Abstract
DegS is a periplasmic Escherichia coli protease, which functions as a trimer to catalyze the initial rate-limiting step in a proteolytic cascade that ultimately activates transcription of stress response genes in the cytoplasm. Each DegS subunit consists of a protease domain and a PDZ domain. During protein folding stress, DegS is allosterically activated by peptides exposed in misfolded outer membrane porins, which bind to the PDZ domain and stabilize the active protease. It is not known whether allostery is conferred by the PDZ domains or is an intrinsic feature of the trimeric protease domain. Here, we demonstrate that free DegSΔPDZ equilibrates between active and inactive trimers with the latter species predominating. Substrate binding stabilizes active DegSΔPDZ in a positively cooperative fashion. Mutations can also stabilize active DegSΔPDZ and produce an enzyme that displays hyperbolic kinetics and degrades substrate with a maximal velocity within error of that for fully activated, intact DegS. Crystal structures of multiple DegSΔPDZ variants, in functional and non-functional conformations, support a two-state model in which allosteric switching is mediated by changes in specific elements of tertiary structure in the context of an invariant trimeric base. Overall, our results indicate that protein substrates must bind sufficiently tightly and specifically to the functional conformation of DegSΔPDZ to assist their own degradation. Thus, substrate binding alone may have regulated the activities of ancestral DegS trimers with subsequent fusion of the protease domain to a PDZ domain, resulting in ligand-mediated regulation.
Keywords: Allosteric Regulation, Bacterial Signal Transduction, Biophysics, Crystal Structure, Enzyme Mechanisms, Enzyme Structure, Intramembrane Proteolysis, Protease
Introduction
Proteases are carefully regulated to ensure that these destructive enzymes are only activated under the proper circumstances. For example, most extracellular proteases are synthesized as inactive proenzymes or zymogens (1). In the trypsin proenzyme, the Ser-His-Asp catalytic triad is properly formed, but the oxyanion hole of the active site is malformed and only rearranges to the active structure following the processing event that produces the mature enzyme (2). The activities of intracellular trypsin-like proteases are also regulated by changes in active site conformation. In the DegS protease, for instance, alternative conformations of the oxyanion hole result in an equilibrium distribution of inactive and active enzymes whose populations are controlled by the binding of substrates and allosteric effectors (3–8).
Escherichia coli DegS is a trimeric HtrA family enzyme consisting of a trypsin-like protease domain, a PDZ domain, and a sequence that anchors the protease to the periplasmic surface of the inner membrane (3, 9). The trimer is stabilized by interactions between the protease domains. Protein folding stress in the periplasm activates DegS cleavage of RseA, a transmembrane protein whose cytoplasmic domain binds and inhibits the σE transcription factor (10, 11). DegS cleavage of RseA initiates a cascade of subsequent cleavages by other proteases, resulting in release of σE and transcriptional activation of genes whose products are needed to repair damage caused by envelope stress. The PDZ domain of DegS plays a critical role in signal transduction. Specifically, C-terminal peptides of misfolded outer membrane porins (OMPs),2 which end with a conserved Tyr-Xaa-Phe motif, bind to the PDZ domain, and activate cleavage of RseA (Fig. 1A and Ref. 3). Indeed, DegS cleavage of RseA in vitro can be accelerated more than 500-fold by OMP peptides (6, 7).
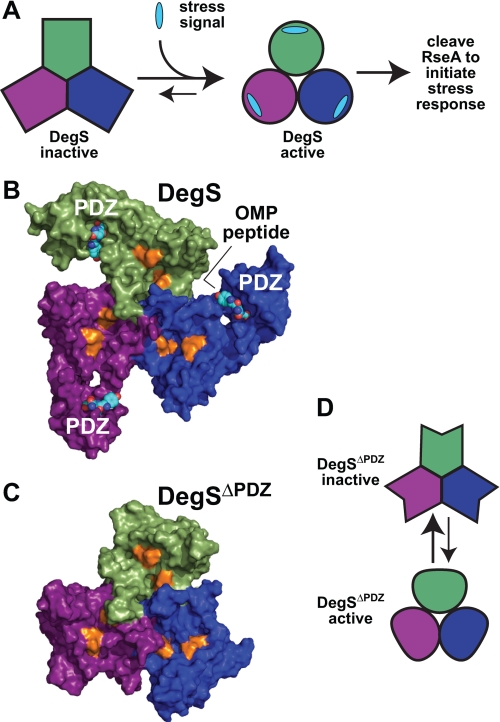
FIGURE 1.
Allosteric control of DegS activity. A, DegS equilibrates between inactive and active structures. Environmental stress produces a signal in the form of exposed OMP peptide sequences that stabilizes active DegS, resulting in cleavage of RseA and initiation of a transcriptional stress response. B, OMP peptides (shown in Corey-Pauling-Koltun representation) bind to the PDZ domains of an intact DegS trimer (surface representation; Protein Data Bank code 3GDV). The proteolytic active sites (gold color) are distant from the peptide-binding sites. C, the protease domains in DegSΔPDZ (surface representation; Protein Data Bank code 2QF3) form a stable trimer with a structure similar to the corresponding regions of peptide-activated DegS. D, model in which DegSΔPDZ also equilibrates between active and inactive conformations.
OMP peptide activation of DegS appears to occur by relieving inhibitory contacts between the PDZ domain and the protease domain. For example, salt bridges between the PDZ domain and the protease domain are observed in crystal structures of peptide-free DegS, which has a malformed oxyanion hole, but these interactions are absent in peptide-bound DegS, which has a functional oxyanion hole (4, 5). Importantly, disrupting inhibitory interactions between the PDZ domain and the protease domain by point mutations or by deleting the PDZ domain entirely (DegSΔPDZ) activates cleavage of RseA to levels observed with some OMP peptides (6, 7). Moreover, crystal structures of DegSΔPDZ closely resemble the active conformation of intact DegS (Fig. 1, B and C) and have a functional oxyanion hole (6, 12). These results support an allosteric model in which the unliganded PDZ domain stabilizes the inactive protease domain conformation, whereas OMP peptide binding to the PDZ domain reduces or eliminates this inhibitory effect.
Although both OMP peptides and the RseA substrate bind more tightly to active DegS than to inactive DegS, neither saturating peptide nor saturating RseA is sufficient to shift the majority of enzymes into the active conformation (7). As a consequence, substrate cleavage is positively cooperative with respect to both RseA concentration and OMP peptide concentration. Moreover, because OMP peptides and RseA bind both to inactive and active DegS, it cannot be assumed that activity measured in the presence of saturating peptide and saturating substrate corresponds to the maximal cleavage rate that would be observed if all enzymes assumed the active conformation (7). In fact, different OMP peptides support very different maximal levels of RseA cleavage. Moreover, cleavage activity can be increased above the wild-type DegS level by mutations that disrupt inhibitory interactions and by mutations that stabilize the active conformation (6, 8).
