion of underivatized N-glycans was dissolved in 100 μl of 50 mm ammonium acetate buffer, pH 6, and incubated with 0.5 units of α-galactosidase (Green Coffee Bean, Glyco) at 37 °C for 48 h. The digested sample was lyophilized, permethylated, and purified by C18 Sep-Pak chromatography (Waters).
Mass Spectrometric Analysis
MALDI-TOF data were acquired on a Voyager-DE STR mass spectrometer (PerSeptive Biosystems, Framingham, MA) in the reflectron positive mode with delayed extraction. Permethylated samples were dissolved in 10 μl of methanol, and 1 μl of dissolved sample was premixed with 1 μl of matrix (20 mg/ml 2,5-dihydroxybenzoic acid in 70% (v/v) aqueous methanol) before being loaded onto the sample plate. MALDI-TOF/TOF experiments were performed on a 4800 Proteomics Analyzer (Applied Biosystems, Framingham MA) operated in the reflectron positive ion mode.
GC/MS Linkage Analysis
GC-MS linkage analysis of partially methylated alditol acetates was carried out on a PerkinElmer Life Sciences Clarus 500 instrument fitted with RTX-5 fused capillary column (30 m × 0.32 mm internal diameter, Restec Corp.). Partially methylated alditol acetates were prepared from permethylated samples as described previously (38). The permethylated glycans were hydrolyzed with 2 m trifluoroacetic acid for 2 h at 121 °C, reduced with 10 mg/ml sodium borodeuteride in 2 m aqueous ammonium hydroxide at room temperature, and acetylated with acetic anhydride at 100 °C for 1 h. The sample was dissolved in hexanes and injected onto the column at 60 °C. The column was maintained at this temperature for 1 min and then heated to 300 °C at a rate of 8 °C/min.
Automated MS and MS/MS Analysis
Annotation of the MS and MS/MS data was achieved with assistance from the Cartoonist algorithm (39) and the GlycoWorkbench software suite (40).
AlexaFluor 488 Virus Labeling and Glycan Array Binding
The Sw/MN and Sw/ONT viruses were labeled with AlexaFluor 488 based on methods described previously (41–43). Viruses were pelleted by ultracentrifugation (20,000 rpm for 1 h in a Beckman L8–70 M centrifuge), resuspended in Tris-buffered saline (TBS), and layered over a discontinuous 20–60% sucrose gradient in TBS. After centrifugation (20,000 rpm for 1 h), the virus bands at the 20–60% interface were isolated, diluted in TBS, and pelleted by centrifugation again. The pellets were resuspended in 0.15 m NaCl + 0.25 mm CaCl2 + 0.8 mm MgCl2, pH 7.2, and hemagglutination units were determined using turkey erythrocytes. The viruses were diluted to 50,000 hemagglutination units/ml, and 10 μl of 1.0 m sodium bicarbonate, pH 9.0, was added to 100 μl of each virus, followed by 0.0005 μg/hemagglutination units of AlexaFluor 488 (Invitrogen, solubilized in DMSO). Following 1 h of incubation at room temperature, labeled viruses were dialyzed into TBS + 0.25 mm CaCl2 + 0.8 mm MgCl2 with Slide-A-Lyzer MINI dialysis units (7,000 MWCO, Thermo Scientific, Waltham, MA) overnight at 4 °C. Following overnight incubation, hemagglutination units on turkey erythrocytes and infectious titer in MDCK cells was examined to ensure that labeling with AlexaFluor 488 did not affect virus binding or infectivity. The viruses were frozen at −80 °C until further analysis. They were then thawed; BSA and Tween 20 were added to final concentrations of 1 and 0.05%, respectively, and virus binding was examined on a printed glycan microarray (Consortium for Functional Glycomics) (44).
Infection of SRECs
Cells were seeded into type VI collagen-coated 24-well plates at 70,000 cells per well in the presence or absence of sialic acid precursors (N-acetylmannosamine and N-glycolylmannosamine (ManNAc and ManNGc)). ManNAc was obtained from Sigma, and peracetylated ManNGc was a gracious gift from Dr. Ron Schnaar. SRECs were grown to greater than 95% confluency (achieved at ∼48 h post seeding) and washed twice with BEGM® prior to infection. Viruses were diluted in minimal essential medium with 0.2% bovine serum albumin, 0.01% FBS, and antimicrobials, and cells were inoculated at a ratio of 3 TCID50 of each virus per cell. After incubation with virus for 1 h at 37 °C, SRECs were washed twice with BEGM® to remove the inoculum and overlaid with BEGM®. At 12 h post-inoculation, the supernatant was removed; the cells were harvested, and the percentage of infected cells was quantified by flow cytometry.
Detection of Infected SRECs by Flow Cytometry
Infected cells were identified by immunofluorescent labeling of viral nucleoprotein expression by flow cytometry as described previously (18). Briefly, cells were detached with trypsin, fixed with 10% formalin, permeabilized with PBS plus 0.1% saponin (Sigma), blocked with normal horse serum, and stained with a 1:6400 dilution of the mouse anti-influenza A virus nucleoprotein antibody 68D2 followed by a 1:50 dilution of FITC-labeled anti-mouse antibody (Zymed Laboratories Inc.). Fluorescence intensity was measured with a FACSCalibur flow cytometer (BD Biosciences), and infected cells were quantified with FLOWJO 8.5.3 (Treestar, Ashland, OR).
Sialidase Treatment of Live SRECs Prior to Virus Infection
As performed previously (31), confluent SRECs in 24-well plates were incubated for 3 h at 37 °C in BEGM® plus 150 units (as defined by the manufacturer) per well of either α2–3 specific sialidase from Salmonella typhimurium LT2 (New England Biolabs, Beverly, MA), or dual α2–3/6 sialidase from Clostridium perfringens (New England Biolabs). Following enzymatic treatment, cells were washed twice with BEGM® and infected as described above.
