Abstract
The basolateral Na+-K+-2Cl− cotransporter (NKCC1) is a key determinant of transepithelial chloride secretion and dysregulation of chloride secretion is a common feature of many diseases including secretory diarrhea. We have previously shown that activation of protein kinase C (PKC) markedly reduces transepithelial chloride secretion in human colonic T84 cells, which correlates with both functional inhibition and loss of the NKCC1 surface expression. In the present study, we defined the specific roles of PKC isoforms in regulating epithelial NKCC1 and chloride secretion utilizing adenoviral vectors that express shRNAs targeting human PKC isoforms (α, δ, ϵ) (shPKCs) or LacZ (shLacZ, non-targeting control). After 72 h of adenoviral transduction, protein levels of the PKC isoforms in shPKCs-T84 cells were decreased by ∼90% compared with the shLacZ-control. Activation of PKCs by phorbol 12-myristate 13-acetate (PMA) caused a redistribution of NKCC1 immunostaining from the basolateral membrane to intracellular vesicles in both shLacZ- and shPKCα-T84 cells, whereas the effect of PMA was not observed in shPKCδ- and shPKCϵ- cells. These results were further confirmed by basolateral surface biotinylation. Furthermore, activation of PKCs by PMA inhibited cAMP-stimulated chloride secretion in the uninfected, shLacZ- and shPKCα-T84 monolayers, but the inhibitory effect was significantly attenuated in shPKCδ- and shPKCϵ-T84 monolayers. In conclusion, the activated novel isoforms PKCδ or PKCϵ, but not the conventional isoform PKCα, inhibits transepithelial chloride secretion through inducing internalization of the basolateral surface NKCC1. Our study reveals that the novel PKC isoform-regulated NKCC1 surface expression plays an important role in the regulation of chloride secretion.
Keywords: Chloride Transport, Epithelial Cell, Protein Kinase C (PKC), shRNA, Trafficking, NKCC1, RNAi, Adenovirus, Endocytosis, Ion Transporter
Introduction
The Na-K-2Cl cotransporter (NKCC1),3 encoded by the SLC12A2 gene, is a member of the cation-chloride cotransporter superfamily and is widely expressed in epithelial and non-epithelial cell types (1). In secretory epithelia, NKCC1 is exclusively expressed at the basolateral membrane (1, 2) and plays a central role in transepithelial chloride (Cl−) transport, the physiological process that largely accounts for mucosal hydration and fluid secretion by the gastrointestinal tract, airway, and exocrine organs (3, 4). Dysregulation of chloride secretion is a common feature of many diseases including cystic fibrosis and secretory diarrhea (3).
Chloride secretion requires the coordinated function of various ion pumps, transporters, and channels that are selectively arrayed in either the basolateral or apical membrane of polarized epithelial cells. Basolateral chloride entry into epithelial cells is accomplished by NKCC1, which transports one Na+, one K+, and two Cl− into the intracellular compartment driven by the favorable Na+ gradient generated by the Na+/K+-ATPase (1). Potassium is recycled across the basolateral membrane by K+ channels, which also serve to maintain the negative intracellular potential that drives electrogenic chloride exit across the apical membrane via various regulated chloride channels (2, 5). NKCC1 plays a pivotal role in setting the net secretory capacity of the epithelium (5–8). NKCC1 is an electroneutral facilitated transporter and, as such, responds to the transmembrane chemical gradients of the cotransported ions. However, overall NKCC1 function has been shown to be determined by a number of additional factors including NKCC1 phosphorylation/dephosphorylation, gene expression, and cell surface expression (6, 9, 10).
Protein kinase C (PKC) is a family of serine-threonine kinases that regulate a variety of physiological processes including ion transport. Eleven isoforms of PKC have been identified to date and are grouped into conventional (cPKC), novel (nPKC), and atypical PKC based on activation requirements and structural characteristics (11). Previous studies from our laboratory showed that activation of PKC by phorbol 12-myristate 13-acetate (PMA) and the non-phorbol PKC activator bryostatin-1 and carbachol (CCh, a M3 muscarinic receptor agonist) inhibits epithelial chloride secretion in the human colonic crypt T84 epithelial cell line (12–14). PKC-dependent inhibition of chloride secretion correlated closely with a marked reduction of NKCC1 function, which in turn was paralleled by a loss of NKCC1 units from the basolateral membrane, and, subsequently, a reduction in gene expression (12, 15).
A number of membrane transporters such as the sodium-dependent phosphate transporter (16), human cationic amino acid transporter hCAT-1 (17), dopamine transporter DAT (18), and glutamate transporter GLT-1 (19) have been shown to be regulated by PKC. Importantly, it has recently been reported that PKC activation regulates membrane transporter trafficking, thereby leading to the regulation of the transporter biological function (20–22). Using a Madin-Darby canine kidney cell line stably transfected with eGFP-tagged NKCC1, we have recently demonstrated that activation of PKC induces rapid internalization of NKCC1 associated with the clathrin-dependent endocytic pathway (23). In most instances, the specific PKC isoform(s) responsible for transporter regulation have not yet been elucidated. We previously used a series of pharmacological inhibitors to indirectly link the novel PKCϵ isoform to PMA inhibition of chloride secretion in T84 cells (9, 13). However, the specificity and selectivity of these agents are imperfect.
The T84 colonic epithelial cell is a widely used and well characterized model for investigating the regulation of transepithelial chloride secretion (6, 24, 25). However, effective delivery of small interference RNA (siRNA) duplex and exogenous genes into T84 cell monolayers has proven to be a difficult technical challenge. In the present study, we developed small hairpin RNA (shRNA)-expressing adenoviral constructs specific to human PKC isoforms and a novel effective adenoviral delivery approach to directly address the specific mechanistic roles of PKCα, PKCδ, and PKCϵ isoforms in regulating chloride secretion in the T84 cell model. Our results demonstrated that the novel adenoviral approach for delivering PKC shRNAs resulted in ∼90% knockdown of PKC proteins in T84 monolayers. Immunofluorescence staining and basolateral surface biotinylation showed that specific knockdown of PKCδ or PKCϵ, but not PKCα, prevented loss of basolateral surface expression of NKCC1 in T84 monolayers during PMA exposure. Electrophysiological studies demonstrated that specific knockdown of novel PKCδ or PKCϵ isoforms, but not the conventional PKCα isoform, prevented PMA inhibition of cAMP-stimulated chloride secretion in T84 monolayers. Our study reveals a critical role for the novel PKC isoforms in regulation of NKCC1 surface expression and, by implication, chloride secretion.
EXPERIMENTAL PROCEDURES
Reagents
Dulbecco's modified Eagle's medium (DMEM) with 4.5 g/liter of d-glucose and l-glutamine, Opti-MEM I reduced serum medium, fetal bovine serum (FBS), penicillin/streptomycin, and Lipofectamine 2000 were purchased from Invitrogen. The DMEM/F-12 50/50 mixture with l-glutamine and 15 mm HEPES was purchased from Mediatech, Inc. (Manassas, VA). Phorbol 12-myristate 13-acetate (PMA) and protease inhibitors mixture set I were from Calbiochem (La Jolla, CA), and EGTA was from Fisher. Forskolin was obtained from Sigma. EZ-Link Sulfo-NHS-Biotin, ImmunoPure-immobilized streptavidin-agarose beads, and ECL Western blotting substrate were purchased from Pierce. Gel electrophoresis reagents and PVDF transfer membrane were obtained from Bio-Rad and GE Healthcare. All other chemicals were obtained from Fisher.
Antibodies
Primary antibodies used for Western blotting were purchased from following companies: rabbit anti-PKCα (catalog number 2056) and rabbit anti-PKCϵ (catalog number 2683S) were purchased from Cell Signaling Technology (Danvers, MA), rabbit anti-PKCδ (catalog number sc-937, 200 μg/ml) and rabbit anti-PKCβ1 (catalog number sc-209, 200 μg/ml) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, catalog number G8795, clone GAPDH-71.1, ∼1 mg/ml) from Sigma, and mouse anti-NKCC1 monoclonal antibody T4, developed by Dr. Lytle (26), was obtained from the Developmental Studies Hybridoma Bank operated under the auspices of the National Institutes of Health, NICHD, and maintained by the University of Iowa (Iowa City, IA). Horseradish peroxidase-conjugated secondary antibodies used for Western blotting were purchased from Jackson ImmunoResearch Laboratories, Inc.: goat anti-rabbit IgG (H+L), code number 111-035-003, 0.8 mg/ml; and donkey anti-mouse IgG (H+L), code number 715-035-150, 0.6 mg/ml. Primary antibodies used for immunofluorescence staining were obtained from Abcam Inc., Cambridge, MA (rabbit monoclonal anti-PKCα, catalog number ab32376, clone Y124), Santa Cruz Biotechnology, Inc. (rabbit anti-PKCδ, catalog number sc-937; and rabbit anti-PKCϵ, catalog number sc-214, 200 μg/ml), as well as from Abnova, Taiwan (rabbit anti-PKCϵ, catalog number PAB4537, 250 μg/ml) and Abgent, San Diego, CA (rabbit anti-PKCϵ, catalog number AP7019a, 250 μg/ml). Mouse anti-β-galactosidase monoclonal antibody (catalog number 2372, clone 14B7) was obtained from Cell Signaling Technology. Fluorescent-conjugated secondary antibodies Alexa Fluor 488 goat anti-mouse IgG (H+L) (catalog number A11029, 2 mg/ml) and Alexa Fluor 555 goat anti-rabbit IgG (H+L) (catalog number A21429, 2 mg/ml) were purchased from Invitrogen.
Cell Lines and Cell Culture
HEK 293A cells were obtained from Invitrogen and cultured in DMEM with 4.5 g/liter of d-glucose and l-glutamine supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin at 37 °C, in a 5% CO2 atmosphere incubator. T84 human colonic crypt epithelial cells (27) were obtained from Dr. James A. McRoberts (University of California, Los Angeles, CA). T84 cells were used from passage 32 to 40 and cultured in a DMEM/F-12 50/50 mixture with l-glutamine and 15 mm HEPES supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin at 37 °C, in a 5% CO2 atmosphere incubator. For use in electrophysiology or immunofluorescence experiments, Transwell® permeable supports with 6.5-mm insert diameter and 0.4-μm pore size polycarbonate membrane (catalog number 3413, Corning Incorporated, Corning, NY) were coated with 50 μl of 70 μg/ml of rat tail collagen (catalog number C3867–1VL, Sigma) in 50% ethanol for 2 h at 37 °C and dried for 1 h in a tissue culture hood by removal of the plate covers. T84 cells were seeded at 4 × 105 cells/cm2 on the collagen-coated supports and grown for 8–14 days with feeding at both the apical and basolateral sides every 3 days until confluent and the transepithelial resistance (TER) had reached at least 1000 Ω·cm2 (28). When used for determining viral multiplicity of infection (m.o.i.) or for cell surface biotinylation, T84 cells were grown until TER was at least 1000 Ω·cm2 on uncoated 24-mm diameter Transwell® permeable supports with 0.4-μm pores (catalog number 3412, Corning Inc.).