Is allostery an intrinsic property of the protease domain of DegS, or is it only conferred by the presence of the PDZ domains? The protease activity of the DegSΔPDZ trimer is substantially lower than that of full-length DegS stimulated by OMP peptides with the highest activities (6, 7, 12). There are two potential explanations for this fact. DegSΔPDZ itself may equilibrate between inactive and active conformations (Fig. 1D), and substrate binding may be insufficient to shift most enzymes into the active state. Alternatively, all DegSΔPDZ enzymes may assume the “functional” conformation, but this structure may have lower activity than peptide-activated full-length DegS because of subtle changes in structure or dynamics. For example, based on differences observed in crystal structures, it was concluded that DegSΔPDZ has “a less ordered active site with reduced proteolytic activity” (12).
To probe mechanism, we characterized structure-activity relationships for variants of E. coli DegSΔPDZ. Importantly, we found that the H198P mutation decreased Km, increased Vmax, and essentially eliminated the positive cooperativity of substrate cleavage when compared with wild-type DegSΔPDZ. These results strongly support a model in which positive cooperativity arises for wild-type DegSΔPDZ because substrates bind more tightly to the active than inactive conformation and are inconsistent with a model in which positive cooperativity is a consequence of substrate-substrate interactions as was suggested previously (6). We also found that the protease activity of DegSΔPDZ could be decreased by many of the same mutations that destabilize the active conformation of full-length DegS. Moreover, crystal structures of DegSΔPDZ, H198P DegSΔPDZ, and inactive DegSΔPDZ mutants define two basic conformations with functional and non-functional oxyanion holes, respectively. Thus, our combined results support a two-state model in which allostery is an intrinsic property of the protease domain of DegS.
EXPERIMENTAL PROCEDURES
Variants of E. coli DegSΔPDZ (residues 27–256) with an N-terminal His6 tag and lacking the wild-type membrane anchor were purified by Ni2+-nitrilotriacetic acid affinity chromatography and gel filtration on a Superdex 200 column (6, 7). An 35S-labeled variant of the periplasmic domain of E. coli RseA (residues 121–216) with a C-terminal His6 tag and a His6-tagged variant of DegS (residues 27–355) were purified as described (6, 7). Mutations were generated in plasmid-borne genes by the QuikChange method (Stratagene). The synthetic YYF peptide was purified by HPLC. Modification with diisopropylfluorophosphate (DFP) was performed as described (8).
Cleavage assays were performed as described (6, 7). Enzymes were incubated with 35S-labeled RseA for different times at room temperature (23 ± 1 °C), and cleavage was quantified by scintillation counting of acid-soluble radioactivity. Michaelis-Menten curves were fitted to the Hill equation using the non-linear least square subroutine in KaleidaGraph (Synergy software).
DegSΔPDZ variants were crystallized at 20 °C by hanging drop vapor diffusion crystallization using protein stocks at 10–20 mg/ml in 50 mm NaHPO4 (pH 8.0), 200 mm NaCl, 5 mm EDTA, and 10% glycerol. All crystals were obtained after 1:1 mixing with 50 mm sodium cacodylate (pH 6.5), 50–150 mm sodium citrate, and 15–20% isopropanol and were cryoprotected by using 30% methylpentanediol.
Diffraction data were collected using a Rigaku MicroMax008-HP rotating source or at the NE-CAT 24-ID-C beamline at the Argonne National Laboratory Advanced Photon Source. Initial phases were obtained by molecular replacement using PHASER (13). Structures were determined by cycles of model building using COOT and refinement using PHENIX (14, 15). To obtain the best possible geometry, all structures were refined with riding hydrogen atoms, which were added using REDUCE (16). This procedure does not result in the introduction of additional parameters during refinement as the positions and B-factors of hydrogens are determined by the atoms to which they were attached. Refinement statistics for all final structures were also determined after stripping the hydrogens (without allowing changes in coordinates or B-factors of other atoms). As shown in Table 1, removing the hydrogens resulted in very small differences in Rwork and Rfree. Model building was guided by MolProbity (17), which was used to identify portions of the model with poor geometry or steric clashes for subsequent correction. In the final structures, fewer than 0.2% of residues had poor side-chain rotamers, none were Ramachandran outliers, and 98% or more were in the most favored regions of the Ramachandran plot (Table 1).
TABLE 1.
Crystallographic data and refinement statistics for DegSΔPDZ structures
Rsym = ShSj|Ij(h) − 〈I(h)〉|/ShS[j〈I(h)〉 where Ij(h) is the jth reflection of index h and I(h) is the average intensity of all observations of I(h). Rwork = Sh|Fobs(h) − Fcalc(h)‖/Sh|Fobs(h)| calculated over the 95% of the data in the working set. Rfree is equivalent to Rwork except that it is calculated over the 5% of the data assigned to the test set. Numbers in parentheses represent values for the highest resolution bin. r.m.s.d., root mean square deviation.