Statistical Analysis
Comparisons of infectivity levels of each virus were analyzed using ANOVA-protected Student's t tests. Analyses were performed using the R statistical software. p values of less than 0.01 were considered significant.
RESULTS
Glycomic Strategy for Profiling SREC Glycans
To prepare samples for mass spectrometry, the cells were sonicated, reduced, carboxymethylated, and digested with trypsin to generate peptides/glycopeptides. N-Glycans were released from extracted glycopeptides by PNGase F digestion, and O-glycans were chemically released by reductive elimination from the glycopeptides remaining after the release of N-glycans. The glycans were permethylated using the sodium hydroxide/DMSO procedure and purified on Sep-Pak C18 cartridges, followed by MALDI-TOF profiling, MALDI-TOF-TOF sequencing, and GC-MS linkage analysis. Data from these experiments were complemented by exo- and endoglycosidase digestions.
MALDI Fingerprinting of SREC Glycans
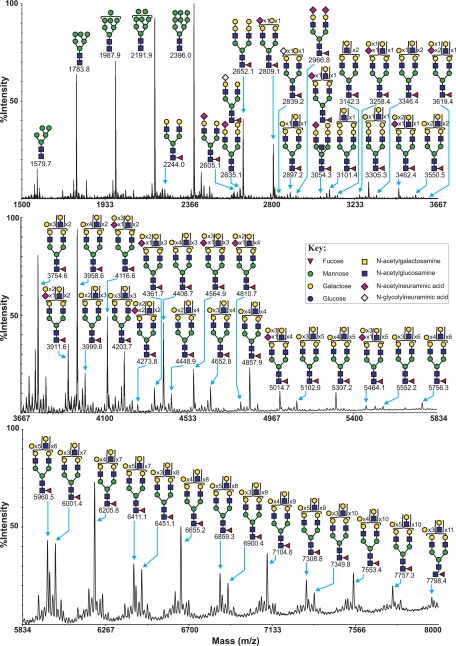
MALDI-TOF profiling of the permethylated N-glycans afforded a spectrum rich in [M + Na]+ molecular ion signals up to m/z 8000 (Fig. 1 and supplemental Table S1). High mannose N-glycans were observed at m/z 1579.7, 1783.8, 1987.9, 2191.9, and 2396.0. In addition, a series of complex glycans with compositions consistent with core fucosylated bi-, tri- and tetra-antennary structures bearing multiple LacNAc extensions were observed. Minor species with compositions consistent with the presence of bisecting GlcNAc are also present (for example m/z 3054.3, NeuAcHex6HexNAc5Fuc). The major nonreducing end-capping groups are either the Galα1–3Gal sequence (for example m/z 2652.1, 3305.3, and 3958.6, Hex7,9,11HexNAc4–6Fuc, respectively, consistent with bi-, tri-, and tetra-antennary core fucosylated structures) or Sia (for example m/z 2605.1 and 2966.8, NeuAc1–2Hex5HexNAc4Fuc, consistent with mono- and disialylated bi-antennary core fucosylated structures). Minor complex N-glycans capped with the NeuGc form of Sia were also observed (for example m/z 2635.1, NeuGcHex5HexNAc4Fuc). It has been demonstrated that relative quantitation based on signal intensities of permethylated glycans analyzed by MALDI-TOF MS is a reliable method, especially when comparing signals over a small mass range within the same spectrum (40). It can therefore be observed that Galα1–3Gal-capped structures are more abundant than Sia-capped structures, as seen, for example, by comparison of the ratios of the signals at m/z 2652.1 and m/z 2966.8 (both bi-antennary core-fucosylated structures capped with either Galα1–3Gal or NeuAc). O-Glycans were chemically released by reductive elimination, and their permethyl derivatives demonstrated the presence of both core 1 and core 2 structures, which are capped with Sia and/or GalαGal (supplemental Fig. S1 and supplemental Table S2). As observed with the N-glycans, NeuAc-terminated glycans are much more abundant than NeuGc.
FIGURE 1.
MALDI-TOF MS profiles of the permethylated N-linked glycans derived from SRECs. For complete annotation of the spectrum, see supplemental Table S1. Data were obtained from the 50% acetonitrile fraction, and all molecular ions are present in sodiated form ([M + Na]+).
Characterization of SREC Glycans by MS/MS Analysis
To confirm the putative structures deduced from the MALDI-MS data, MS/MS analyses were performed on a majority of SREC complex N-glycan molecular ions between m/z 2500 and m/z 5800. All molecular ions whose compositions indicated the presence of at least one fucose residue gave rise to a Y-ion at m/z 474, which is indicative of monofucosylated cores (Fucα1–6GlcNAc). All molecular ions whose compositions indicated the presence of a Galα1–3Gal-capping group gave rise to a B-ion at m/z 690, whereas all molecular ions whose compositions indicated the presence of a NeuAc-capping group gave rise to a B-ion at m/z 847. These data provide additional confirmation of these capping groups (supplemental Fig. S2). MS/MS analysis of molecular ions above m/z 3000 produced structurally informative fragment ions consistent with a heterogeneous mixture of bi-, tri-, and tetraantennary glycans with varying lengths of poly-LacNAc extension. Therefore, the annotations in Fig. 1 are simplified by using biantennary structures with the total number of LacNAc moieties listed in parentheses. Rigorous MS/MS analysis of the mono-sialylated core 1 structures of O-linked glycans (m/z 895 NeuAcHexHexNAc and m/z 925 NeuGcHexHexNAc) indicated that the sialic acid can be attached either to the Gal or GalNAc residue of the core 1 structure (supplemental Fig. S5).