Construction and Screening of PKC shRNA-expressing U6 Entry Clones
To generate small hairpin RNA (shRNA)- expressing pENTRTM/U6 entry constructs, 21-bp sequences targeting mRNA sequences of human PKCα, -δ, and -ϵ isoforms were selected based on an siRNA design algorithm as previously reported (29, 30) and BLAST search against GenBankTM. Selected target sequences include human PKCα (PRKCA, GenBank accession number NM_002737), target 1, 5′-GCCTCCATTTGATGGTGAAGA-3′, target 2, 5′-GCGTCCTGTTGTATGAAATGC-3′, and target 3, 5′-GGGATCGAACAACAAGGAATG-3′; human PKCδ (PRKCD, GenBank accession number NM_006254), target 1, 5′-GCAAGAAGAACAATGGCAAGG-3′ and target 2, 5′-GCTTCAAGGTTCACAACTACA-3′; and human PKCϵ (PRKCE, GenBank accession number NM_005400), target 1, 5′-GCAGGGATACCAGTGTCAAGT-3′, target 2, 5′-GCAGGGTTTGCAGTGTAAAGT-3′ and target 3, 5′-GGACGTCATCCTTCAGGATGA-3′. Single-stranded DNA Oligo sequences encoding the shRNA of interest were designed following the guidelines of the BLOCK-it U6 RNAi Entry Vector kit (catalog number K4945-00, Invitrogen). Non-targeting lacZ control DNA oligo sequences were provided in the user manual. The single-stranded DNA oligos were annealed and cloned into a Gateway-adapted pERTR/U6 vector included in the kit following the manufacturer's instructions. All clones were sequenced by using both a forward primer 5′-GGACTATCATATGCTTACCG-3′ and reverse primer 5′-CAGGAAACAGCTATGAC-3′.
To screen PKC shRNA-expressing U6 entry clones for efficient knockdown of target genes, appropriate numbers of HEK 293A cells were seeded into 6-well plates so that the cells would be ∼90% confluent at the time of transfection. The cells were then transfected with 4 μg of DNA/well of PKC shRNA entry clones or non-targeting LacZ shRNA control entry clone, respectively, using Lipofectamine 2000 following the manufacturer's instructions. Cell lysates were prepared 48 h after transfection and analyzed for PKC isoform and GAPDH protein expression by Western blotting. The specificity of knockdown to particular PKC isoforms was determined by re-probing the same membranes with antibodies against other PKC isoforms.
Generation of shRNA-expressing Adenoviral Destination Clones
The BLOCK-iT Adenoviral RNAi Expression System (catalog number K4941-00, Invitrogen) was used to generate adenoviral shRNA destination clones. To transfer the U6 RNAi cassette from the entry clones generated above into adenoviral pAd/BLOCK-iT-DEST RNAi Gateway vector, LR recombination reactions were performed using the Gateway LR Clonase II enzyme mixture. Subsequently, transformation and selection of the appropriate transformants were conducted following the manufacturer's instructions. The resulting constructs for recombinant shRNA-expressing adenoviral destination clones were prepared using the EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA) and named pAd/shPKCα, pAd/shPKCδ, pAd/shPKCϵ, and pAd/shLacZ, respectively.
Adenovirus Production and Titration
To produce shRNA-adenoviral stocks, PacI-digested pAd/shPKCα, pAd/shPKCδ, pAd/shPKCϵ, and pAd/shLacZ plasmids were prepared and purified. Following the BLOCK-iT manufacturer's instructions, HEK 293A cells were seeded at 5 × 105 cells/well in a 6-well plate 1 day before transfection. The next day the cells were transfected with PacI-digested plasmids, cultured, and the medium was replaced with fresh complete growth medium every 2–3 days until visible regions of cytopathic effect were evident (typically 5–7 days post-transfection). The cells were fed fresh complete growth medium and cultured until ∼80% cytopathic effect of the culture was observed (typically 7–9 days post-transfection). Both adenovirus-containing cells and medium were harvested and transferred to a sterile-capped tube. Adenovirus-containing supernatants (crude adenovirus stocks) were prepared by repeating a cycle of freezing and thawing 3 times and by centrifuging to remove cell debris. The crude virus stocks were further amplified by infection of HEK 293A cells grown in 10-cm dishes. The amplified viral stocks were named shLacZ-, shPKCα-, shPKCδ-, and shPKCϵ-adenovirus, respectively, and viral stocks were stored in aliquots at −80 °C.
To determine the adenovirus titer, the Adeno-X Rapid Titer immunoassay from Clontech (Mountain View, CA) was conducted following the manufacturer's instructions. Viral stocks with titers of 2 × 109 to 5.5 × 109 infectious units/ml were used in subsequent experiments.
Adenoviral Transduction, PKC Knockdown Efficiency, and Determination of Multiplicity of Infection in T84 Cells
Initial experiments indicated that infection of confluent monolayers of T84 cells was very inefficient (5–10% of cells infected), possibly because the Coxsackie virus and adenovirus receptor is shielded in confluent cells by the junctional complexes. To circumvent this, T84 cells were incubated in sterile phosphate-buffered saline (PBS) without Ca2+/Mg2+ (PBS−) at 37 °C to open tight junctions and expose Coxsackie virus and adenovirus receptor. A 20-min optimal incubation time in PBS− was determined by measuring the decrease in the TER. The cells were then infected with a β-galactosidase-expressing adenovirus (Adeno-X-LacZ, Clontech, m.o.i. = 20) in Opti-MEM I reduced serum medium containing 10 mm EGTA (to deplete Ca2+ and Mg2+ from the medium) for 2 h, then supplemented with fresh completed growth medium, and incubated for an additional 72 h at 37 °C in a 5% CO2 incubator. Using these conditions, ∼90% of the cells expressed β-galactosidase, and the monolayer TER was recovered to normal values (>1000 Ω·cm2) at 48–72 h post-supplement with fresh growth medium.
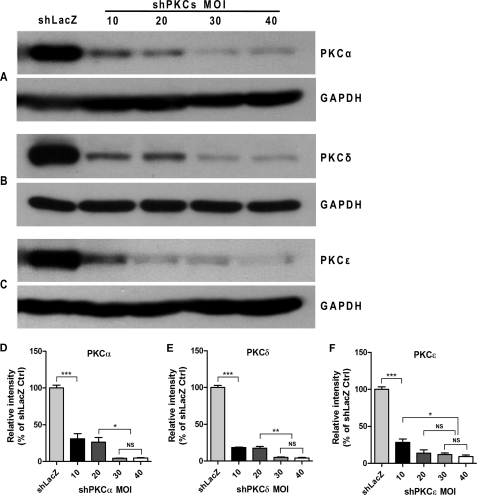
To determine adenoviral shPKCs-mediated knockdown efficiency and m.o.i., T84 cells grown to confluence on 24-mm insert diameter Transwell permeable supports were infected after opening the junctions with shLacZ-adenovirus (m.o.i. = 30) or with shPKCs-adenoviruses at different m.o.i. for 72 h. The knockdown efficiency in PKC proteins was then assessed in comparison with the non-targeting shLacZ control by Western blotting of cell extracts with specific antibodies (see Fig. 2).
FIGURE 2.
Determination of shPKC-adenoviral m.o.i. and PKC knockdown efficiency in T84 destination cells. T84 cell monolayers grown on Transwell supports (24-mm insert diameter) were infected with shLacZ-adenovirus (m.o.i. = 30) serving as non-targeting shRNA control or with shPKCα-, shPKCδ-, and shPKCϵ-adenovirus with increasing m.o.i. ranging from 10 to 40 for 72 h, respectively. The cell lysates were collected and subjected to Western blotting for PKC α, δ, and ϵ proteins. GAPDH protein was probed as a sample loading control. A, PKCα knockdown by shPKCα-adenovirus transduction; B, PKCδ knockdown by shPKCδ-adenovirus transduction; and C, PKCϵ knockdown by shPKCϵ-adenovirus transduction in T84 cells at m.o.i. of 10, 20, 30, and 40. Quantitative analysis of the blot densitometric values from three independent experiments was performed using one-way ANOVA with Tukey's multiple comparison tests after normalized to the values of respective GAPDH blots (D–F). Data are expressed as the percentage of shLacZ control and values represent mean ± S.E. (n = 3). ***, p < 0.001; **, p < 0.01; *, p < 0.05; NS, not significant. As m.o.i. of 30 was the minimum that resulted in ∼90% knockdown of each PKC isoform in T84 cells compared with shLacZ control, we chose it for use in the subsequent experiments.
Cell Lysate Preparation and Western Blotting
HEK 293A and T84 cells were lysed in modified RIPA buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.25% sodium deoxycholate) containing 1 mm phenylmethylsulfonyl fluoride and protease inhibitors mixture set I. The cell lysates were centrifuged at 13,000 × g for 10 min at 4 °C, and the protein concentration of the supernatants determined using a BCA protein assay kit (Pierce). For Western blotting, 25–50 μg of each protein sample was mixed with Laemmli sample buffer (20 mm Tris, 0.2 m DTT, 0.01% bromphenol blue, and 1.0% SDS) and heated at 95 °C for 10 min before being loaded and separated on 7 or 10% SDS-PAGE gels, and subsequently transferred onto PVDF membranes. The membranes were blocked in TBS-T buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20) with 5% nonfat dry milk (Labscientific Inc., Livingston, NJ) for 1 h at room temperature. The membranes were then incubated at 4 °C overnight with primary antibodies diluted, respectively, in 2.5% milk/TBS-T at the following concentrations: anti-PKCα or anti-PKCϵ, 1:1,000; anti-PKCδ or anti-PKCβ1, 1:600; anti-NKCC1, T4 1:2000; and anti-GAPDH, 1:10,000. After extensive washing in TBS-T, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies at 1:10,000 dilutions for 1 h at room temperature. Proteins were detected using the ECL Western blotting substrate (Pierce) and an X-Ray Film Processor (AlphaTek AX 200, Broadview, IL). The blots were scanned on a flatbed scanner and analyzed using Image J software (National Institutes of Health, Bethesda, MD).