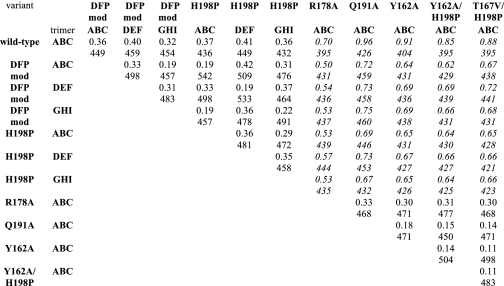
| Wild type | DFP-modified | H198P | R178A | Q191A | Y162A | Y162A/H198P | T167V/H198P | |
|---|---|---|---|---|---|---|---|---|
| Protein Data Bank code | 3LGI | 3LH3 | 3LGV | 3LGY | 3LH1 | 3LGU | 3LGT | 3LGW |
| Space group | P212121 | P212121 | P212121 | H3 | H3 | H3 | H3 | H3 |
| Unit cell (Å) | a = 70.38, b = 72.76, c = 128.87 | a = 71.53, b = 132.74, c = 231.20 | a = 71.54, b = 133.56, c = 230.27 Å | a = 70.22, b = 70.22, c = 119.23 Å | a = 70.89, b = 70.89, c = 120.84 Å | a = 70.56, b = 70.56, c = 118.32 Å | a = 70.65, b = 70.65, c = 120.05 Å | a = 70.66, b = 70.66, c = 121.03 Å |
| Subunits/asymmetric unit | 3 | 9 | 9 | 1 | 1 | 1 | 1 | 1 |
| Resolution (Å) | 1.65 | 2.35 | 2.73 | 2.70 | 2.51 | 2.46 | 2.69 | 2.50 |
| Wavelength (Å) | 0.97933 | 0.97949 | 1.54178 | 1.54178 | 0.97949 | 1.54178 | 1.54178 | 1.54178 |
| Rsym | 0.069 (0.481) | 0.070 (0.401) | 0.097 (0.502) | 0.058 (0.201) | 0.058 (0.163) | 0.043 (0.131) | 0.035 (0.212) | 0.034 (0.138) |
| Unique reflections | 79450 (2513) | 84470 (2046) | 55770 (2337) | 5394 (2556) | 6637 (905) | 7790 (2648) | 5868 (2933) | 7320 (3759) |
| Completeness (%) | 99.4 (89) | 91.0 (64) | 94.1 (80) | 89.3 (85) | 85.4 (59) | 97.3 (99) | 93.5 (93) | 94.0 (97) |
| Data redundancy | 12.9 (4.2) | 6.4 (5.5) | 5.6 (5.3) | 10.2 (5.6) | 7.1 (5.5) | 5.5 (5.3) | 4.5 (3.7) | 4.2 (2.5) |
| Average I/σI | 35.2 (3.4) | 18.4 (4.6) | 12.8 (3.4) | 33.0 (4.6) | 26.7 (6.7) | 26.9 (5.5) | 25.1 (4.9) | 27.8 (5.5) |
| Rwork (%) | 15.2 (19.5) | 18.5 (20.5) | 22.0 (25.8) | 23.6 (24.2) | 22.2 (29.5) | 21.0 (25.1) | 22.0 (24.5) | 21.2 (22.2) |
| Rfree (%) | 18.2 (22.4) | 22.7 (26.9) | 27.1 (34.5) | 25.2 (29.7) | 25.9 (36.0) | 24.6 (31.2) | 26.2 (32.1) | 24.1 (30.8) |
| Rwork/Rfree (no hydrogen) | 15.9/18.9 | 18.5/22.9 | 21.7/27.0 | 23.5/25.4 | 21.9/25.3 | 20.9/22.6 | 21.6/24.8 | 21.0/24.8 |
| r.m.s.d. bond length (Å) | 0.005 | 0.004 | 0.004 | 0.004 | 0.005 | 0.003 | 0.004 | 0.004 |
| r.m.s.d. bond angle (°) | 1.043 | 0.883 | 0.800 | 0.803 | 0.825 | 0.767 | 0.822 | 0.844 |
| Total atoms including hydrogen | 11199 | 28642 | 27132 | 2599 | 2894 | 2839 | 2666 | 2784 |
| Solvent atoms | 836 | 737 | 211 | 12 | 10 | 28 | 10 | 21 |
| Average B-value | 19.9 | 46.0 | 52.1 | 87.7 | 82.0 | 74.9 | 92.8 | 79.8 |
| Ramachandrana favored/allowed (%) | 99.1/100 | 98.8/100 | 98.2/100 | 98.8/100 | 99.0/100 | 98.3/100 | 98.2/100 | 98.3/100 |
| Favorable rotamersa (%) | 100 | 99.9 | 100 | 100 | 100 | 100 | 100 | 100 |
| All-atom clash scorea | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.36 | 0.0 | 0.0 |
a Allowed/favorable Ramachandran angles, allowed rotamers, and the all-atom clash score (number of steric overlaps ≥0.4 Å/1000 atoms) were calculated using MolProbity (17).
For the structural comparisons shown in Fig. 5, individual subunits were initially aligned to a single reference structure using PyMOL. For each pairwise combination of structures, a displacement between corresponding Cα positions (e.g. residue 60) was calculated, and then these values were averaged over the entire set of comparisons involving that residue. If the Cα atom was missing in one or both structures, a displacement between 5 and 7 Å was assigned using a random number algorithm.
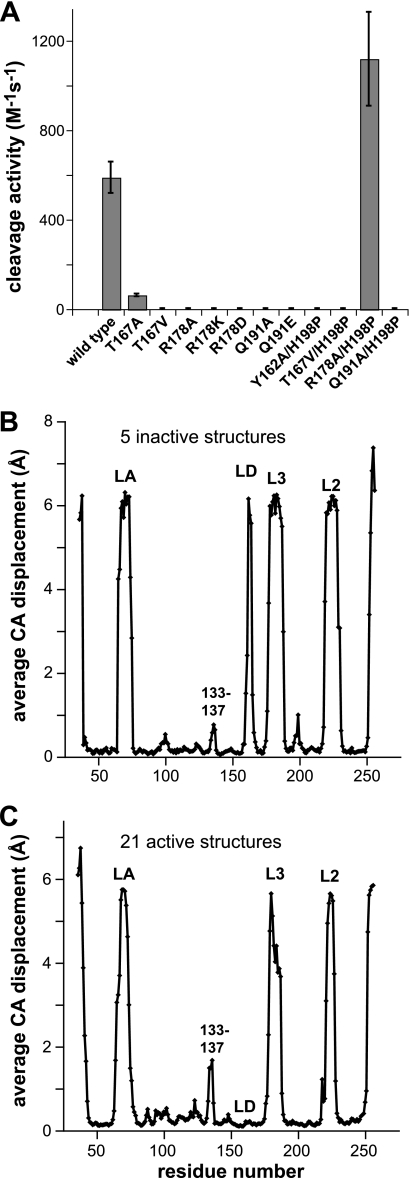
FIGURE 5.
Activities and structural properties of DegSΔPDZ mutants and variants. A, steady-state rate of cleavage of a sub-Km concentration of 35S-labeled RseA by wild-type DegSΔPDZ and single and double mutant variants. For variants with detectable activity, values are averages (n = 3) ± 1 S.D. For variants with undetectable activity, error bars were set to a value of 5 m−1 s−1, which represents the approximate sensitivity of the assay. Assay conditions were the same as in Fig. 2A. B, subunits from the inactive R178A, Q191A, Y162A, Y162A/H198P, and T167V/H198P DegSΔPDZ structures were aligned, and an average displacement for each Cα (CA) position in all pairwise combinations was calculated and plotted. High values represent regions that are disordered or have different conformations in different subunits. Low values represent regions that are very similar in all structures. C, average Cα (CA) displacements following alignment of 21 active subunits from the structures of wild-type DegSΔPDZ, DFP-modified DegSΔPDZ, and H198P DegSΔPDZ.