GC-MS Linkage Analysis of SREC N-Glycans
The complexity of the glycan pools precluded the assignment of each detected component to an individual glycan. However, important conclusions can be drawn from the linkage data (supplemental Table S3). High levels of 3,6-linked mannose and 4-linked GlcNAc were in accordance with these residues being constituents of the core of all N-glycans. Fucosylated cores in the N-glycan pool were confirmed by the presence of 4,6-linked GlcNAc, which is consistent with MS/MS data (see above). The presence of 3,4,6-linked mannose confirms bisecting GlcNAc structures. The high abundance of 2-linked 2,4- and 2,6-linked Man revealed the presence of bi-, tri-, and tetra-antennary complex N-glycans. The presence of high levels of 3-linked Gal (which can also be produced from other structural features, see below) and 4-linked GlcNAc supports LacNAc extensions, whereas the presence of 3,6-linked Gal indicates that some of the LacNAc extensions are branched. The presence of both 3- and 6-linked Gal indicate the presence of both α2–3- and α2–6-sialylated glycans. Finally, the high levels of terminal Gal and 3-linked Gal are consistent with the high levels of Galα1–3Gal capping.
Exo- and Endoglycosidase Digestions of SREC N-Glycans
The presence of poly-LacNAc sequences was confirmed by digestion with endo-β-galactosidase (supplemental Fig. S3). This enzyme cleaves the β1–4 linkage of unbranched, repeating poly-LacNAc structures (GlcNAcβ1–3Galβ1–4)n (45). Endo-β-galactosidase treatment gave rise to signals at m/z 518.3, 722.4, 896.5, 926.5, and 1083.5, corresponding to the release of internal HexNAcHex from poly-LacNAc repeats and nonreducing HexHexNAcHex, HexHexNAcFucHex, Hex2HexNAcHex, and NeuAcHexHexNAcHex structures derived from poly-LacNAc. Signals at m/z 1835.9, 2081.5, and 2326.2 correspond to the enzyme-trimmed cores and indicate that SRECs can extend all four of their antennae with LacNAc repeats. Even after exhaustive digestion with endo-β-galactosidase, high molecular weight signals with multiple LacNAc repeats still remained. Taken together with the presence of 3,6-linked Gal in the GC-MS linkage analysis, these data indicate the presence of branched LacNAc repeats on SREC N-glycans. The presence of Galα1–3Gal capping of N-glycans was confirmed by digestion with α-galactosidase. This caused a loss of hexose units as exemplified by the peak at m/z 2652.3 (Hex7HexNAc4Fuc), which shifted to m/z 2244.1 (by the loss of two hexose units [Hex5HexNAc4Fuc]), thus confirming the biantennary core fucosylated structure with both antennae capped with Galα1–3Gal (data not shown).
Virus Infection of SRECs
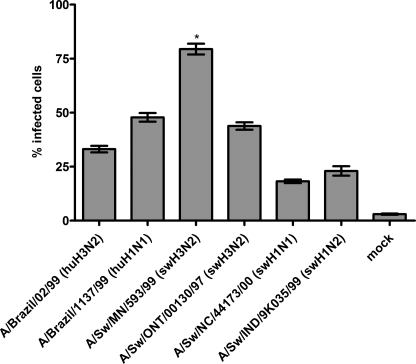
To compare the infectivity of Sw/MN (representative of the reassortant H3N2 viruses that spread throughout the North American swine population) with Sw/ONT (a nonreassortant human-lineage H3N2 virus isolated from a single pig) and other swine viruses, SRECs were infected with Sw/MN, Sw/ONT, and viruses representing the two other predominant subtypes circulating in North American pigs (H1N1 and H1N2). In addition, because Sw/ONT is a human-lineage virus and Sw/MN contains human-lineage HA, NA, and PB1 genes, representative viruses of the two major subtypes circulating in humans (H1N1 and H3N2) were examined. Each of these viruses infected the SRECs, but individual viruses exhibited different infectivity levels (Fig. 2). In particular, whereas Sw/ONT exhibited a similar infectivity level as the human viruses, Sw/MN infected a significantly higher percentage of SRECs than Sw/ONT (consistent with previous studies (18)). The Sw/MN virus also exhibited a significantly higher infectivity level than the H1N1 or H1N2 swine viruses or the reference human viruses.
FIGURE 2.
Flow cytometry-based quantification of virus infectivity levels in SRECs. The data are mean ± S.E. of three independent experiments performed in triplicate. *, p < 0.01 compared with infectivity level of each of the other viruses.
Virus Binding to Glycan Microarray
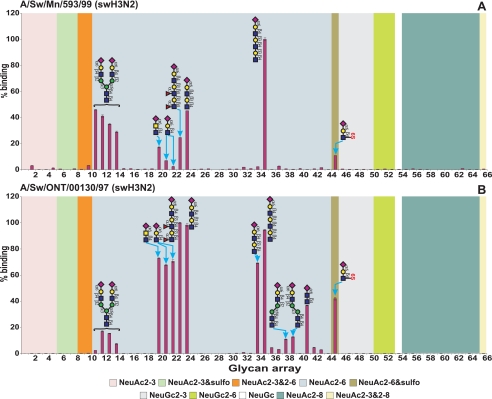
To compare the receptor-binding profiles of the highly infectious Sw/MN virus and the lower infectivity Sw/ONT virus, we used a microarray covering a wide range of nonsialylated and sialylated glycans (44). Binding of fluorescent-labeled viruses revealed that both viruses bound specifically to α2–6-sialylated glycans but did not bind to either asialo-, α2–3-, or α2–8-sialylated glycans (Fig. 3 and supplemental Table S4). Most Sia on the microarray were NeuAc, so a thorough examination of virus binding preference for NeuAc versus NeuGc was not possible. However, both viruses showed very low binding to NeuGc-containing glycans suggesting that these viruses preferentially bind NeuAc-containing glycans. Sw/MN bound with highest specificity to NeuAcα2–6-sialylated polylactosamine glycans (glycan 35) and to α2–6 bi-antennary N-glycans (glycans 11–14). Sw/ONT had similar binding profile, but with enhanced binding to glycans with additional structural features such as fucosylation, sulfation and subterminal type 1/2 lactosamine linkages (glycans 23, 24, 34, and 45). Regardless, the fact that both viruses bound to glycans containing NeuAcα2–6-sialylated polylactosamine and sialylated N-glycan components demonstrates the potential importance of these elements for virus binding.