Immunofluorescence and Confocal Microscopy
To test the effect of PKC isoform depletion on the plasma membrane NKCC1, T84 monolayers grown to confluence on 6.5-mm insert diameter Transwell supports were infected with non-targeting shLacZ-adenovirus and shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h. Subsequently, PKC isoforms and NKCC1 in the cell monolayers were detected by dual immunofluorescence staining and confocal microscopy. The dual immunofluorescent staining was conducted as previously described (12, 13) and the staining was done while the filters remained in their plastic supports. Briefly, the cells were rinsed twice with ice-cold K+-free PBS and fixed in 100% methanol at −20 °C for 10 min. The cells were then rinsed for an additional two times before incubation in blocking buffer (1% BSA and 10% normal goat serum in K+-free PBS) for 1 h at room temperature, and subsequently incubated with one of PKC isoform-specific primary antibodies (rabbit anti-PKCα, 1:300; rabbit anti-PKCδ, 1:300; or rabbit anti-PKCϵ, 1:50 diluted in blocking buffer) along with the mouse anti-NKCC1 T4 monoclonal antibody (1:200) in a humidified chamber overnight at 4 °C. The cells were then washed three times in K+-free PBS and once in blocking buffer, and then incubated with Alexa Fluor 555 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse secondary antibodies (1:500 diluted in blocking buffer) for 1 h at room temperature in the dark. The filters were washed, excised, and mounted before confocal images of the cells were acquired with a Zeiss LSM510 confocal scanning microscope using a ×63 magnification/1.4 numerical aperture Plan Apochromat objective. To assess the effect of PKC isoform depletion on NKCC1 internalization during PKC activation, the T84 monolayers that were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus were treated with vehicle (DMSO) or 100 nm PMA in T84 basic medium (without FBS) for 1 h at 37 °C in a 5% CO2 atmosphere incubator, followed by immunofluorescent staining utilizing mouse anti-NKCC1 T4 primary antibody and Alexa Fluor 488 goat anti-mouse secondary antibody.
Cell Surface Protein Biotinylation
To further confirm the effect of PKC isoform depletion on surface expression of the basolateral membrane NKCC1, T84 monolayers grown to confluence on 24-mm insert diameter Transwell permeable supports were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) as described before. After a 72-h adenoviral transduction, the cells were treated with 100 nm PMA or vehicle (DMSO) in T84 basic medium for 1 h at 37 °C, in a 5% CO2 atmosphere incubator, followed by basolateral surface biotinylation as previously reported (12). Briefly, the cells were washed 3 times with ice-cold PBS supplemented with 0.1 mm CaCl2 and 1.0 mm MgCl2 (PBS+), then incubated with fresh 1 mg/ml of EZ-Link Sulfo-NHS-Biotin in cold PBS+ at the basolateral side for 20 min on ice with constant gentle shaking. The biotinylation step was then repeated with fresh EZ-Link Sulfo-NHS-Biotin, and the cells were then washed 5 times with ice-cold PBS+ supplemented with 100 mm glycine as a quenching agent (31). The filters were excised and incubated in 1 ml of modified RIPA buffer containing 1 mm phenylmethylsulfonyl fluoride and protease inhibitors mixture set I for 50 min on ice. To remove residual cellular protein, the filters were scraped in the extraction buffer, and the complete lysates centrifuged at 14,000 × g at 4 °C for 10 min. Supernatants were collected and protein concentrations determined. Equal amounts of protein samples (∼800 μg/sample) were incubated with 100 μl of ImmunoPure-immobilized streptavidin-agarose beads overnight with constant mixing at 4 °C. The beads were collected using brief centrifugation and washed three times with ice-cold RIPA buffer, twice with a high-salt wash buffer (0.1% Triton X-100, 500 mm NaCl, and 50 mm Tris, pH 7.5), and once with a low-salt wash buffer (10 mm Tris, pH 7.5). Each pellet was mixed with 40 μl of Laemmli protein sample buffer, heated at 95 °C for 10 min, and subjected to Western blotting. To confirm specific distribution of NKCC1 in the cell surface membrane, biotinylation assays at the apical or basolateral surface of uninfected T84 monolayers were performed. The procedure for apical surface biotinylation assay was similar to that for basolateral surface biotinylation, but EZ-Link Sulfo-NHS-Biotin in cold PBS+ was added to the apical side.
Electrophysiological Measurement in T84 Cell Monolayers
Short-circuit currents (Isc), which approximates the net chloride secretion, was measured with a voltage/current clamp (710C-1 Dual voltage clamp, Bioengineering, the University of Iowa) and Ag/AgCl and calomel electrodes interfaced via chopstick KCl-agar bridges. The TER was calculated by the Ohm law from the voltage change measured during the injection of a 25-μA current pulse (E0-E25) (6, 13). Isc was defined as the external current required to nullify the spontaneous potential difference (E0) across the monolayer. To assess the impact of PKC isoform-specific knockdowns on chloride secretion, T84 monolayers grown to confluence on collagen-coated Transwell were infected with shLacZ- and shPKCs-adenoviruses (m.o.i. = 30) for 72 h. On the day of the experiment, the infected cultures or uninfected controls were washed three times and subsequently incubated for 1 h at 37 °C in a HEPES phosphate-buffered saline solution (HPBSS) containing 140 mm NaCl, 5 mm KCl, 1.2 mm MgSO4, 1 mm CaCl2, 2 mm NaH2PO4, 10 mm HEPES, 10 mm glucose, 2 mm Na-pyruvate, and 2 mm l-glutamine, pH 7.40 ± 0.03, at 37 °C and an osmolarity of 300 ± 5 milliosmole (measured with a vapor-pressure osmometer, model 5520C, Wescor Inc., Logan, UT, and adjusted with either NaCl or H2O). After the pre-equilibration period, baseline measurements of TER and Isc in HPBSS were taken every 5 min over a period of 15 to 20 min. Next, the monolayers were pre-treated for 1 h with 100 nm PMA or vehicle (DMSO) in HPBSS added to the basolateral side, and then another set of measurements were taken. Finally, the monolayers were transferred into HPBSS containing DMSO or PMA supplemented with 10 μm forskolin (FSK, a cAMP inducer) at the basolateral side, and the TER and cAMP-dependent Isc were measured every 5 min over a period of 30 min.
Statistical Analysis
Data are presented as mean ± S.E. along with the number of observations (n). Data were analyzed by Student's t test or by one-way ANOVA with Tukey's post test for multiple comparisons and Dunnett's post test for comparison with control, as appropriate. The value of p < 0.05 was considered statistically significant.
RESULTS
Screening of PKC shRNA-expressing pENTR/U6 Entry Clones Using HEK 293A Cells
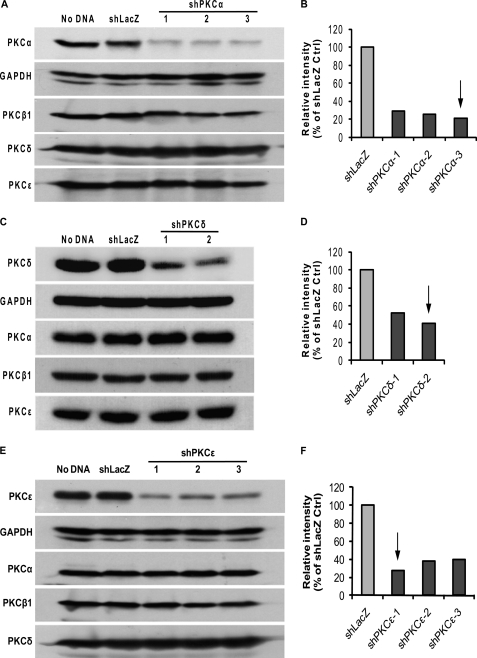
Three targets for human PKCα, two for human PKCδ, and three for human PKCϵ were selected and the corresponding oligos encoding PKC shRNAs (shPKCs) were cloned into U6 entry vector, respectively. Entry clone expressing LacZ shRNA (shLacZ) was made as a non-targeting shRNA control. All U6 entry clones used were verified by sequencing. Knockdown efficiency and specificity of the different targeting sequences were determined by transient transfection into HEK 293A cells, which express both conventional and novel PKC isoforms and have a transfection efficiency of ∼85% (data not shown). For the purpose of screening, the protein levels of PKC isoforms were examined at 48 h after transfection. As shown in Fig. 1, shLacZ did not affect the expression of PKCs compared with Lipofectamine transfection reagent control (no DNA) after 48 h transfection, it was thus used as a control for shPKCs. Fig. 1, A and B, show PKCα knockdown by three shPKCα- expressing entry clone constructs. The average level of PKCα protein in the cells transfected with the three shPKCα- entry constructs was reduced by 70, 74, and 78% compared with shLacZ control, respectively. On the other hand, knockdown of PKCα by these shPKCα constructs did not affect protein expression of PKCβ1, PKCδ, and PKCϵ, therefore specificity of the knockdown by shPKCα was verified. Similarly, the average PKCδ protein level in the cells expressing shPKCδ was decreased by 47 and 59% compared with shLacZ control, respectively. No cross-knockdown by shPKCδ was found of PKCα, PKCβ1, and PKCϵ in the same lysates (Fig. 1, C and D). In the cells with three shPKCϵ- entry constructs, PKCϵ protein was decreased on average by 73, 62, and 60%, respectively, and PKCϵ knockdown did not alter the expression of PKCα, PKCβ1, and PKCδ (Fig. 1, E and F). Based on these results, target 3 for shPKCα, target 2 for shPKCδ, and target 1 for shPKCϵ were chosen to be subcloned into the adenoviral Gateway vector, respectively, to produce shPKCs-expressing adenoviruses.
FIGURE 1.