RESULTS
H198P Mutation Increases DegSΔPDZ Activity and Active Site Reactivity
Each wild-type or mutant variant of DegS (residues 27–355) or DegSΔPDZ (residues 27–256) lacked the wild-type membrane anchor and contained His6 affinity tags as described (3, 6). We previously found that the H198P substitution in DegS increased basal RseA cleavage in the absence of OMP peptide and enhanced peptide-stimulated cleavage under conditions of limiting substrate (8). These results were explained by crystal structures that revealed packing contacts between Pro198 and Tyr162, which stabilize the active oxyanion hole and thus shift the conformational equilibrium to favor active DegS (8). As a result, the H198P mutation should only increase DegSΔPDZ activity if this truncated enzyme also exists as an equilibrium mixture of inactive and active species.
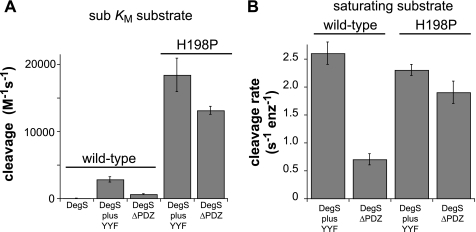
Earlier studies comparing DegSΔPDZ and peptide-activated DegS used non-quantitative assays or utilized OMP peptides that are now known to be suboptimal for activation (6, 7, 12). To eliminate these problems, we measured cleavage of a sub-Km concentration of 35S-labeled RseA and used saturating concentrations of an YYF tripeptide, which is as potent as any known OMP peptide (7). Under these conditions, DegSΔPDZ cleaved RseA ∼4.5-fold more slowly than peptide-activated DegS (Fig. 2A). Importantly, however, H198P DegSΔPDZ cleaved RseA much faster than either wild-type DegSΔPDZ or YYF-activated DegS and at ∼70% the rate of YYF-activated H198P DegS (Fig. 2A). When we determined the steady-state rate of RseA cleavage at saturating substrate concentrations, peptide-activated DegS was slightly more active than peptide-activated H198P DegS, and H198P DegSΔPDZ had ∼85% the activity of the latter enzyme (Fig. 2B). These results confirm that the H198P mutation increases DegS activity only by increasing the concentration of active relative to inactive species and also strongly support an allosteric model in which this mutation also stabilizes an active conformation of DegSΔPDZ.
FIGURE 2.
Proteolytic activities of wild-type and mutant DegS variants. A, steady-state rates of cleavage of a sub-Km concentration of 35S-labeled RseA(121–216) with a C-terminal His6 tag were determined using wild-type DegS (0.9 μm trimer), H198P DegS (0.3 μm trimer), wild-type DegSΔPDZ (0.9 μm trimer), or H198P DegSΔPDZ (0.3 μm trimer). The concentration of the YYF tripeptide, if present, was 300 μm. Rates were divided by the total enzyme (enz) and substrate concentrations to generate the values shown on the y axis, which are averages (n = 3) ± 1 S.D. B, Vmax values for cleavage of 35S-labeled RseA(121–216) were calculated by fitting the substrate dependence of cleavage to the Hill form of the Michaelis-Menten equation. Error bars represent the error of the non-linear least square fit. When present, the YYF peptide concentration was 300 μm.
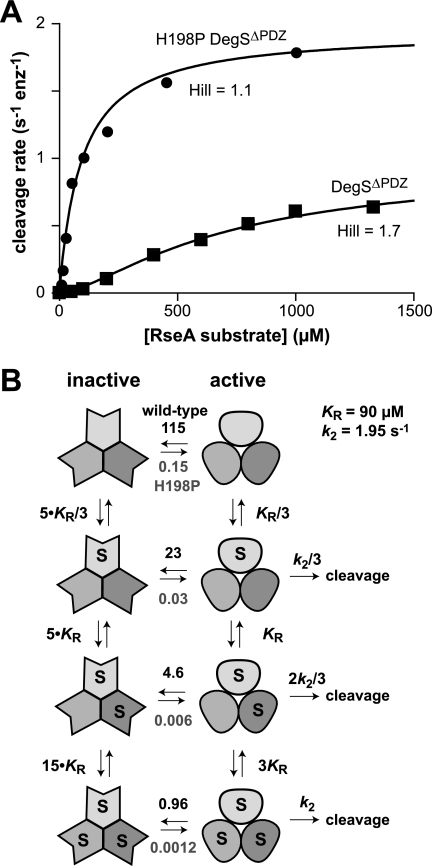
If the H198P mutation stabilizes an active conformation relative to an inactive conformation of DegSΔPDZ, then both the Hill constant and apparent Km (substrate concentration at half-maximal activity) should be lower for the mutant than for the wild-type truncated enzyme (6). To test this prediction, we assayed the steady-state rate of cleavage as a function of RseA concentration (Fig. 3A). Cleavage by DegSΔPDZ showed substantial positive cooperativity (Hill constant, 1.7 ± 0.1), and half-maximal degradation required a substrate concentration of ∼650 μm as expected from previous studies (6). By contrast, cleavage by H198P DegSΔPDZ showed almost no cooperativity (Hill constant, 1.1 ± 0.1), and half-maximal degradation was observed at a concentration of ∼90 μm. These results support a model in which the majority of unliganded DegSΔPDZ molecules assume an inactive conformation in solution with substrate binding and the H198P mutation acting to shift the equilibrium to favor the active structure. A model of Monod-Wyman-Changeux allostery (Fig. 3B) in which substrate binds 5-fold more tightly to active than inactive DegSΔPDZ with a conformational equilibrium constant of 115 for the unliganded wild-type species and 0.15 for the H198P mutant produced good fits of the experimental data in Fig. 3A (R2 > 0.986). For wild-type DegSΔPDZ, this model predicts that 1% of the enzymes are active in the absence of the substrate, and 48% assume the active conformation in the presence of saturating RseA substrate. For H198P DegSΔPDZ, these values are 87 and 99%, respectively.
FIGURE 3.
Allosteric model for DegSΔPDZ cleavage. A, substrate dependence of the steady-state rate of cleavage of RseA(121–216) by wild-type DegSΔPDZ (0.9 μm trimer) or H198P DegSΔPDZ (0.3 μm trimer). The solid lines were calculated from the model, equilibrium constants, and kinetic parameters shown in B. enz, enzyme. B, Monod-Wyman-Changeux allosteric model for DegSΔPDZ activity. The conformational equilibrium constants ([active]/[inactive]) are shown above the equilibrium arrows for wild-type DegSΔPDZ and below the equilibrium arrows for wild-type H198P DegSΔPDZ.