FIGURE 3.
Glycan microarray analysis of Sw/MN and Sw/ONT viruses. Sw/MN (A) and Sw/ONT (B) binding to glycans was performed on microarray version 4.1 from the Consortium for Functional Glycomics. Results shown are the average of four replicate spots ± S.E. after the highest and lowest readings of six were excluded, with the highest value set to 100. As the binding of all asialo- and α2–3-sialylated glycans was below 1.5%, the structures of only five α2–3-sialylated glycans are plotted on the graph for clarity of presentation. For complete glycan sequences and relative luciferase units of viruses binding to all glycans see supplemental Table S4.
Strategy for Examining Functionally Important Glycans on SRECs
The Sw/MN and Sw/ONT viruses preferentially bound to NeuAcα2–6-terminated structures on the glycan microarray, suggesting that these glycan components are important for virus binding and infection. To further investigate functionally important glycans (i.e. which Sia species and linkages influenza viruses utilize for infection), prior to infection, SREC glycans were modulated to alter relative NeuAc versus NeuGc content and relative α2–3- versus α2–6-linked Sia.
Incorporation of Sia Precursors into SRECs
To examine Sia species utilized for virus infection, Sia precursors were used to alter the relative levels of Sia species on SRECs. These mannosamine precursors of Sia were added to culture media, entered cells, converted into sialic acids, and displayed on glycoconjugates on the cell surface (46–48). We utilized ManNAc (a precursor to NeuAc) and ManNGc (a precursor to NeuGc) and determined the lowest concentrations that had substantial effects on virus infectivity. ManNGc is peracetylated, whereas ManNAc is not, and others have found that peracetylated precursors are 10–100 times more potent than nonperacetylated molecules (46).6 The optimal concentrations were found to be 5 mm ManNAc and 0.1 mm ManNGc, which are within the concentration ranges used by others (46, 49). Treatment with precursors did not alter SREC morphology, proliferation, or viability (data not shown).
To ensure that the precursors had the expected modulatory effects on the expression of Sia species, we analyzed SREC N- and O-glycans by MALDI-TOF MS (supplemental Fig. S4). Treatment with ManNGc led to an increase in NeuGc and a decrease in NeuAc, relative to untreated cells. For example, the peak at m/z 2809.4 shifts 30 Da to m/z 2839.9 after ManNGc treatment, indicative of a NeuAc to NeuGc conversion (supplemental Fig. S4, A and B; the mass increment of permethylated NeuGc compared with NeuAc is 30 Da). Simultaneous treatment with both precursors partially reversed the effect of ManNGc alone, decreasing the amount of NeuGc and increasing the amount of NeuAc, relative to cells treated with ManNGc alone. For example, the relative intensity of the NeuAc peak at m/z 2809.4 increases and the NeuGc peak at m/z 2840.0 decreases (supplemental Fig. S4, B and C). Unexpectedly, in addition to incorporation of NeuGc, a series of ions indicating species 30 Da larger than the fully methylated N-glycans were also detected in the MS spectra of the samples treated with ManNGc alone or with both ManNGc and ManNAc precursors (see supplemental Figs. S4 and S5). These data indicate that for nonsialylated glycans the +30 addition was associated with HexNAc residues, and for sialylated glycans the +30 addition was associated with both a switch from NeuAc to NeuGc and modification of HexNAc residues.
Sialic Acid Species and Virus Infection
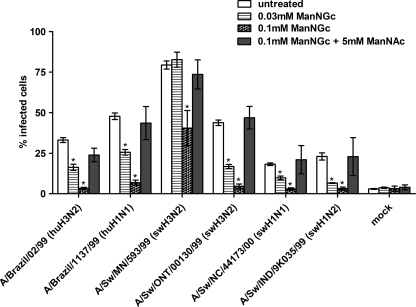
Culturing SRECs in the presence of ManNAc alone had little to no effect on virus infectivity levels (data not shown). However, when SRECs were grown in the presence of ManNGc and infected with the same viruses used in Fig. 2, each virus was significantly inhibited in a dose-dependent manner (p < 0.01, see Fig. 4). To ensure that the decrease in virus infectivity after ManNGc treatment was not due to nonspecific effects of the precursor, SRECs were simultaneously treated with ManNGc and ManNAc. In cells treated with both precursors, virus infectivity levels were not significantly different from the levels seen in untreated cells (Fig. 4). To confirm that virus infectivity in SRECs treated with both precursors was Sia-dependent and that the ManNAc was not simply allowing viruses to enter SRECs by an alternative non-Sia-dependent mechanism, cells were grown with both precursors and treated with Vibrio cholerae sialidase prior to infection with three viruses. The sialidase treatment decreased the infectivity of Sw/MN, Sw/ONT, and Sw/NC/44173/00 to 5–10% cells infected. In each case, these are statistically significant (p < 0.01) decreases, demonstrating that virus infection in SRECs treated with both precursors is Sia-dependent. To gain a deeper understanding of the dynamics of the precursor effects on infectivity, addition of the precursors was temporally modulated, followed by infection. Relatively slow changes in infectivity were seen following addition of precursors (for example, virus infection was not inhibited significantly until 8 h after addition of ManNGc), strongly suggesting that the ManNGc is metabolically incorporated into the cell, rather than simply directly binding to cell surfaces (data not shown). Taken together, these data indicate that infectivity is inhibited by ManNGc, and the inhibitory effects of ManNGc can be reversed with ManNAc treatment and that each of the influenza viruses tested selectively utilize NeuAc over NeuGc for infection.
FIGURE 4.