Screening of shPKC targets in HEK 293A. HEK 293A cells were transfected with pENTRY clone constructs expressing shPKCα, -δ, and -ϵ for 48 h, and cell lysates were then examined for specific PKC knockdown and cross-knockdown with other PKC isoforms by Western blotting. No DNA was a transfection reagent control and shLacZ was set up as a non-targeting shRNA control. GAPDH was used as a loading control for normalization. A, screening blots of three shPKCα targets and cross-knockdown assay for PKCβ1, PKCδ, and PKCϵ. B, relative intensity (% of shLacZ control) of blots shown in A. All the three shPKCα targets decreased PKCα expression by more than 70% without affecting the expression of PKCβ1, -δ, and -ϵ. The arrow points to the shPKCα target giving the highest knockdown efficiency that was chosen to be subcloned into adenoviral vector. C, screening blots of two shPKCδ targets and cross-knockdown assay for PKCα, PKCβ1, and PKCϵ. D, relative intensity (% of shLacZ control) of blots shown in C. The two shPKCδ targets decreased PKCδ expression by ∼50% without affecting the expression of PKCα, -β1, or -ϵ. The arrow points to the shPKCδ target giving the higher knockdown efficiency that was chosen to be subcloned into adenoviral vector. E, screening blots of three shPKCϵ and cross-knockdown assay for PKCα, PKCβ1, and PKCδ. F, relative intensity (% of shLacZ control) of blots shown in E. The three shPKCϵ targets reduced PKCϵ expression by more than 60% without affecting the expression of PKCα, -β1, and -δ. The arrow points to the shPKCϵ target giving the highest knockdown efficiency that was chosen to be subcloned into adenoviral vector. The blots shown are representative of two independent experiments.
Determination of Adenoviral m.o.i. for shPKCs Maximal Knockdown Efficiency in T84 Cells
To determine an optimal m.o.i. for shPKCs-adenovirus transduction leading to the most effective knockdown in T84 cells, the cells grown to confluence on Transwell permeable supports (24-mm insert diameter) were infected with shPKCα-, shPKCδ-, or shPKCϵ-adenovirus at an increasing m.o.i. ranging from 10 to 40 as indicated in Fig. 2, and with shLacZ-adenovirus (m.o.i. = 30) as a non-targeting shRNA control, respectively. Because the screening experiments showed ∼70% knockdown in the PKC proteins at 48 h post-transfection with shPKCs (Fig. 1), to reach further depletion levels of the targets, the knockdown levels of the PKC isoforms at 72 h in T84 cells were tested. As shown in Fig. 2, the increasing m.o.i. (10 to 30) of shPKCα-, shPKCδ-, and shPKCϵ- adenovirus caused a progressive decrease in PKCα (Fig. 2A), PKCδ (Fig. 2B), and PKCϵ (Fig. 2C) proteins as compared with shLacZ control after 72 h adenovirus transduction. Furthermore, the transduction with m.o.i. of 40 for shPKCα-, shPKCδ-, and shPKCϵ- adenoviruses did not cause further significant reduction in the proteins of interest. Quantitative data obtained from three separate experiments indicated that m.o.i. of 30 was the minimum that caused ∼90% knockdown in all the three PKC isoforms (Fig. 2, D–F). Taken together, an optimal m.o.i. of 30 for all the three viruses was thus chosen to be used in subsequent experiments.
Depletion of PKC Isoforms Does Not Affect Expression of Total Cellular NKCC1
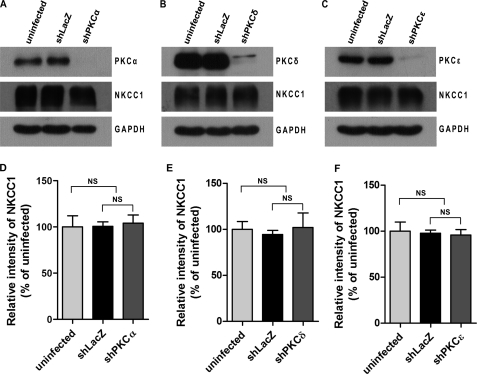
To test whether depletion of PKCs in T84 cells affects the expression of total cellular NKCC1 protein, T84 cell monolayers grown on 24-mm insert diameter Transwell supports were uninfected or infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h, respectively. The cell lysates were then immunoblotted for proteins of PKC isoforms, total NKCC1 protein, and GAPDH by Western blotting. As shown in Fig. 3, the protein levels of PKCα (Fig. 3A), PKCδ (Fig. 3B), and PKCϵ (Fig. 3C) were knocked down by more than 90% in shPKCα-, shPKCδ-, and shPKCϵ-adenovirus-transduced cells compared with either shLacZ non-targeting control or the uninfected control cells, respectively. The expression levels of NKCC1 protein were similar among the uninfected, shLacZ- and shPKC- cell lysates when the same membranes were re-probed using NKCC1-specific T4 antibody. GAPDH was re-probed as a sample loading control. Quantitative data analyzed from three independent experiments demonstrated that there was no significant difference in the expression of total cellular NKCC1 protein between the three cell lysates (Fig. 3, D–F). These results show that depletion of PKCα, PKCδ, or PKCϵ in T84 cells does not affect the expression of total cellular NKCC1 protein.
FIGURE 3.
Effect of PKC isoform-specific knockdown on the expression of total cellular NKCC1 in T84 cells. T84 cell monolayers grown on Transwell supports (24-mm insert diameter) were uninfected or infected with non-targeting shLacZ-adenovirus or shPKCα-, shPKCδ-, and shPKCϵ-adenoviruses (m.o.i. = 30) for 72 h. The cell lysates were probed for proteins of PKC isoforms, NKCC1, and GAPDH by Western blotting. A, effect of PKCα knockdown on total NKCC1 protein expression in T84 cells. The top blot shows the expression levels of PKCα protein, the middle blot shows the expression levels of total NKCC1 protein, and the lower blot indicates the expression of GAPDH serving as sample loading control. B, effect of PKCδ knockdown on total NKCC1 protein expression in T84 cells. The top blot shows the expression levels of PKCδ protein. C, effect of PKCϵ knockdown on total NKCC1 protein expression in T84 cells. The top blot shows the expression levels of PKCϵ protein. The blots shown are representative of three separate experiments. Quantitation of three blots from triplicate experiments was conducted by densitometry and data were analyzed using one-way ANOVA with Tukey's multiple comparison tests after normalized to the respective GAPDH blots (D–F). Data are presented as the percentage of uninfected cells (mean ± S.E., n = 3). NS, not significant.
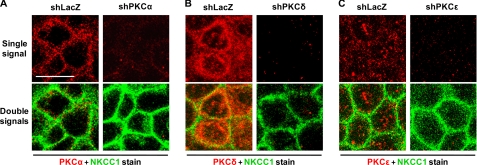
Depletion of PKC Isoforms Does Not Alter Distribution of the Plasma Membrane NKCC1
T84 monolayers grown on 6.5-mm insert diameter Transwell supports were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h. Subsequently, PKC isoforms and NKCC1 in the cell monolayers were detected by immunofluorescence and confocal microscopy. As shown in Fig. 4, upper panels, the staining signal (red) for PKCα, PKCδ, and PKCϵ was observed in the cytoplasm of shLacZ-adenovirus-transduced T84 cells. PKCα was concentrated near the plasma membrane (the upper left panel in Fig. 4A), PKCδ was highly expressed and concentrated close to the plasma membrane (the upper left panel in Fig. 4B), and PKCϵ was not concentrated, instead, dotted throughout the cytoplasm (the upper left panel in Fig. 4C). As compared with shLacZ-adenovirus-transduced controls, the staining signal for PKCα, PKCδ, and PKCϵ was depleted in T84 monolayers transduced with the corresponding shPKCα- (the upper right panel in Fig. 4A), shPKCδ- (the upper right panel in Fig. 4B), and shPKCϵ- (the upper right panel in Fig. 4C) adenovirus, respectively. Thus, depletion of the PKC isoforms in T84 monolayers by shPKCs-adenovirus transduction was confirmed. As seen in the lower panels (the merged images) in Fig. 4, the staining signal (green) for NKCC1 was visualized in the plasma membrane of all shLacZ-, shPKCα-, shPKCδ-, and shPKCϵ-T84 cells, which is consistent with NKCC1 distribution in normal T84 cells (data not shown), and the NKCC1 staining signal was similar between shLacZ non-targeting control and the individual PKC isoform-depleted cells. Together, the results indicate that depletion of the PKC isoforms does not alter distribution of the plasma membrane NKCC1. Similar results were observed in three separate experiments.
FIGURE 4.
Effect of PKC knockdown on distribution of NKCC1 in T84 cells. T84 cell monolayers grown to confluence on Transwell supports (6.5-mm insert diameter) were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h. The cells were fixed in methanol and dually stained using mouse anti-NKCC1 primary antibody T4 (labeled with Alexa Fluor 488 goat anti-mouse secondary antibody, green) and rabbit anti-PKCα, -δ, or -ϵ (labeled with Alexa Fluor 555 goat anti-rabbit secondary antibody, red). The images of the cells were captured by confocal microscopy. The upper panels display staining signal of individual PKC isoforms (red) and the lower panels represent the PKC staining signal (red) merged with NKCC1 staining signal (green). A, knockdown of PKCα in shPKCα-T84 monolayers. Staining signals for PKCα and NKCC1 are shown in both shLacZ- and shPKCα- cells. B, knockdown of PKCδ in shPKCδ-T84 monolayers. Staining signals for PKCδ and NKCC1 are shown in both shLacZ- and shPKCδ- cells. C, knockdown of PKCϵ in shPKCϵ-T84 monolayers. Staining signals for PKCϵ and NKCC1 are shown in both shLacZ- and shPKCϵ- cells. Scale bar, 10 μm.