Active DegSΔPDZ Structures
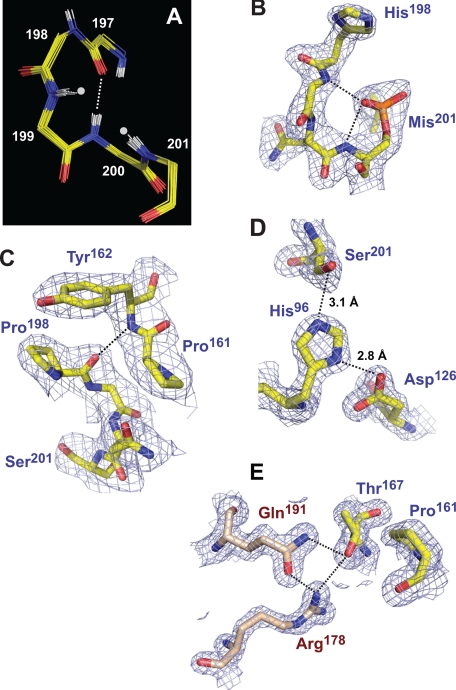
We solved crystal structures of DegSΔPDZ at 1.65-Å resolution, of H198P DegSΔPDZ at 2.7-Å resolution, and of DegSΔPDZ modified by reaction with DFP at 2.35-Å resolution (Table 1). The latter structure is a mimic for the substrate-bound acyl enzyme. In each of the 21 crystallographically independent subunits in these three structures, the oxyanion hole was in the active conformation (Fig. 4A). In the structure of DFP-treated DegSΔPDZ, the active site serine (Ser201) was modified to monoisopropylphosphorylserine (Mis201), and the O1P oxygen of this residue occupied the oxyanion hole and accepted hydrogen bonds from the backbone -NH groups of Gly199 and Ser201 (Fig. 4B). The side chain of Pro198 in H198P DegSΔPDZ packed against the aromatic ring of Tyr162 (Fig. 4C) as observed previously in the structure of a full-length DegS variant containing the H198P substitution (8). This interaction should increase the stability of the functional conformation of the oxyanion hole by helping to stabilize the hydrogen bond between the backbone -NH of Tyr162 and the carbonyl oxygen of Pro198 (Fig. 4C).
FIGURE 4.
Structural features of active DegSΔPDZ structures. A, overlay of the polypeptide backbones of residues 197–201 following global alignment of 21 crystallographically independent active subunits from the wild-type, DFP-modified, and H198P structures. The backbone -NH groups of residues 199 and 201 (marked by gray dots) form the oxyanion hole of the active site. The dashed line represents a hydrogen bond between the -NH of residue 200 and the carbonyl oxygen of residue 197. B, model and electron density (contoured at 1.5 σ) for residues 198–201 of subunit B of the DFP-modified structure (Protein Data Bank code 3LH3). The dashed lines represent hydrogen bonds between the O1P atom of Mis201 (a proxy for the backbone oxygen of a substrate in the acyl enzyme intermediate) and the oxyanion hole. For simplicity, hydrogens are not shown in this and subsequent panels. C, model and electron density (contoured at 1.1 σ) for residues 161–162 and 198–201 of subunit A of the H198P structure (Protein Data Bank code 3LGV). The pyrrolidine ring of Pro198 packs against the aromatic ring of Tyr162 and stabilizes a backbone hydrogen bond (dashed line) between these residues. D, model and electron density (contoured at 1.5 σ) for the catalytic triad of subunit A in the wild-type structure (Protein Data Bank code 3LGI). The hydrogen bond lengths are typical of active serine proteases. E, model and electron density (contoured at 1.5 σ) for Thr167 (chain C), Arg178 (chain A), and Gln191 (chain A) in the wild-type structure (Protein Data Bank code 3LGI). The intersubunit hydrogen bond network linking these side chains (dashed lines) is present in all active DegSΔPDZ subunits. Packing between Thr167 and Pro161 and between Pro161 and residues that form the oxyanion hole (see C) connects this network to the proteolytic active site.
Based on a DegSΔPDZ structure, Hasselblatt et al. (12) concluded that Asp126 in the catalytic triad was too far from His96 to allow efficient activation of Ser201 for nucleophilic attack on the RseA substrate. Our results do not support this model. In the catalytic triad shown in Fig. 4D, for example, the hydrogen bond between the Oδ2 oxygen of Asp126 and Nδ2 nitrogen of His96 is 2.8 Å. This value is very similar to the distances observed in peptide-activated DegS and other serine proteases (4, 8, 12, 18). Similarly, the hydrogen bond between the Nϵ2 nitrogen of His96 and Oγ oxygen of Ser201 is 3.1 Å (Fig. 4D), which is typical of serine proteases and active DegS. We did find that the catalytic triads in many DegSΔPDZ subunits were disrupted by rotation of the His96 side chain away from Ser201 and Asp126. Importantly, however, there was no correlation between solution activity and the number of irregular active sites in various structures. For example, His96 assumed a rotamer incompatible with catalysis in seven of nine subunits in the asymmetric unit of the H198P DegSΔPDZ crystal. However, this enzyme variant was substantially more active than wild-type DegSΔPDZ in which only two of 15 rotamers of His96 were aberrant in the structures reported here and previously (Protein Data Bank codes 2QF0 and 2QF3; Ref. 6). Moreover, the irregular rotamer of His96 was observed in six of nine subunits in DFP-modified DegSΔPDZ, and yet these active sites had to be functional for modification of Ser201 to occur in the first place. His96 probably equilibrates between active and inactive rotamer conformations in all of these structures with small changes in crystal packing influencing the rotamer observed.
Mutations That Reduce DegSΔPDZ Activity
Previously, we found that the R178A mutation severely reduced the activity of DegS and DegSΔPDZ (6). In our active DegSΔPDZ structures, the side chains of Arg178 and Gln191 were hydrogen-bonded to each other and to the side chain of Thr167 from an adjacent subunit (Fig. 4E). The Thr167-Arg178-Gln191 network is not present in the structures of DegSΔPDZ and peptide-bound DegS reported by Hasselblatt et al. (12) and Wilken et al. (4). However, this absence is likely to reflect model-building errors as the hydrogen bonds we observed could be introduced into those structures by changing the Thr167 rotamer with good fits to the electron density. Formation of the hydrogen bond network also sets up a packing interaction between the side chain of Thr167 and Pro161 (Fig. 4E), which in concert with Tyr162 helps stabilize the functional conformation of the oxyanion hole (Fig. 4C). In prior studies, the Y162A DegSΔPDZ variant was found to be inactive in RseA cleavage (6).