ManNGc and ManNAc treatment of SRECs followed by virus infection. SRECs were grown in the presence of ManNGc and/or ManNAc for 2 days prior to infection. Precursor molecules were solubilized in DMSO and added to a final concentration of 0.05% DMSO. A DMSO control showed no change in infectivity (data not shown). The data shown are the means ± S.E. of three independent experiments performed in triplicate. *, p < 0.01 compared with untreated cells.
Sialic Acid Linkages of SREC N-Glycans
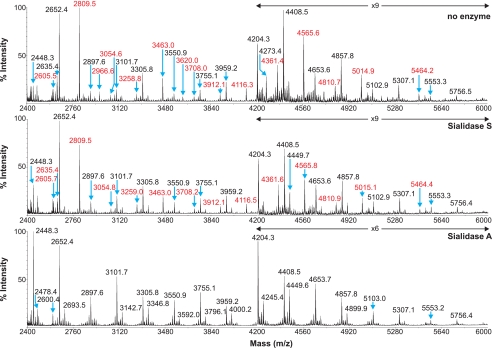
We have previously shown by lectin binding experiments that SRECs express both α2–3- and α2–6-linked Sia (18, 31). To confirm and extend these findings, sensitivity to digestion with linkage-specific sialidases was examined by subsequent MALDI-TOF and GC/MS analysis. Sialidase S was used for the specific release of α2–3-linked Sia and sialidase A for release of both α2–3- and α2–6-linked Sia. The partial MALDI-TOF spectra of SREC N-glycans after digestion with sialidase S or sialidase A, and control undigested glycans, are presented in Fig. 5. Digestion of the SREC N-glycans with sialidase A causes a complete loss of all Sia-containing glycans (m/z 2605.5, 2809.5, 2966.6, 3054.6, 3258.8, 3463.0, 3620.0, 3708.0, 3912.1, 4116.3, 4361.4, 4565.6, 4810.7, 5014.9, and 5464.2). Concurrent with these observations, the glycans at m/z 2448.3, 2693.5, 2897.6, 3101.7, 3346.8, 3550.9, 3755.1, 4000.2, 4204.3, 4449.6, 4653.7, and 5103.0, which would be produced by de-sialylating the above glycans, increased in abundance, confirming the presence of Sia-containing N-glycans in the SRECs. In contrast, digestion of the N-glycans with sialidase S caused a complete loss of disialylated glycans (m/z 2966.6 and 3620.0) and partial loss of Sia by monosialylated glycans, again with concurrent increase in abundance of the de-sialylated products. These data confirm that SREC N-glycans contain both α2–3- and α2–6-linked Sia, with a higher abundance of α2–6-Sia. The GC-MS data of the samples before and after sialidase digestion corroborate the results from the MALDI-TOF analysis. Peaks for 3- and 6-linked Gal were detected in untreated SRECs. After sialidase S treatment, the 6-linked Gal peak was unaffected, confirming the specificity of the enzyme. The 3-linked Gal peak was also still prominent after sialidase S digestion. This can be rationalized by the fact that large amounts of Galα1–3Gal-capping and poly-LacNAc extensions remain following sialidase S treatment, which contribute to the 3-linked Gal peak. After sialidase A, the 6-linked Gal peak was completely lost, and again the 3-linked Gal peak remained prominent (data not shown). These data confirm the presence of both α2–3- and α2–6-linked Sia, with the latter being of higher abundance.
FIGURE 5.
Partial MALDI-TOF MS profiles of the permethylated N-linked glycans derived from SRECs after digestion with sialidase S or sialidase A. Data were obtained from the 50% acetonitrile fraction and all molecular ions are present in sodiated form ([M + Na]+). Sialylated species are annotated in red (see supplemental Table S1).
Sialic Acid Linkages and Virus Infectivity
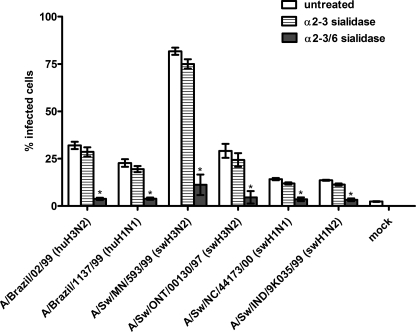
To determine which Sia linkage influenza viruses utilize to infect SRECs, prior to infection cells were treated with α2–3-specific sialidase (from Salmonella enterica serovar Typhimurium) or dual α2–3/6-sialidase (from C. perfringens). Confirmation of sialidase specificity by flow cytometry analysis of lectin binding has been reported previously (31). Specifically, although sialidases do not remove all of the lectin binding signal, treatment with either sialidase substantially decreased the Maackia amurensis agglutinin lectin (α2–3-recognizing) signal to approximately equal low levels, whereas only the α2–3/6-sialidase dramatically decreased the Sambucus nigra agglutinin (α2–6-recognizing) signal. We have previously demonstrated that infectivity levels of H4N6 viruses were unchanged following treatment with α2–3-specific sialidase but that treatment with α2–3/6 sialidase inhibited virus infectivity (31). Here, SRECs were treated with sialidase and infected with the same viruses used in Figs. 2 and 4. After treatment with α2–3-specific sialidase, infectivity was either unchanged or decreased slightly (Fig. 6). However, after treatment with α2–3/6-sialidase, the infectivity level of each virus tested was significantly lower (p < 0.01). Taken together, the data suggest that the viruses examined here preferentially utilize α2–6-sialylated glycans to infect SRECs.
FIGURE 6.
Sialidase treatment of SRECs prior to virus infection. The data shown are the mean ± S.E. of three independent experiments performed in triplicate. *, p < 0.01 compared with untreated cells.