Depletion of Novel PKC Isoforms δ or ϵ, but Not Conventional PKCα, Prevents Basolateral Surface NKCC1 Internalization Mediated by PMA
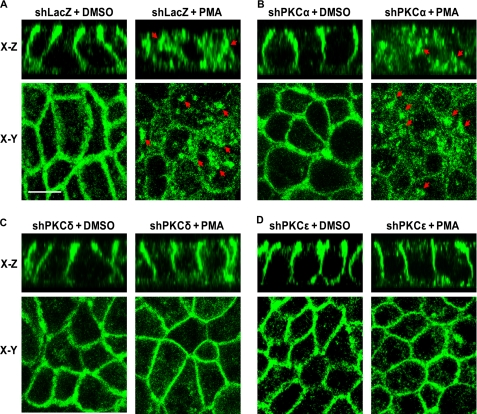
Our previous studies demonstrated that PMA increased activity of all three PKC isoforms (α, δ, and ϵ) and activation of the PKCs by PMA caused internalization of cell membrane surface NKCC1 (12, 13, 23). Based on our previous findings, a protocol with a dose of 100 nm PMA and a 1-h treatment was chosen for the present study as it was most effective for activation of PKCs, leading to NKCC1 internalization and inhibition of chloride secretion. To determine the specific PKC isoforms implicated in NKCC1 internalization mediated by PMA, T84 monolayers, which were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus in the same way as described in Fig. 4, were treated with 100 nm PMA or DMSO (vehicle) for 1 h at 37 °C in a 5% CO2 atmosphere incubator. The cells were then stained with NKCC1 T4 antibody and NKCC1 staining signal was detected by immunofluorescence and confocal microscopy. As shown in Fig. 5, NKCC1 staining (green) in vehicle-treated shLacZ-, shPKCα-, shPKCδ-, and shPKCϵ-T84 cells (Fig. 5, A–D, left panels) was similar and it appeared in the basolateral membrane in both x-z and x-y sections, which is consistent with the localization of NKCC1 in normal T84 cells. These results suggest that depletion of PKCα, PKCδ, and PKCϵ in cells by the corresponding shPKC adenovirus transduction does not alter distribution of the membrane NKCC1, verifying the observations shown in Fig. 4 (lower panels). On the other hand, NKCC1 staining in the basolateral membrane of PMA-treated shLacZ- and shPKCα-T84 cells was dramatically reduced, instead, many NKCC1 staining (arrows) vesicles were visualized in the cytoplasm, indicating internalization of NKCC1 (Fig. 5, A and B, right panels). In contrast, as seen in PMA-treated shPKCδ- and shPKCϵ-T84 cells (Fig. 5, C and D, right panels), NKCC1 staining was retained in the basolateral membrane and was similar to the observations in vehicle-treated controls, suggesting that PMA treatment failed to induce internalization of NKCC1 in shPKCδ- and shPKCϵ-T84 cells. Similar results were obtained in triplicate experiments. These results demonstrate that depletion of novel PKC isoforms δ or ϵ, but not conventional isoform PKCα, prevents NKCC1 internalization mediated by PKC activation.
FIGURE 5.
PKCδ or PKCϵ knockdown prevents NKCC1 internalization during PMA exposure. T84 cell monolayers grown to confluence on Transwell supports (6.5-mm insert diameter) were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h. The cells were then exposed to vehicle (DMSO) or 100 nm PMA for 1 h. Subsequently, the cells were fixed in methanol, stained, and visualized by confocal microscopy. A, NKCC1 distribution detected by immunofluorescence with Alexa Fluor-488 (green) in shLacZ- cells treated with DMSO (left panel) or 100 nm PMA (right panel). The upper two panels represent X-Z sections and the lower two panels represent X-Y sections. B, same as A except the cells were transduced with shPKCα-adenovirus. C, same as A except the cells were transduced with shPKCδ-adenovirus. D, same as A except the cells were transduced with shPKCϵ-adenovirus. Scale bar, 10 μm.
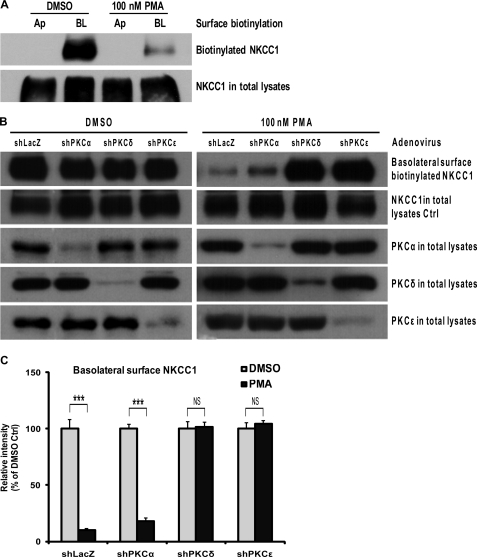
Depletion of PKCδ or PKCϵ Prevents Reduction of the Surface NKCC1 Assayed by Basolateral Surface Biotinylation
To analyze surface distribution of NKCC1 in T84 monolayers, biotinylation assays at the apical or basolateral sides were performed. As shown in Fig. 6A, NKCC1 was localized in the basolateral surface, but not in the apical surface. PMA treatment did not cause translocation of NKCC1 to the apical surface. To confirm that depletion of novel isoforms PKCδ and PKCϵ prevents PKC activation-mediated reduction in the basolateral surface NKCC1, basolateral surface biotinylation assays were performed. Confluent T84 monolayers grown on 24-mm insert diameter Transwell supports were infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus for 72 h, respectively, and then exposed to DMSO or 100 nm PMA for 1 h before biotinylation was conducted. In Fig. 6, B and C, as compared with the DMSO-treated group (left panel), PMA treatment (right panel) led to a dramatic reduction in the amount of basolateral surface biotinylated NKCC1 protein (the first row blot) in shLacZ- and shPKCα-T84 cells (p < 0.001, unpaired t test; PKCα knockdown is shown in the third row blot). On the other hand, PMA treatment failed to decrease surface NKCC1 in both shPKCδ- and shPKCϵ-T84 cells as compared with the DMSO-treated group (PKCδ and PKCϵ knockdown are shown in the fourth and fifth row blots). Whereas, as a sample control, the amount of NKCC1 in the total lysates was similar (the second row blots) among shLacZ-, shPKCα-, shPKCδ-, and shPKCϵ- cells. These data demonstrate that depletion of novel PKC isoforms δ or ϵ, but not conventional isoform PKCα, prevents reduction of the basolateral surface NKCC1 mediated by PKC activation, which is consistent with observations by immunofluorescent staining in Fig. 5. These results suggested that PMA-activated PKCδ and PKCϵ caused a reduction in the surface NKCC1, but did not affect expression of total cellular NKCC1 protein in T84 cells.
FIGURE 6.
Surface biotinylation assay for the membrane surface NKCC1 expression. T84 cell monolayers were grown to confluence on Transwell supports (24-mm insert diameter). A, cells were treated with vehicle (DMSO) or 100 nm PMA for 1 h, and then cell surface biotinylation assays were performed at either the apical (Ap) or basolateral side (BL) of Transwell supports and the cell lysates were subjected to Western blotting using primary mouse anti-NKCC1 T4 antibody. Shown are representative blots of three independent experiments. B, cells were infected with non-targeting shLacZ-adenovirus and with shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h before treatment with vehicle (DMSO, left panel) or 100 nm PMA (right panel) for 1 h. The basolateral surface biotinylation assay was then conducted and the cell lysates were analyzed by Western blotting. The first row blots from the top represent the dynamic change of biotinylated NKCC1 protein after PMA or DMSO treatment in the four groups of shLacZ-, shPKCα-, shPKCδ-, and shPKCϵ- cells. The second row blots show the total NKCC1 protein expression. The third, fourth, and fifth row blots indicate the expression levels of PKCα, PKCδ, and PKCϵ, respectively, in the total cell lysates. The blots shown are representative of three independent experiments. C, blot bands of basolateral surface biotinylated NKCC1 were quantified by densitometry, and data from the three experiments were analyzed with unpaired t test after normalization to respective NKCC1 in total lysates. The data are presented as the percentage of DMSO control (mean ± S.E., n = 3). ***, p < 0.001; NS, not significant.
Specific Knockdown of Novel PKC Isoforms δ or ϵ, but Not Conventional PKCα, Prevents Inhibition of cAMP-stimulated Chloride Secretion (Isc) by PMA
To define the specific roles for individual PKC isoforms in the regulation of epithelial chloride secretion (Isc), the effects of PKC isoform-specific knockdown on PMA inhibition of T84 chloride secretion were assessed. First, the effects of the adenovirus transduction and PMA (a PKC activator) pre-treatment on the TER and Isc were examined. As shown in Table 1, all the monolayers before DMSO/PMA pre-treatment had a baseline TER > 1000 Ω·cm2 and no significant differences in TER were found between the uninfected and the adenovirus transduced groups (p > 0.05, one-way ANOVA), indicating that transduction with either shLacZ- or shPKCs-adenovirus does not affect the TER under steady-state conditions. Although the baseline Isc values were slightly different (p < 0.01, one-way ANOVA), probably reflecting small differences in baseline ion transport activity, no biologically meaningful differences among the groups were found. DMSO was not associated with consistent changes in baseline TER. After a 1-h PMA pre-treatment, the TERs of the monolayers increased in each group as compared with that of the before PMA treatment, which is consistent with as previously observed (13), and a slight change in Isc in shLacZ and shPKCδ was observed (Table 1).
TABLE 1.
Effect of vehicle (DMSO) and PMA pre-treatment on T84 epithelial TERs and Isc
Confluent T84 monolayers grown on 6.5-mm insert diameter Transwell supports were uninfected or infected with shLacZ-, shPKCα−, shPKCδ−, or shPKCϵ−adenovirus (m.o.i. = 30) for 72 h, respectively, as described under “Experimental Procedures.” The cells were washed and equilibrated in a HEPES phosphate-buffered saline solution for ∼1 h at 37 °C in a water bath. TER and Isc were computed as described under “Experimental Procedures.” The data are expressed as mean ± S.E. of TERs (Ω·cm2) and Isc (μA/cm2). Numbers (n) of samples indicate the number of Transwell inserts. One-way ANOVA was performed for analysis of significant difference before and after DMSO or PMA treatments.