To test the importance of the Thr167-Arg178-Gln191 network, we constructed, purified, and assayed the cleavage activity of DegSΔPDZ variants with T167A, T167V, R178K, R178D, Q191A, and Q191E mutations. All of these variants formed stable trimers as determined by gel filtration chromatography (data not shown). Nevertheless, only one of these mutants (T167A) had detectable activity, which was ∼10% the value of wild-type DegSΔPDZ (Fig. 5A). Importantly, an isosteric substitution at the same position (T167V) produced an inactive enzyme (Fig. 5A), suggesting that a water molecule may partially substitute for the missing side-chain hydroxyl in T167A DegSΔPDZ. We conclude that interactions between the side chains of Arg178, Gln191, and Thr167 play important roles in maintaining DegSΔPDZ activity.
To probe for potential rescue of activity, we combined the H198P substitution with the Y162A, T167V, R178A, or Q191A mutations in DegSΔPDZ. The Y162A/H198P, T167V/H198P, and Q191A/H198P DegSΔPDZ mutants had no detectable activity (Fig. 5A). Surprisingly, however, R178A/H198P DegSΔPDZ was more active than wild-type DegSΔPDZ (Fig. 5A). Because the Y162A, T167V, and Q191A mutations were not rescued by the H198P substitution, it appears that Arg178 plays a less important role than Tyr162, Thr167, or Gln191 in stabilizing the active protease domain.
Structural Features of Inactive Mutants and Allosteric Transition
In principle, different mutations might inactivate DegSΔPDZ in diverse ways or by stabilizing a common inactive conformation. To evaluate these possibilities, we carried out crystallization trials using inactive DegSΔPDZ mutants and obtained structures for the Y162A, Q191A, T167V/H198P, and Y162A/H198P variants (Table 1). We also rerefined a previously reported structure of R178A DegSΔPDZ (Table 1). As described below, these structures had malformed oxyanion holes and resembled the protease domains in peptide-free DegS (4, 5).
Pairwise comparisons of aligned subunits from the five inactive DegSΔPDZ structures produced an average displacement of corresponding Cα atoms of 0.18 ± 0.11 Å for 168 residue positions but also revealed substantial structural variations at the N and C termini, in residues 133–137, and in parts of the LA, LD, L3, and L2 loops (Fig. 5B). Parts or all of these regions were disordered or assumed different structures in different subunits. Similar variations were observed in pairwise comparisons of the 21 active DegSΔPDZ subunits (Fig. 5C) with the notable exception of the LD loop, which was well ordered in all active structures and showed little structural variation. It seems likely that the remaining highly variant regions play little if any role in stabilizing either the active or inactive proteolytic conformation. In pairwise alignments of trimers (Table 2), the average root mean square deviations were 0.32 ± 0.08 Å between active trimers (21 comparisons), 0.21 ± 0.09 Å between inactive trimers (10 comparisons), and 0.68 ± 0.10 Å for active versus inactive trimers (35 comparisons).
TABLE 2.
Pairwise alignments of active and inactive DegSΔPDZ trimers
The top number is the root mean square deviation (Å), and the bottom number is the number of Cα comparisons. Italics mark comparisons of active with inactive structures. DFP mod, DFP-modified.
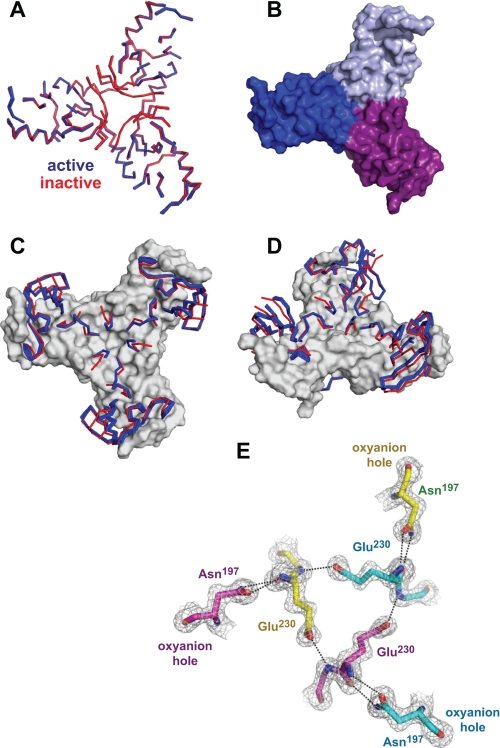
Careful comparisons of the active and inactive DegSΔPDZ subunits revealed a common set of backbone positions, which aligned extremely well in individual subunits as well as in intact trimers. These amino acids (45–58, 82–84, 90–93, 117–121, 139–146, 151–159, 167–175, 189–194, 204–216, and 233–249) form a solid triangular base (Fig. 6, A and B), which remains essentially invariant in the conversion of the inactive to the active enzyme. Most of the remaining residues, which switch conformations between the active and inactive structures, project upward from this base (Fig. 6, C and D).
FIGURE 6.
Common and varied features in active and inactive DegSΔPDZ structures. A, ribbon representation of backbone positions in the “base” (residues 45–58, 82–84, 90–93, 117–121, 139–146, 151–159, 167–175, 189–194, 204–216, and 233–249) that are highly conserved following structural alignment of seven active (blue) and five inactive (red) DegSΔPDZ trimers. B, surface representation of the trimer base from the wild-type structure (Protein Data Bank code 3LGI). C and D, ordered backbone positions that switch conformation between the active (blue) and inactive (red) structures are shown in ribbon representation. The conserved base is shown in surface representation. E, model and electron density (1.5 σ) for residues Asn197, Glu230, and Gly231 in the wild-type DegSΔPDZ trimer (Protein Data Bank code 3LGI): yellow carbons, chain A; cyan carbons, chain B; purple carbons, chain C. All active trimers contain the same network of hydrogen bonds (dashed lines), which link the oxyanion holes of different subunits.