DISCUSSION
We report here the first detailed characterization of swine respiratory epithelial cell glycans. The MALDI-TOF profiles revealed a wide variety of complex glycans with extensive linear and branched poly-LacNAc sequences (Fig. 1 and supplemental Fig. S2). The majority of SREC glycans are terminated with sialic acids and/or GalαGal. Although the GalαGal moiety is not present on human cells (29, 50), it is not unexpected to find it on pig cells, given the history of immune reactions to the GalαGal moiety of pig organs among human patients receiving pig organ transplants (51).
We have previously shown that Sw/MN is more infectious for pigs and has higher infectivity levels in SRECs than Sw/ONT and that infectivity in SRECs depends on the HA gene (17, 18). To determine which glycans these viruses bound, and to begin to define differences between these viruses that may account for virus infectivity, binding profiles of these viruses were analyzed via glycan microarray analysis. Both Sw/MN and Sw/ONT solely bound to NeuAcα2–6-sialylated glycans and showed high binding to sialylated polylactosamine glycans and sialylated N-glycans (Fig. 3), structural features confirmed to be present on SRECs by our glycomic analysis. Because NeuAcα2–6 polylactosamine glycans are known to play major roles in human virus infection (29, 52, 53), the importance of these receptor components in influenza virus infection of swine cells has implications for transmission of influenza viruses between humans and pigs. In addition, the common receptor components may be a molecular basis for pigs as an adaptation host, as replication in pigs could cause virus mutations leading to binding and utilization of NeuAcα2–6 polylactosamine, thus increasing the ability of the virus to infect humans.
Although Sw/MN and Sw/ONT binding profiles have similarities, the specific binding profiles of the viruses do differ. The viruses have differential binding affinity for some glycans (for example glycans 11, 22, and 44), and Sw/ONT binds to a wider range of glycans (for example glycans 34, 39, and 41) than Sw/MN. Because we have shown previously that SREC infectivity levels of Sw/MN and Sw/ONT are controlled by the HA gene (18), the differences in binding profiles could account for infectivity levels. However, it has to be borne in mind that the glycan array does not contain multiantennary N-glycans, glycan probes capped with Galα1–3Gal, or branched polylactosamine sequences, all of which the glycomic profiling indicated as prominent features of SRECs. Therefore, the effects of these structural features on virus binding were not observed.
Many lines of evidence indicate that α2–6-sialylated glycans are crucial for influenza virus infection in swine cells. First, avian-lineage H4N6 viruses isolated from pigs contained HA mutations from the avian consensus to amino acids that allow for preferential binding to α2–6 glycans, suggesting a selective pressure in pigs toward α2–6-linked Sia binding and utilization (31, 54). Second, removing α2–3-linked Sia had minimal effect on infectivity, whereas removal of both α2–3- and α2–6-linked Sia significantly inhibited the infectivity of each virus examined (Fig. 6), suggesting that the influenza viruses examined preferentially utilize α2–6-linked sialic acids for infection. Third, others have shown that swine viruses preferentially bind α2–6-sialylated glycans (21, 22). Our microarray analysis extends these findings, as Sw/MN and Sw/ONT bound to biantennary and polylactosamine α2–6-sialylated glycans (Fig. 3).
The SRECs express higher levels of NeuAc than NeuGc, which is somewhat unexpected, given a report that NeuAc and NeuGc were found in roughly equal amounts in swine trachea by Suzuki et al. (27). The discrepancy between the data presented here and the previous report may be due to the specific cell types analyzed in each study. A pure population of epithelial cells (18) was exclusively studied here, whereas the previous report likely included connective tissue and other cell types contained in the harvested ex vivo tracheal samples. With regard to influenza viruses, Suzuki et al. (27) suggested that NeuAc and NeuGc could both be important in virus infection of pigs, whereas Gambaryan et al. (22) found that swine viruses bound NeuAc- but not NeuGc-containing polymers. Our data help to clarify this discrepancy and indicate that NeuAc is the Sia species preferentially bound and utilized for infection by the influenza viruses examined here. This finding is consistent with structures of the virus HA protein complexed with Sia, which demonstrate that the terminal methyl group of the NeuAc N-acyl side chain fits into a space between amino acids 153 and 155 of the H3 HA protein and makes van der Waals contact with the HA (55–57). The NeuAc to NeuGc conversion introduces polarity and extends the N-acyl side chain, which breaks the nonpolar interaction and may make the NeuGc too large to effectively fit (49). Thus, in terms of both steric hindrance and polarity, conversion from NeuAc to NeuGc likely reduces the binding affinity of HA/Sia interactions.
The structural glycan analysis, virus glycan microarray binding, and virus infection data presented here provide unprecedented molecular detail of influenza virus infection of swine cells. The structural characterization of the actual glycans in the SREC cellular system described here provide valuable information for the design of custom glycan microarrays for monitoring swine influenza virus infectivity. Virus binding and infection results demonstrate the importance of NeuAcα2–6 glycans, suggest that sialylated polylactosamine glycans and sialylated N-glycans are also crucial for virus infection, and further our understanding of the unique infectivity characteristics of the triple reassortant H3N2 viruses that emerged and spread widely throughout the North American swine population at the end of the 20th century. The similarity of these glycan components to glycans involved in human virus infection also expands our understanding of the molecular basis for virus transmission between pigs and humans and the ability of pigs to serve as possible adaptation hosts and/or intermediate virus reassortment hosts in the creation of novel viruses for humans.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Alexander Karasin for excellent technical assistance; Dr. Ron Schnaar of The Johns Hopkins University for providing ManNGc; Dr. M. Suresh of the University of Wisconsin-Madison for flow cytometry assistance; Dr. Werner Reutter of Freie Universität Berlin, Germany, for helpful discussions regarding the mannosamine precursors; and Drs. David Smith and Jamie Heimburg-Molinaro of the Consortium for Functional Glycomics for performing the glycan array binding. We also acknowledge Dr. Yoshihiro Kawaoka and the University of Wisconsin-Madison Influenza Virus Repository; Dr. Gene Erickson of the Rollins Veterinary Diagnostic Laboratory, Raleigh, NC; and Dr. Alexander Klimov of the Centers for Disease Control and Prevention for providing some of the influenza virus isolates used.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI060646 (to C. W. O.). This work was also supported by Grant 082098 from the Wellcome Trust (to S. M. H. and A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S5.