| Cell inserts | n | TER |
p value | Isc |
p value | ||
|---|---|---|---|---|---|---|---|
| Before DMSO/PMA | After DMSO/PMA | Before DMSO/PMA | After DMSO/PMA | ||||
| Ω·cm2 | μA/cm2 | ||||||
| Uninfected control | |||||||
| DMSO group | 29 | 1135 ± 44 | 1194 ± 58 | 0.2131 | 2.53 ± 0.34 | 2.95 ± 0.46 | 0.059 |
| PMA group | 32 | 1207 ± 56 | 1413 ± 53 | 0.0006 | 2.02 ± 0.23 | 2.04 ± 0.21 | 0.7202 |
| shLacZ control | |||||||
| DMSO group | 37 | 1196 ± 51 | 1336 ± 50 | 0.0001 | 2.73 ± 0.27 | 3.30 ± 0.31 | 0.0002 |
| PMA group | 36 | 1219 ± 60 | 1515 ± 62 | <0.0001 | 2.54 ± 0.26 | 2.65 ± 0.26 | 0.043 |
| shPKCα | |||||||
| DMSO group | 15 | 1344 ± 61 | 1551 ± 74 | 0.1255 | 3.45 ± 0.54 | 4.07 ± 0.64 | 0.00729 |
| PMA group | 14 | 1318 ± 94 | 1628 ± 95 | 0.0294 | 2.85 ± 0.57 | 2.99 ± 0.60 | 0.309 |
| shPKCδ | |||||||
| DMSO group | 43 | 1137 ± 43 | 1237 ± 52 | 0.1427 | 2.14 ± 0.11 | 2.88 ± 0.18 | <0.0001 |
| PMA group | 40 | 1116 ± 41 | 1284 ± 48 | 0.0093 | 2.16 ± 0.10 | 2.33 ± 0.11 | 0.0053 |
| shPKCϵ | |||||||
| DMSO group | 17 | 1305 ± 90 | 1480 ± 78 | 0.1522 | 3.84 ± 1.08 | 4.03 ± 0.73 | 0.8397 |
| PMA group | 17 | 1303 ± 82 | 1555 ± 84 | 0.0388 | 3.91 ± 0.62 | 3.87 ± 0.57 | 0.8615 |
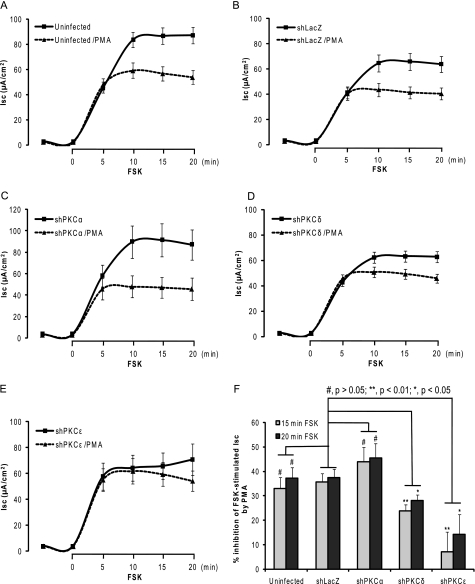
Second, whether specific depletion of PKCs can modify cAMP-stimulated Isc was assessed in T84 monolayers. As shown in Fig. 7, Isc values of all the monolayers exposed to vehicle (solid lines) were markedly increased in response to forskolin (FSK, a cAMP inducer) exposure and followed by a plateau phase after 10 min FSK exposure. No significant difference in Isc (μA/cm2, mean ± S.E.) measured after a 15-min FSK exposure (in the middle of the plateau) was found in the uninfected (83.72 ± 6.18, n = 29), shPKCα (89.76 ± 15.06, n = 13), shPKCδ (62.16 ± 4.45, n = 38), and shPKCϵ (64.29 ± 10.05, n = 16) as compared with shLacZ control (64.63 ± 6.85, n = 33)(p > 0.05 in all groups, one-way ANOVA with Dunnett's post test). The data reveal that specific depletion of PKCα, PKCδ, or PKCϵ in T84 monolayers by shPKCs did not modify cAMP-stimulated Isc.
FIGURE 7.
Impact of PKC isoform-specific knockdown on PMA inhibition of cAMP-stimulated Isc. T84 cell monolayers grown to confluence on Transwell supports (6.5-mm insert diameter) were uninfected or infected with shLacZ-, shPKCα-, shPKCδ-, or shPKCϵ-adenovirus (m.o.i. = 30) for 72 h, respectively. The procedure for Isc measurement was carried out as described under “Experimental Procedures.” The cells of each group were exposed to vehicle (solid line) or 100 nm PMA (dashed line) for 1 h and then exposed to 10 μm forskolin (FSK). The Isc (μA/cm2) was measured every 5 min before and after FSK exposure. The data shown here were obtained from uninfected control cells (A), non-targeting shLacZ-adenovirus transduced cells (B), shPKCα-adenovirus transduced cells (C), shPKCδ-adenovirus transduced cells (D), and shPKCϵ-adenovirus transduced cells (E), respectively. F, summary of % inhibition of FSK-stimulated Isc by PMA at 15 and 20 min FSK exposure. The data are presented as mean ± S.E. % of inhibition of Isc by PMA.
Finally, it was addressed whether specific PKC depletion can prevent PMA inhibition of cAMP-stimulated Isc in T84 monolayers. In Fig. 7, the curves of Isc (μA/cm2) in the absence (solid lines) and presence (dashed lines) of PMA are shown. As summarized in Fig. 7F, after exposure for 15 and 20 min to FSK, PMA reduced FSK-stimulated Isc (mean ± S.E. % of Isc inhibition) by 35.05 ± 4.48 (15 min) and 37.34 ± 4.18% (20 min) in the uninfected control (n = 29) as well as 35.58 ± 3.63 and 37.38 ± 3.46% in shLacZ non-targeting control (n = 33). No significant difference was found between the uninfected and shLacZ (p > 0.05, unpaired t test). PMA inhibited FSK-stimulated Isc by 43.9 ± 6.02 and 45.44 ± 5.9% in shPKCα (n = 13), which was not different from PMA inhibition in shLacZ (p > 0.05, unpaired t test). On the other hand, PMA inhibited FSK-stimulated Isc by 23.91 ± 2.42 and 28.18 ± 2.2% in shPKCδ (n = 38, p < 0.01 and p < 0.05) as well as 7.06 ± 8.13 and 14.23 ± 8.22% in shPKCϵ (n = 16, p < 0.01 and p < 0.05) compared with shLacZ control (unpaired t test), respectively. These data demonstrate that specific knockdown of novel PKC isoforms δ or ϵ, but not conventional PKCα, prevents PMA inhibition of cAMP-stimulated Isc (chloride secretion), suggesting an inhibitory role for novel PKC isoforms PKCδ and PKCϵ in regulation of epithelial chloride secretion. Taken together, our study demonstrates that activated novel PKC isoforms δ and ϵ play a critical role in regulating the surface NKCC1 internalization, hence leading to the inhibition of epithelial chloride secretion in T84 cells.
DISCUSSION
In the present study, a novel effective adenoviral approach delivering PKC shRNAs to confluent T84 monolayers grown on Transwell supports was successfully established and resulted in ∼90% knockdown of PKC proteins in T84 monolayers. By using this novel approach it was found that depletion of novel PKC isoforms δ or ϵ, but not conventional isoform PKCα, prevented PMA-mediated internalization of the basolateral surface NKCC1 in T84 cell monolayers. Furthermore, electrophysiological results demonstrated that depletion of the novel PKC isoforms δ or ϵ rather than the conventional isoform PKCα prevented PMA inhibition of cAMP-stimulated transepithelial chloride secretion in T84 monolayers. Together, our study reveals that activated novel PKC isoforms δ and ϵ play critical roles in regulating surface NKCC1 internalization and, hence, the inhibition of epithelial chloride secretion.
T84 cells are perhaps the most widely used model for studying epithelial chloride transport and barrier function (12–14, 24, 25, 32), and in the vast majority of studies, a pharmacological approach has been used to interrogate signaling pathways and other mechanistic aspects of the underlying biological processes. This approach has its limitations because of the lack of specificity of various pharmacological activators and inhibitors such as those of Gö6850 selective for cPKC and nPKC, Gö6976 for cPKC, and rottlerin for PKCδ. It has been recognized that selectivity of these inhibitors of Gö6850, Gö6976, and rottlerin toward other members of PKC family is concentration-dependent, and that these inhibitors have also been shown to inhibit activity of other protein kinases (33–36). Because activity of all PKC isoforms is suppressed by the intramolecular interaction between the pseudosubstrate binding sites at their N-terminal regulatory domain and substrate binding region of the catalytic domain, several groups have reported that pseudosubstrate peptides designed based on the pseudosubstrate sequence of PKC isoforms are able to selectively inhibit specific PKC isoforms (37–40). However, like most peptides, these pseudosubstrate peptide inhibitors are plasma membrane impermeable. Although it has been shown that cell permeability of the pseudosubstrate peptide inhibitors can be improved through modification by attachment of a fatty acid such as myristoylation, modification itself of the peptides confers new properties and altered selectivity to them beyond their original function (40–42). In addition, the pseudosubstrate peptides derived from individual PKC isoforms have cross-inhibitory actions. For example, it has been reported that the pseudosubstrate peptide based on PKCϵ inhibits activity of PKCα (39).
RNA interference (RNAi) represents a highly specific and powerful tool to silence the gene expression of interest and has been extensively used for investigating the consequences for loss-of-function of individual genes and identifying key regulatory components of complex signaling pathways (43–47). By transient transfection of HEK 293A cells, the PKC isoform-specific shRNA entry construct caused an efficient knockdown of target without affecting other PKC isoforms, indicating the knockdown is highly specific (Fig. 1). However, effective delivery of siRNA duplex and exogenous genes into confluent T84 monolayers is difficult to date and is still challenging to those who use T84 monolayers (48) although some researchers have applied siRNA duplexes to subconfluent T84 cells that were grown on plastic (49–51). One report indicated that siRNA was introduced into 70–75% confluent rat cholangiocytes that were then trypsinized and seeded onto Transwell supports (52). This approach is problematic in T84 cells because the duration required for T84 cells to reach full confluence and functional differentiation is typically around 7 days, and it is beyond the time frame for studying functions of protein kinases by knockdown. Moreover, this approach is compromised when depleting target proteins that are essential for growth, differentiation, or formation of the epithelial monolayer itself. Other alternatives to introduce siRNA into confluent epithelia include a magnetofection approach, which has been reported in a renal proximal tubule cell line (53). However, this technique has not been applied to a tight epithelium such as the T84 line. One study reported knockdown of Toll-like receptor 9 in T84 monolayers but transfection efficiency was not reported (54), whereas others have used electroporation to transfer exogenous genes into T84 monolayers (48). In the latter study, the efficiency of gene transfer was still poor and electroporation on Transwell-grown monolayers requires a custom-designed electroporation chamber. To deplete PKC isoforms in T84 monolayers, a novel effective approach delivering specific shRNA or siRNA to T84 monolayers is required to be developed.