Importantly, the inactive structures showed a common set of differences when compared with active DegSΔPDZ structures. For example, in each inactive variant, residues 229–231 in the L2 loop had different conformations than in active structures, the oxyanion hole assumed a non-functional conformation, residues 161–164 in the LD loop were disordered or assumed different conformations than in active structures, Gln191 and Thr167 were not hydrogen-bonded, and side-chain density for Arg178 was absent. Except for the final feature, these differences are also observed in the protease domains of full-length DegS in active and inactive conformations (4, 5, 8, 12). In active structures, by contrast, the side chains of Glu230 form a network of polar interactions around the 3-fold axis, which is coupled via interactions with Asn197 to the oxyanion hole of the active sites (Fig. 6E). The oxyanion hole, in turn, interacts with residues in the LD loop and the Thr167-Arg178-Gln191 network (Fig. 4, C and E). These connections, which are lost in the non-functional structures, collectively link the active sites of different DegSΔPDZ subunits and are probably responsible for the apparent all-or-none character of the allosteric transition.
The foregoing results support a model in which trimers of DegSΔPDZ can assume two basic structures. Mutations such as H198P stabilize the active conformation, whereas mutations such as Y162A, T167V, R178A, and Q191A stabilize the inactive conformation. Combining stabilizing and destabilizing mutations in the T167V/H198P and Y162A/H198P DegSΔPDZ variants resulted in enzymes that were inactive in solution and assumed the non-functional conformation in crystals. Thus, the H198P mutation only stabilizes the active conformation of the protease domain in concert with interactions mediated by other residues.
DISCUSSION
The studies presented here provide strong evidence that the protease domain of DegS in the absence of the PDZ domain equilibrates between inactive and active conformations. Degradation of the RseA substrate by the DegSΔPDZ trimer is positively cooperative (Hill constant, 1.7 ± 0.1), supporting a model in which substrate binding stabilizes the active conformation relative to the inactive conformation. In principle, positive cooperativity could also be caused by favorable substrate-substrate interactions (see Ref. 6). However, a substrate interaction model does not account for our finding that the H198P mutation in DegSΔPDZ almost completely abolished positive cooperativity in RseA cleavage. The H198P mutation also reduced Km for RseA cleavage by DegSΔPDZ from ∼650 μm to less than 100 μm. This result provides additional support for the allosteric model, which predicts that some of the energy of RseA substrate binding to wild-type DegSΔPDZ is used to drive conversion of inactive to active enzymes. If most molecules of H198P DegSΔPDZ are already active, then RseA should bind more tightly to this variant than to wild-type DegSΔPDZ as was observed.
The crystal structures of H198P DegSΔPDZ and wild-type DegSΔPDZ are essentially the same with oxyanion holes characteristic of functional proteases. Pro198 in the mutant makes new packing interactions with Tyr162 that should stabilize the active fold relative to the inactive fold. Moreover, these structures are almost identical to that of DFP-modified DegSΔPDZ, which mimics the acyl intermediate in peptide bond cleavage, and to the protease domains in crystal structures of peptide-activated full-length DegS (4, 8, 12). Because functional studies indicate that the majority of wild-type DegSΔPDZ enzymes assume an inactive conformation in solution, crystallization probably traps the active conformation of this variant. We did observe an inactive conformation of DegSΔPDZ in crystal structures of mutant with substitutions that destabilize the active enzyme, strongly supporting the idea that wild-type DegSΔPDZ can also exist in active and inactive conformations.
How do the proteolytic activities of the functional conformations of DegSΔPDZ and full-length DegS compare? Because both enzymes can equilibrate between active and inactive conformations, answering this question is not straightforward. For example, depending on the OMP peptide used, the maximal activity of peptide-stimulated full-length DegS can be higher or lower than that of DegSΔPDZ (6, 7). When we activated full-length H198P DegS with the YYF peptide, which has stimulatory activity as high as any reported allosteric effector, Vmax for RseA cleavage was only ∼20% higher than that of H198P DegSΔPDZ. Moreover, YYF-stimulated H198P DegS appears to be completely in the functional conformation as we previously found that its activity was not increased further by mutations that allosterically increase the activity of wild-type DegS (7). Thus, although small differences may exist, the preponderance of evidence indicates that the catalytic activities of the functional conformations of DegS and DegSΔPDZ are similar. This result supports a model in which the principal function of the PDZ domain is to stabilize the inactive conformation of unliganded DegS and to modulate allosteric activation by allowing OMP peptides to bind with higher affinity to active DegS than to inactive DegS (7, 8). Indeed, some OMP peptides bind only marginally more tightly to active DegS, and thus even saturating concentrations of these peptides result in lower activity than observed with DegSΔPDZ (7). For example, full-length DegS in the presence of a saturating OMP peptide with the sequence KRRKGKVYYF cleaved a sub-Km concentration of RseA ∼8-fold more slowly than did DegSΔPDZ, which our results show is only partially active because of conformational equilibration.
Variants of DegSΔPDZ with substitutions for Tyr162, Thr167, Arg178, or Gln191 have very low activity and crystallize in similar conformations in which the oxyanion hole of the active site is malformed. These conformations resemble those observed in peptide-free full-length DegS (4, 5). In the parlance of two-state allostery, these inactive structures correspond to the “tense” enzyme conformation, whereas the active structures define the “relaxed” conformation (19). Notably, side-chain contacts mediated by Tyr162, Thr167, Arg178, or Gln191, which appear to stabilize active DegSΔPDZ, were not observed in any of the inactive structures. By definition, the unliganded tense conformation must be thermodynamically more stable in solution than the unliganded relaxed conformation. Thus, we searched for structural features of the “inactive” DegSΔPDZ structures that might account for increased stability relative to the “active” conformation. Surprisingly, there were no obvious tertiary or quaternary interactions in the inactive structures that were absent in the active structures. In fact, the active structures had more hydrogen bonds, salt bridges, and buried surface area than the inactive structures. The inactive structures did have more disordered residues, including parts of the LD and L2 loops, and it is possible, therefore, that the entropic cost of ordering these residues makes the active conformation less stable than the inactive conformation.
By comparing our collection of active and inactive protease domain structures, numerous features of the active structures were identified that were absent in the inactive structures. In addition to the contacts made by Tyr162, Thr167, Arg178, and Gln191, the active structures contained additional polar interactions mediated by the side chains of Asn197 and Glu230 and packing interactions mediated by Pro161, Leu164, Phe220, Pro229, Ile232, and Phe234. Important roles in allosteric switching have also been proposed for Ile179, Pro183, Gln187, Asp221, and Glu227 (see Refs. 4 and 12), but we found no consistent interactions that were mediated by these side chains and that differed between our active and inactive structures. Moreover, we previously found that DegS variants bearing the P183A and E227A mutations were as active or more active than wild-type DegS (6). Additional mutagenesis experiments will be required to determine whether any of the remaining candidate residues play important roles in allosteric switching.