Werner Reutter, personal communication.
- SREC
- swine respiratory epithelial cell
- NeuGc
- N-glycolylneuraminic acid
- MDCK
- Madin-Darby canine kidney cells
- BEGM
- bronchial epithelial cell growth media
- ManNAc
- N-acetylmannosamine
- ManNGc
- N-glycolylmannosamine.
REFERENCES
- 1.Kida H., Ito T., Yasuda J., Shimizu Y., Itakura C., Shortridge K. F., Kawaoka Y., Webster R. G. (1994) J. Gen. Virol. 75, 2183–2188 [DOI] [PubMed] [Google Scholar]
- 2.Landolt G. A., Olsen C. W. (2007) Anim. Health Res. Rev. 8, 1–21 [DOI] [PubMed] [Google Scholar]
- 3.Brown I. H. (2000) Vet. Microbiol. 74, 29–46 [DOI] [PubMed] [Google Scholar]
- 4.Scholtissek C. (1990) Med. Princ. Pract. 2, 65–71 [Google Scholar]
- 5.Ito T., Couceiro J. N., Kelm S., Baum L. G., Krauss S., Castrucci M. R., Donatelli I., Kida H., Paulson J. C., Webster R. G., Kawaoka Y. (1998) J. Virol. 72, 7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scholtissek C., Bürger H., Kistner O., Shortridge K. F. (1985) Virology 147, 287–294 [DOI] [PubMed] [Google Scholar]
- 7.Karasin A. I., Schutten M. M., Cooper L. A., Smith C. B., Subbarao K., Anderson G. A., Carman S., Olsen C. W. (2000) Virus Res. 68, 71–85 [DOI] [PubMed] [Google Scholar]
- 8.Olsen C. W., Karasin A. I., Carman S., Li Y., Bastien N., Ojkic D., Alves D., Charbonneau G., Henning B. M., Low D. E., Burton L., Broukhanski G. (2006) Emerg. Infect. Dis. 12, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou N. N., Senne D. A., Landgraf J. S., Swenson S. L., Erickson G., Rossow K., Liu L., Yoon K., Krauss S., Webster R. G. (1999) J. Virol. 73, 8851–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karasin A. I., Landgraf J., Swenson S., Erickson G., Goyal S., Woodruff M., Scherba G., Anderson G., Olsen C. W. (2002) J. Clin. Microbiol. 40, 1073–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma W., Gramer M., Rossow K., Yoon K. J. (2006) J. Virol. 80, 5092–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent A. L., Ma W., Lager K. M., Gramer M. R., Richt J. A., Janke B. H. (2009) Virus Genes, [DOI] [PubMed] [Google Scholar]
- 13.Ma W., Vincent A. L., Gramer M. R., Brockwell C. B., Lager K. M., Janke B. H., Gauger P. C., Patnayak D. P., Webby R. J., Richt J. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20949–20954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawood F. S., Jain S., Finelli L., Shaw M. W., Lindstrom S., Garten R. J., Gubareva L. V., Xu X., Bridges C. B., Uyeki T. M. (2009) N. Engl. J. Med. 360, 2605–2615 [DOI] [PubMed] [Google Scholar]
- 15.Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., Sessions W. M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C. B., Emery S. L., Hillman M. J., Rivailler P., Smagala J., de Graaf M., Burke D. F., Fouchier R. A., Pappas C., Alpuche-Aranda C. M., López-Gatell H., Olivera H., López I., Myers C. A., Faix D., Blair P. J., Yu C., Keene K. M., Dotson P. D., Jr., Boxrud D., Sambol A. R., Abid S. H., St George K., Bannerman T., Moore A. L., Stringer D. J., Blevins P., Demmler-Harrison G. J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H. F., Belongia E. A., Clark P. A., Beatrice S. T., Donis R., Katz J., Finelli L., Bridges C. B., Shaw M., Jernigan D. B., Uyeki T. M., Smith D. J., Klimov A. I., Cox N. J. (2009) Science 325, 197–20119465683 [Google Scholar]
- 16.Smith G. J., Vijaykrishna D., Bahl J., Lycett S. J., Worobey M., Pybus O. G., Ma S. K., Cheung C. L., Raghwani J., Bhatt S., Peiris J. S., Guan Y., Rambaut A. (2009) Nature 459, 1122–1125 [DOI] [PubMed] [Google Scholar]
- 17.Landolt G. A., Karasin A. I., Phillips L., Olsen C. W. (2003) J. Clin. Microbiol. 41, 1936–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busch M. G., Bateman A. C., Landolt G. A., Karasin A. I., Brockman-Schneider R. A., Gern J. E., Suresh M., Olsen C. W. (2008) Virus Res. 133, 269–279 [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich M. N., Gambaryan A. S., Teneberg S., Piskarev V. E., Yamnikova S. S., Lvov D. K., Robertson J. S., Karlsson K. A. (1997) Virology 233, 224–234 [DOI] [PubMed] [Google Scholar]
- 20.Rogers G. N., Paulson J. C., Daniels R. S., Skehel J. J., Wilson I. A., Wiley D. C. (1983) Nature 304, 76–78 [DOI] [PubMed] [Google Scholar]
- 21.Rogers G. N., D'Souza B. L. (1989) Virology 173, 317–322 [DOI] [PubMed] [Google Scholar]
- 22.Gambaryan A. S., Karasin A. I., Tuzikov A. B., Chinarev A. A., Pazynina G. V., Bovin N. V., Matrosovich M. N., Olsen C. W., Klimov A. I. (2005) Virus Res. 114, 15–22 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki Y., Ito T., Suzuki T., Holland R. E., Jr., Chambers T. M., Kiso M., Ishida H., Kawaoka Y. (2000) J. Virol. 74, 11825–11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo C. T., Takahashi N., Yagi H., Kato K., Takahashi T., Yi S. Q., Chen Y., Ito T., Otsuki K., Kida H., Kawaoka Y., Hidari K. I., Miyamoto D., Suzuki T., Suzuki Y. (2007) Glycobiology 17, 713–724 [DOI] [PubMed] [Google Scholar]