Adenoviral vector delivery of shRNAs has been used to introduce RNAi into a variety of mammalian cells (45, 55–57) and adenoviral shRNA-induced RNAi can achieve a relatively long lasting knockdown of target (58), thus this virus vector was chosen in the present study. Given that adenoviral high transduction efficiency is a crucial step for shRNA adenoviral delivery to evoke RNAi, we tested transduction efficiency of adenovirus in T84 monolayers using a β-galactosidase-expressing adenovirus. Our initial experiments showed only 5–10% transduction efficiency for adenovirus in T84 monolayers (data not shown), possibly because the Coxsackie virus and adenovirus receptor, a component of the tight junction complex (59), is shielded in confluent cells. We therefore optimized adenovirus infection conditions by a 20-min incubation of T84 monolayers in sterile PBS without Ca2+/Mg2+ (PBS−) at 37 °C to open tight junctions and expose Coxsackie virus and adenovirus receptor, and by subsequent infection for 2 h with adenovirus (m.o.i. = 30) in Ca2+/Mg2+-depleted Opti-MEM I reduced serum medium. This novel adenoviral delivery approach resulted in ∼90% efficiency of the transduction with β-galactosidase-expressing adenovirus (data not shown), and thereafter, achieved ∼90% knockdown of PKC proteins in T84 monolayers transduced with the corresponding shPKCs-adenovirus (Figs. 2 and 3). In most of the cases, the infection-reduced TERs were recovered to normal values (more than 1000 Ω·cm2) after 2–3 days incubation with fresh complete growth medium, and the cell monolayers were not damaged by adenovirus infection. Thus, this study provides for the first time a novel effective approach for adenovirus-based delivery of shRNA to confluent T84 monolayers grown on Transwell permeable supports, which also has implications for studies using epithelial monolayers with poor gene delivery. Our study showed that depletion of individual PKCα, PKCδ, or PKCϵ did not affect expression of total cellular NKCC1 (Fig. 3) and did not alter NKCC1 distribution in the plasma membrane (Figs. 4 and 5) in T84 cells that abundantly express proteins of these PKC isoforms, suggesting that PKC does not constitutively regulate NKCC1.
The mechanisms by which PKC regulates NKCC1 activity are unclear. PMA-induced down-regulation of NKCC1 gene expression has previously been reported (15), but our data in Fig. 3 indicate that PKC knockdown did not affect expression of total cellular NKCC1, which was further supported by our biotinylation data (Fig. 6B), which show similar expression levels of total NKCC1 between shPKC- and shLacZ- cells in the absence of and presence of PMA. Alternatively, PKC may regulate NKCC1 activity through inducing internalization of the basolateral surface NKCC1. Indeed, emerging evidence has pointed to a global role of PKC in regulating the processes of internalization and trafficking of plasma membrane transporters (20, 22, 60–62). PKC activation down-regulates or up-regulates cell surface expression of a transporter depending on the transporter type (20–22, 63). We have recently shown that activation of PKC by PMA induced internalization of the basolateral surface NKCC1 in T84 cells (12) and eGFP-tagged NKCC1-expressing MDCK cells (23). Direct phosphorylation by PKC has been reported to mediate internalization of other transporters such as norepinephrine transporter (20). It has also been reported that PKCδ acts upstream of SPAK, which phosphorylates NKCC1 during hyperosmotic stress (64). Thus, the question for whether PKC-induced phosphorylation contributes to NKCC1 internalization in T84 cells remains to be addressed. On the other hand, internalization through dynamin- and clathrin-dependent pathways has been reported for organic anion transporter OAT1 (22) and eGFP-tagged NKCC1 (23). How does PKC regulate dynamin and clathrin pathways leading to NKCC1 internalization requires further investigation. In the current study, we defined the specific PKC isoforms responsible for regulation of NKCC1. Our immunofluorescence studies (Fig. 5) indicate that activated PKCδ and PKCϵ, but not activated PKCα, are the PKC isoforms that induce internalization of NKCC1 in T84 cells. These observations were further supported by our surface biotinylation study (Fig. 6, B and C). To our surprise, PKCδ knockdown blocked NKCC1 internalization to an extent similar to that observed in PKCϵ knockdown. This is a novel finding because our previous pharmacological studies revealed that PMA-induced internalization of NKCC1 is dependent upon isoform PKCϵ (12). In other studies, nPKC and cPKCs have been suggested to regulate membrane protein surface expression, but in all these instances the identity of the specific PKC isoform is only inferable through the use of pharmacological inhibitors (19, 60, 61).
Knockdown of the PKC isoforms did not change steady-state TER (Table 1), suggesting it does not affect the constitutive maintenance of tight junction integrity under these conditions. However, we did not investigate whether other PKC members such as PKCη play such roles, as has been suggested in Caco2 intestinal epithelial monolayers (65). We also did not investigate whether the actions of various growth factors and other PKC-dependent agents that are reported to affect junctional integrity could be abrogated by depletion of the PKC isoforms. These important questions remain to be investigated. For example, PKCϵ and PKCβ1 have been suggested to participate in epidermal growth factor-mediated protection of tight junctions from acetaldehyde-induced disruption (66).
We did not find any difference in the magnitude of the FSK-elicited current in any PKC knockdown group compared with shLacZ controls. This observation was somewhat unexpected in the case of PKCδ given reports by other groups in the lung epithelial Calu-3 line that PKCδ is required to activate NKCC1 during osmotic shock and α1-adrenergic receptor stimulation (64, 67). One possible explanation is that in Calu-3 cells, PKCδ is required for agonist-regulated phosphorylation of NKCC1; whereas in T84 cells, baseline levels of NKCC1 phosphorylation are relatively high and FSK does not appear to increase this further.4 In PMA-treated cells, we found that knockdown of PKCδ significantly attenuated PMA-associated inhibition of FSK-Isc (Fig. 7D) and knockdown of PKCϵ completely abolished PMA inhibition of FSK-elicited Isc (Fig. 7E), confirming our previous inference from a purely pharmacological inhibitor approach (12). These results suggest that PKCϵ, and to a lesser extent PKCδ, but not conventional PKCα, play a critical role in PMA-mediated inhibition of the epithelial chloride secretion. It is possible that other components of the chloride secretory apparatus may be affected by PKC activation. Long-term exposure of HT-29 cells to PMA (24 h) decreases cystic fibrosis transmembrane conductance regulator expression (68, 69), but no short-term exposure data have been reported. In Calu-3 cells, a 30-min exposure to vasoactive intestinal polypeptide leads to cystic fibrosis transmembrane conductance regulator accumulation at the apical membrane, an effect mimicked by phorbol ester (70).
In conclusion, the activated novel PKC isoforms δ or ϵ, but not the activated conventional isoform PKCα, inhibits T84 epithelial chloride secretion through inducing internalization of the basolateral surface NKCC1. Our study reveals that the novel PKC isoform-dependent regulation of NKCC1 surface expression represents an important mechanism of overall regulation of chloride and fluid secretion, which has important implications not only in the development of novel therapeutics for those diseases associated with dysregulated chloride and fluid secretion, but also in other fields such as epilepsy (71) and pain management in which cation chloride transporters are involved (72, 73).
Acknowledgments
We are grateful to Dr. Carrie Rinker-Schaeffer, Dr. Kathleen H. Goss, and Dr. Donald J. Vander Griend, University of Chicago, for critical comments on the manuscript. We also thank Drs. Martin ter Beest and Mirjam M. P. Zegers for technical help with confocal microscopy and Xu Tang for a stock of T84 cell line.
This work was supported, in whole or in part, by National Institutes of Health Grant DK-48010 from the NIDDK (to J. B. M.).
J. Tang, P. Bouyer, A. Mykoniatis, M. Buschmann, K. S. Matlin, and J. B. Matthews, unpublished observations.
- NKCC1
- Na+-K+-2Cl− cotransporter-1
- cPKC
- conventional PKC
- nPKC
- novel PKC
- TER
- transepithelial resistance
- m.o.i.
- multiplicity of infection
- FSK
- forskolin
- DMSO
- dimethyl sulfoxide
- ANOVA
- analysis of variance.