The proteolytic activities of DegS and DegSΔPDZ are modeled well by the Monod-Wyman-Changeux mechanism of allostery (19) in which all subunits of any given trimer assume either the inactive or the active conformation as is observed in structures of DegS and DegSΔPDZ. Moreover, a strictly sequential allosteric model (20) in which only ligand-bound subunits have the active conformation is ruled out by the fact that unliganded DegSΔPDZ adopts the active conformation in crystals. A generalized allosteric model has also been proposed in which all possible combinations of symmetric and asymmetric oligomers are allowed (21). This general model provides an equally good description of DegS allostery with the stipulation that asymmetric trimers are poorly populated relative to symmetric trimers and may provide a more realistic pathway for switching between fully inactive and fully active trimers. Indeed, examination of active and inactive DegS trimers revealed no obvious structural features that would exclude the existence of trimers with a mixture of active and inactive subunits.
The ability of a DegSΔPDZ variant to induce the envelope stress response in vivo was found to be lower than that of peptide-stimulated DegS (3). Although this observation was originally interpreted as evidence that peptide binding is required for some positive stimulatory role in the intact enzyme (4, 5), our current results suggest that the reduced intracellular activity of DegSΔPDZ reflects the fact that substrate binding alone is not sufficient to fully switch an entire population of truncated enzymes into the active conformation. We anticipate, therefore, that a variant like H198P DegSΔPDZ would have higher activity in vivo and thus could be useful in probing potential OMP-independent inputs into the envelope stress pathway (11).
Our finding that the isolated protease domain of DegS can function as an allosteric enzyme raises the possibility that an evolutionary precursor functioned in exactly this fashion. DegS can cleave peptide bonds with a wide variety of P1 and P1′ residues flanking the scissile peptide bond (22), and its active sites are highly exposed. Thus, an intracellular variant that was constitutively active and unregulated might function as a rogue protease and damage numerous cellular proteins. By having the inactive conformation be the stable form, however, only substrates that bind sufficiently tightly and specifically to the active conformation would be cleaved. Strong positive cooperativity would also ensure that most active protease trimers were fully saturated with the proper substrate, minimizing the number of unoccupied but functional active sites. By fusing an allosteric protease of this type to a PDZ domain, an extra layer of ligand-mediated regulation could then be added to generate enzymes of the current DegS family.
Most DegS homologs consist of the protease domain and one or more PDZ domains. Modern biology may also have found uses for the trimeric protease domain in the absence of the PDZ domain as human HtrA proteases have isoforms that lack parts or all of the PDZ domain due to alternative splicing (23). It remains to be seen whether the allosteric potential of these natural ΔPDZ enzymes is utilized and, if so, whether allosteric effectors other than protein substrates are involved in regulation. The allosteric nature of proteases has long been recognized as an important component of many zymogen activation reactions. Reversible allosteric regulation of protease conformation is an essential component of DegS function, and a growing number of studies suggest that other mature proteolytic enzymes without additional regulatory domains can also be allosterically regulated (24–26).
Acknowledgment
We thank Seokhee Kim for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Postdoctoral Fellowship F32AI-074245-03 (to J. S.), Grant AI-16892, and Award RR-15301 from the National Center for Research Resources. This work was also supported by the United States Department of Defense Office of Basic Energy Sciences under Contract DE-AC02-06CH11357.
- OMP
- outer membrane porin
- DFP
- diisopropylfluorophosphate
- Mis
- monoisopropylphosphorylserine.
REFERENCES
- 1.Neurath H., Walsh K. A. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stroud R. M., Kossiakoff A. A., Chambers J. L. (1977) Annu. Rev. Biophys. Bioeng. 6, 177–193 [DOI] [PubMed] [Google Scholar]
- 3.Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. (2003) Cell 113, 61–71 [DOI] [PubMed] [Google Scholar]
- 4.Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T. (2004) Cell 117, 483–494 [DOI] [PubMed] [Google Scholar]
- 5.Zeth K. (2004) FEBS Lett. 569, 351–358 [DOI] [PubMed] [Google Scholar]
- 6.Sohn J., Grant R. A., Sauer R. T. (2007) Cell 131, 572–583 [DOI] [PubMed] [Google Scholar]
- 7.Sohn J., Sauer R. T. (2009) Mol. Cell 33, 64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohn J., Grant R. A., Sauer R. T. (2009) Structure 17, 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D. Y., Kim K. K. (2005) J. Biochem. Mol. Biol. 38, 266–274 [DOI] [PubMed] [Google Scholar]
- 10.Alba B. M., Gross C. A. (2004) Mol. Microbiol. 52, 613–619 [DOI] [PubMed] [Google Scholar]
- 11.Ades S. E. (2008) Curr. Opin. Microbiol. 11, 535–540 [DOI] [PubMed] [Google Scholar]
- 12.Hasselblatt H., Kurzbauer R., Wilken C., Krojer T., Sawa J., Kurt J., Kirk R., Hasenbein S., Ehrmann M., Clausen T. (2007) Genes Dev. 21, 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 14.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 16.Word J. M., Lovell S. C., Richardson J. S., Richardson D. C. (1999) J. Mol. Biol. 285, 1733–1747 [DOI] [PubMed] [Google Scholar]
- 17.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leskovac V., Trivic S., Pericin D., Popovic M., Kandrac J. (2008) J. Serb. Chem. Soc. 73, 393–403 [Google Scholar]
- 19.Monod J., Wyman J., Changeux J. P. (1965) J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 20.Koshland D. E., Jr., Némethy G., Filmer D. (1966) Biochemistry 5, 365–385 [DOI] [PubMed] [Google Scholar]
- 21.Hammes G. G., Wu C. W. (1971) Science 172, 1205–1211 [DOI] [PubMed] [Google Scholar]
- 22.Li X., Wang B., Feng L., Kang H., Qi Y., Wang J., Shi Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie G. Y., Li Y., Minoura H., Batten L., Ooi G. T., Findlay J. K., Salamonsen L. A. (2003) Mol. Hum. Reprod. 9, 279–290 [DOI] [PubMed] [Google Scholar]
- 24.Hardy J. A., Lam J., Nguyen J. T., O'Brien T., Wells J. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheer J. M., Romanowski M. J., Wells J. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy J. A., Wells J. A. (2009) J. Biol. Chem. 284, 26063–26069 [DOI] [PMC free article] [PubMed] [Google Scholar]