- 25.Ito T., Suzuki Y., Suzuki T., Takada A., Horimoto T., Wells K., Kida H., Otsuki K., Kiso M., Ishida H., Kawaoka Y. (2000) J. Virol. 74, 9300–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou H. H., Hayakawa T., Diaz S., Krings M., Indriati E., Leakey M., Paabo S., Satta Y., Takahata N., Varki A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11736–11741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T., Horiike G., Yamazaki Y., Kawabe K., Masuda H., Miyamoto D., Matsuda M., Nishimura S. I., Yamagata T., Ito T., Kida H., Kawaoka Y., Suzuki Y. (1997) FEBS Lett. 404, 192–196 [DOI] [PubMed] [Google Scholar]
- 28.Nicholls J. M., Chan R. W., Russell R. J., Air G. M., Peiris J. S. (2008) Trends Microbiol. 16, 149–157 [DOI] [PubMed] [Google Scholar]
- 29.Chandrasekaran A., Srinivasan A., Raman R., Viswanathan K., Raguram S., Tumpey T. M., Sasisekharan V., Sasisekharan R. (2008) Nat. Biotechnol. 26, 107–113 [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan A., Viswanathan K., Raman R., Chandrasekaran A., Raguram S., Tumpey T. M., Sasisekharan V., Sasisekharan R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2800–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bateman A. C., Busch M. G., Karasin A. I., Bovin N., Olsen C. W. (2008) J. Virol. 82, 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Poucke S. G., Nicholls J. M., Nauwynck H. J., Van Reeth K. (2010) Virol. J. 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelli R. K., Kuchipudi S. V., White G. A., Perez B. B., Dunham S. P., Chang K. C. (2010) BMC Vet. Res. 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landolt G. A., Karasin A. I., Schutten M. M., Olsen C. W. (2006) J. Clin. Microbiol. 44, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karasin A. I., Olsen C. W., Anderson G. A. (2000) J. Clin. Microbiol. 38, 2453–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed L., Muench H. (1938) Am. J. Hyg. 27, 493–497 [Google Scholar]
- 37.Jang-Lee J., North S. J., Sutton-Smith M., Goldberg D., Panico M., Morris H., Haslam S., Dell A. (2006) Methods Enzymol. 415, 59–86 [DOI] [PubMed] [Google Scholar]
- 38.Albersheim P., Nevis D. J., English P. D., Karr A. (1967) Carbohydr. Res. 5, 340–345 [Google Scholar]
- 39.Goldberg D., Sutton-Smith M., Paulson J., Dell A. (2005) Proteomics 5, 865–875 [DOI] [PubMed] [Google Scholar]
- 40.Ceroni A., Maass K., Geyer H., Geyer R., Dell A., Haslam S. M. (2008) J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 41.Kumari K., Gulati S., Smith D. F., Gulati U., Cummings R. D., Air G. M. (2007) Virol. J. 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lugovtsev V. Y., Smith D. F., Weir J. P. (2009) Virology 394, 218–226 [DOI] [PubMed] [Google Scholar]
- 43.Gulati S., Smith D. F., Air G. M. (2009) Virol. J. 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuda M. N., Matsumura G. (1976) J. Biol. Chem. 251, 6218–6225 [PubMed] [Google Scholar]
- 46.Collins B. E., Fralich T. J., Itonori S., Ichikawa Y., Schnaar R. L. (2000) Glycobiology 10, 11–20 [DOI] [PubMed] [Google Scholar]
- 47.Keppler O. T., Horstkorte R., Pawlita M., Schmidt C., Reutter W. (2001) Glycobiology 11, 11R–18R [DOI] [PubMed] [Google Scholar]
- 48.Bardor M., Nguyen D. H., Diaz S., Varki A. (2005) J. Biol. Chem. 280, 4228–4237 [DOI] [PubMed] [Google Scholar]
- 49.Keppler O. T., Herrmann M., von der Lieth C. W., Stehling P., Reutter W., Pawlita M. (1998) Biochem. Biophys. Res. Commun. 253, 437–442 [DOI] [PubMed] [Google Scholar]
- 50.Galili U., Shohet S. B., Kobrin E., Stults C. L., Macher B. A. (1988) J. Biol. Chem. 263, 17755–17762 [PubMed] [Google Scholar]
- 51.Sandrin M. S., Vaughan H. A., Dabkowski P. L., McKenzie I. F. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11391–11395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller-Podraza H., Johansson L., Johansson P., Larsson T., Matrosovich M., Karlsson K. A. (2000) Glycobiology 10, 975–982 [DOI] [PubMed] [Google Scholar]
- 53.Ogata M., Hidari K. I., Kozaki W., Murata T., Hiratake J., Park E. Y., Suzuki T., Usui T. (2009) Biomacromolecules, [DOI] [PubMed] [Google Scholar]
- 54.Karasin A. I., Brown I. H., Carman S., Olsen C. W. (2000) J. Virol. 74, 9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauter N. K., Hanson J. E., Glick G. D., Brown J. H., Crowther R. L., Park S. J., Skehel J. J., Wiley D. C. (1992) Biochemistry 31, 9609–9621 [DOI] [PubMed] [Google Scholar]
- 56.Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. (1988) Nature 333, 426–431 [DOI] [PubMed] [Google Scholar]
- 57.Skehel J. J., Wiley D. C. (2000) Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.