REFERENCES
- 1.Russell J. M. (2000) Physiol. Rev. 80, 211–276 [DOI] [PubMed] [Google Scholar]
- 2.Haas M., Forbush B., 3rd (2000) Annu. Rev. Physiol. 62, 515–534 [DOI] [PubMed] [Google Scholar]
- 3.Lebenthal E., Duffey M. E. (1990) Textbook of Secretory Diarrhea, Raven Press, New York [Google Scholar]
- 4.Halm D. R., Raymond F. A. (1990) in Textbook of Secretory Diarrhea (Lebenthal E., Duffey M. E. eds) pp. 47–58, Raven Press, New York [Google Scholar]
- 5.Matthews J. B. (2002) World J. Surg. 26, 826–830 [DOI] [PubMed] [Google Scholar]
- 6.Matthews J. B., Hassan I., Meng S., Archer S. Y., Hrnjez B. J., Hodin R. A. (1998) J. Clin. Investig. 101, 2072–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geibel J. P. (2005) Annu. Rev. Physiol. 67, 471–490 [DOI] [PubMed] [Google Scholar]
- 8.Flagella M., Clarke L. L., Miller M. L., Erway L. C., Giannella R. A., Andringa A., Gawenis L. R., Kramer J., Duffy J. J., Doetschman T., Lorenz J. N., Yamoah E. N., Cardell E. L., Shull G. E. (1999) J. Biol. Chem. 274, 26946–26955 [DOI] [PubMed] [Google Scholar]
- 9.Song J. C., Hrnjez B. J., Farokhzad O. C., Matthews J. B. (1999) Am. J. Physiol. Cell Physiol. 277, C1239–1249 [DOI] [PubMed] [Google Scholar]
- 10.Darman R. B., Forbush B. (2002) J. Biol. Chem. 277, 37542–37550 [DOI] [PubMed] [Google Scholar]
- 11.Steinberg S. F. (2008) Physiol. Rev. 88, 1341–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Castillo I. C., Fedor-Chaiken M., Song J. C., Starlinger V., Yoo J., Matlin K. S., Matthews J. B. (2005) Am. J. Physiol. Cell Physiol. 289, C1332–1342 [DOI] [PubMed] [Google Scholar]
- 13.Song J. C., Hanson C. M., Tsai V., Farokhzad O. C., Lotz M., Matthews J. B. (2001) Am. J. Physiol. Cell Physiol. 281, C649–661 [DOI] [PubMed] [Google Scholar]
- 14.Matthews J. B., Awtrey C. S., Hecht G., Tally K. J., Thompson R. S., Madara J. L. (1993) Am. J. Physiol. Cell Physiol. 265, C1109–1117 [DOI] [PubMed] [Google Scholar]
- 15.Farokhzad O. C., Sagar G. D., Mun E. C., Sicklick J. K., Lotz M., Smith J. A., Song J. C., O'Brien T. C., Sharma C. P., Kinane T. B., Hodin R. A., Matthews J. B. (1999) J. Cell. Physiol. 181, 489–498 [DOI] [PubMed] [Google Scholar]
- 16.Jobbagy Z., Olah Z., Petrovics G., Eiden M. V., Leverett B. D., Dean N. M., Anderson W. B. (1999) J. Biol. Chem. 274, 7067–7071 [DOI] [PubMed] [Google Scholar]
- 17.Rotmann A., Strand D., Martiné U., Closs E. I. (2004) J. Biol. Chem. 279, 54185–54192 [DOI] [PubMed] [Google Scholar]
- 18.Miranda M., Wu C. C., Sorkina T., Korstjens D. R., Sorkin A. (2005) J. Biol. Chem. 280, 35617–35624 [DOI] [PubMed] [Google Scholar]
- 19.González M. I., Susarla B. T., Robinson M. B. (2005) J. Neurochem. 94, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 20.Jayanthi L. D., Annamalai B., Samuvel D. J., Gether U., Ramamoorthy S. (2006) J. Biol. Chem. 281, 23326–23340 [DOI] [PubMed] [Google Scholar]
- 21.Lee H. H., Walker J. A., Williams J. R., Goodier R. J., Payne J. A., Moss S. J. (2007) J. Biol. Chem. 282, 29777–29784 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q., Hong M., Duan P., Pan Z., Ma J., You G. (2008) J. Biol. Chem. 283, 32570–32579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mykoniatis A., Shen L., Chaiken M. F., Tang J., Tang X., Worrell R. T., Delpire E., Turner J. R., Matlin K. S., Bouyer P., Matthews J. B. (2010) Am. J. Physiol. Cell Physiol. 298, C85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Mahony F., Toumi F., Mroz M. S., Ferguson G., Keely S. J. (2008) Am. J. Physiol. Cell Physiol. 294, C1362–1370 [DOI] [PubMed] [Google Scholar]
- 25.Chappell A. E., Bunz M., Smoll E., Dong H., Lytle C., Barrett K. E., McCole D. F. (2008) FASEB J. 22, 2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lytle C., Xu J. C., Biemesderfer D., Forbush B., 3rd (1995) Am. J. Physiol. Cell Physiol. 269, C1496–1505 [DOI] [PubMed] [Google Scholar]
- 27.Dharmsathaphorn K., McRoberts J. A., Mandel K. G., Tisdale L. D., Masui H. (1984) Am. J. Physiol. Gastroentest. Liver Physiol. 246, G204–208 [DOI] [PubMed] [Google Scholar]
- 28.Dharmsathaphorn K., Mandel K. G., Masui H., McRoberts J. A. (1985) J. Clin. Investig. 75, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiscornia G., Singer O., Verma I. M. (2006) Nat. Protocols 1, 234–240 [DOI] [PubMed] [Google Scholar]
- 30.Paddison P. J., Cleary M., Silva J. M., Chang K., Sheth N., Sachidanandam R., Hannon G. J. (2004) Nat. Methods 1, 163–167 [DOI] [PubMed] [Google Scholar]
- 31.Gottardi C. J., Dunbar L. A., Caplan M. J. (1995) Am. J. Physiol. Renal Physiol. 268, F285–295 [DOI] [PubMed] [Google Scholar]
- 32.Keely S. J., Barrett K. E. (2003) Am. J. Physiol. Cell Physiol. 284, C339–348 [DOI] [PubMed] [Google Scholar]
- 33.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gschwendt M., Müller H. J., Kielbassa K., Zang R., Kittstein W., Rincke G., Marks F. (1994) Biochem. Biophys. Res. Commun. 199, 93–98 [DOI] [PubMed] [Google Scholar]
- 36.McGovern S. L., Shoichet B. K. (2003) J. Med. Chem. 46, 1478–1483 [DOI] [PubMed] [Google Scholar]
- 37.Eichholtz T., de Bont D. B., de Widt J., Liskamp R. M., Ploegh H. L. (1993) J. Biol. Chem. 268, 1982–1986 [PubMed] [Google Scholar]
- 38.Hvalby O., Hemmings H. C., Jr., Paulsen O., Czernik A. J., Nairn A. C., Godfraind J. M., Jensen V., Raastad M., Storm J. F., Andersen P., et al. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4761–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson J. A. (2004) Life Sci. 74, 3153–3172 [DOI] [PubMed] [Google Scholar]
- 40.Krotova K., Hu H., Xia S. L., Belayev L., Patel J. M., Block E. R., Zharikov S. (2006) Br. J. Pharmacol. 148, 732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris T. E., Persaud S. J., Jones P. M. (1999) Mol. Cell. Endocrinol. 155, 61–68 [DOI] [PubMed] [Google Scholar]
- 42.Lim S., Choi J. W., Kim H. S., Kim Y. H., Yea K., Heo K., Kim J. H., Kim S. H., Song M., Kim J. I., Ryu S. H., Suh P. G. (2008) Life Sci. 82, 733–740 [DOI] [PubMed] [Google Scholar]
- 43.Brummelkamp T. R., Bernards R., Agami R. (2002) Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 44.Abbas-Terki T., Blanco-Bose W., Déglon N., Pralong W., Aebischer P. (2002) Hum. Gene Ther. 13, 2197–2201 [DOI] [PubMed] [Google Scholar]
- 45.Shen C., Buck A. K., Liu X., Winkler M., Reske S. N. (2003) FEBS Lett. 539, 111–114 [DOI] [PubMed] [Google Scholar]
- 46.Takeshita F., Ochiya T. (2006) Cancer Sci. 97, 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- 48.Ghartey-Tagoe E. B., Babbin B. A., Nusrat A., Neish A. S., Prausnitz M. R. (2006) Int. J. Pharm. 315, 122–133 [DOI] [PubMed] [Google Scholar]
- 49.Spitzner M., Ousingsawat J., Scheidt K., Kunzelmann K., Schreiber R. (2007) FASEB J. 21, 35–44 [DOI] [PubMed] [Google Scholar]
- 50.Kuwano Y., Kawahara T., Yamamoto H., Teshima-Kondo S., Tominaga K., Masuda K., Kishi K., Morita K., Rokutan K. (2006) Am. J. Physiol. Cell Physiol. 290, C433–443 [DOI] [PubMed] [Google Scholar]
- 51.Irnaten M., Blanchard-Gutton N., Harvey B. J. (2008) Cell Calcium 44, 441–452 [DOI] [PubMed] [Google Scholar]
- 52.Sheth P., Delos Santos N., Seth A., LaRusso N. F., Rao R. K. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 293, G308–318 [DOI] [PubMed] [Google Scholar]
- 53.Bataille A. M., Goldmeyer J., Renfro J. L. (2008) Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R2024–2033 [DOI] [PubMed] [Google Scholar]
- 54.Ghadimi D., Vrese M., Heller K. J., Schrezenmeir J. (2010) Inflammatory Bowel Dis. 16, 410–427 [DOI] [PubMed] [Google Scholar]
- 55.Huang B., Kochanek S. (2005) Hum. Gene Ther. 16, 618–626 [DOI] [PubMed] [Google Scholar]
- 56.Chen Y., Chen H., Hoffmann A., Cool D. R., Diz D. I., Chappell M. C., Chen A. F., Chen A., Morris M. (2006) Hypertension 47, 230–237 [DOI] [PubMed] [Google Scholar]
- 57.Rinne A., Littwitz C., Bender K., Kienitz M. C., Pott L. (2009) Methods Mol. Biol. 515, 107–123 [DOI] [PubMed] [Google Scholar]
- 58.Yue W., Abe K., Brouwer K. L. (2009) Mol. Pharm. 6, 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergelson J. M., Cunningham J. A., Droguett G., Kurt-Jones E. A., Krithivas A., Hong J. S., Horwitz M. S., Crowell R. L., Finberg R. W. (1997) Science 275, 1320–1323 [DOI] [PubMed] [Google Scholar]
- 60.Perry C., Baker O. J., Reyland M., Grichtchenko I. I. (2009) Am. J. Physiol. Cell Physiol. 297, C1403–C1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hassan H. A., Mentone S., Karniski L. P., Rajendran V. M., Aronson P. S. (2007) Am. J. Physiol. Cell Physiol. 292, C1485–1492 [DOI] [PubMed] [Google Scholar]
- 62.Alvi F., Idkowiak-Baldys J., Baldys A., Raymond J. R., Hannun Y. A. (2007) Cell Mol. Life Sci. 64, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karatas-Wulf U., Koepsell H., Bergert M., Sönnekes S., Kugler P. (2009) Neuroscience 161, 794–805 [DOI] [PubMed] [Google Scholar]
- 64.Smith L., Smallwood N., Altman A., Liedtke C. M. (2008) J. Biol. Chem. 283, 22147–22156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki T., Elias B. C., Seth A., Shen L., Turner J. R., Giorgianni F., Desiderio D., Guntaka R., Rao R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki T., Seth A., Rao R. (2008) J. Biol. Chem. 283, 3574–3583 [DOI] [PubMed] [Google Scholar]
- 67.Liedtke C. M., Papay R., Cole T. S. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 282, L1151–1159 [DOI] [PubMed] [Google Scholar]
- 68.Broughman J. R., Sun L., Umar S., Scott J., Sellin J. H., Morris A. P. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G318–330 [DOI] [PubMed] [Google Scholar]
- 69.Broughman J. R., Sun L., Umar S., Sellin J. H., Morris A. P. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G331–344 [DOI] [PubMed] [Google Scholar]
- 70.Chappe F., Loewen M. E., Hanrahan J. W., Chappe V. (2008) J. Pharmacol. Exp. Ther. 327, 226–238 [DOI] [PubMed] [Google Scholar]
- 71.Sen A., Martinian L., Nikolic M., Walker M. C., Thom M., Sisodiya S. M. (2007) Epilepsy Res. 74, 220–227 [DOI] [PubMed] [Google Scholar]
- 72.Pitcher M. H., Price T. J., Entrena J. M., Cervero F. (2007) Mol. Pain 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price T. J., Cervero F., de Koninck Y. (2005) Curr. Top. Med. Chem. 5, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]