Abstract
Amyotrophic lateral sclerosis (ALS) is an incurable neurodegenerative disease that preferentially targets motor neurons. It was recently found that dominant mutations in two related RNA-binding proteins, TDP-43 (43-kDa TAR DNA-binding domain protein) and FUS/TLS (fused in sarcoma/translated in liposarcoma) cause a subset of ALS. The convergent ALS phenotypes associated with TDP-43 and FUS/TLS mutations are suggestive of a functional relationship; however, whether or not TDP-43 and FUS/TLS operate in common biochemical pathways is not known. Here we show that TDP-43 and FUS/TLS directly interact to form a complex at endogenous expression levels in mammalian cells. Binding was mediated by an unstructured TDP-43 C-terminal domain and occurred within the context of a 300–400-kDa complex that also contained C-terminal cleavage products of TDP-43 linked to neuropathology. TDP-43 C-terminal fragments were excluded from large molecular mass TDP-43 ribonucleoprotein complexes but retained FUS/TLS binding activity. The functional significance of TDP-43-FUS/TLS complexes was established by showing that RNAi silencing of either TDP-43 or FUS/TLS reduced the expression of histone deacetylase (HDAC) 6 mRNA. TDP-43 and FUS/TLS associated with HDAC6 mRNA in intact cells and in vitro, and competition experiments suggested that the proteins occupy overlapping binding sites. The combined findings demonstrate that TDP-43 and FUS/TLS form a functional complex in intact cells and suggest that convergent ALS phenotypes associated with TDP-43 and FUS/TLS mutations may reflect their participation in common biochemical processes.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig's disease), Protein-Protein Interactions, Ribonuclear Protein (RNP), RNA-binding Protein, RNA Silencing, FUS/TLS, HDAC6, TDP-43
Introduction
Amyotrophic lateral sclerosis (ALS)2 is an adult onset, typically fatal, neurodegenerative disorder of poorly understood etiology that destroys motor neurons (1). Dominant mutations in superoxide dismutase 1 were the first established cause of ALS (2, 3); however, ∼90% of cases occur sporadically with no clear genetic link, and there is no effective treatment for the condition (3).
An important breakthrough toward understanding ALS etiology was made by Neumann et al. (4), who showed that TDP-43 is a major constituent of cytoplasmic ubiquitin-positive inclusions that accumulate in the degenerating motor neurons of ALS patients and individuals with ubiquitin-positive fronto-temporal lobar degeneration. TDP-43 is an essential nuclear RNA-binding protein that participates in transcriptional repression, exon splicing inhibition, and mRNA stabilization (5–8). More recently, dominant mutations in the TARDBP gene encoding TDP-43 were found to cause a subset of inherited ubiquitin-positive fronto-temporal lobar degeneration and ALS cases (9–11), which strongly supports a direct role for TDP-43 aggregation in ALS pathogenesis.
The mechanisms whereby mutations in TDP-43 cause neurodegeneration are not known, but models invoking toxic gain of function and loss of critical nuclear function are equally plausible. Virtually all ALS-associated mutations in TDP-43 occur in an unstructured Gly-rich domain that binds to heterogeneous ribonucleoprotein A/B complexes (12–14). ALS-associated mutants of TDP-43 are hyperphosphorylated, ubiquitylated, aggregation-prone, and cleaved into 25- and 35-kDa C-terminal fragments that exhibit cytotoxicity in cellulo (15–18). TDP-43 is degraded by proteasome and autophagy-dependent pathways, which may be mediated in part through its association with the ubiquitin-binding protein Ubiquilin (19–22).
Recent studies have shown that expression of mutant or wild-type TDP-43 is neurotoxic to zebrafish, Drosophila, and mice (22–26). In several of these studies, TDP-43-induced neurodegeneration occurred in the absence of detectable cytosolic TDP-43 aggregation (22, 23, 25). Combined with the finding that TDP-43 loss of function induces motor neuron deficits in Drosophila (27), it is plausible that too much or too little nuclear TDP-43 disrupts RNA processing events critical for motor neuron function.
Remarkably, mutations in a structurally related RNA-binding protein, FUS/TLS, also cause dominantly inherited ALS (28, 29). Like TDP-43, FUS/TLS forms cytosolic aggregates in degenerating neurons of ALS patients, and FUS/TLS mutants exhibit increased cytosolic localization in transfected cells (28, 29). The mechanisms linking FUS/TLS mutation to motor neuron degeneration are unclear; however, FUS/TLS localizes to dendritic spines following mGluR activation, where it may function in the localized protein translation important for neuron function (30, 31).
Here we provide evidence that TDP-43 and FUS/TLS function in a biochemical complex to modulate expression of HDAC6, a recently identified mRNA substrate of TDP-43 (32). We further show that TDP-43 exists in two macromolecular complexes: a 300–400-kDa complex that overlaps with FUS/TLS and a much larger complex with properties of a ribonucleoprotein (RNP) particle. TDP-43 C-terminal fragments were excluded from TDP-43 RNP complexes but retained interaction with FUS/TLS, suggesting that the accumulation of such fragments may compromise TDP-43-FUS/TLS complex function. These findings provide evidence that ALS proteins TDP-43 and FUS/TLS operate together in a common biochemical pathway.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HeLa, HEK 293T, NSC-34, and mouse embryo fibroblast cell lines were maintained in DMEM containing 10% FBS. siGENOME siRNAs SMARTpool (Dharmacon) were used for knockdown of TDP-43 (M-012394-01), and FUS/TLS (L-009397-00) and siGENOME single siRNA were used for knockdown of TDP-43 (D-012394-03). HeLa and HEK 293T cells were transfected with siRNAs using the calcium phosphate method.
Protein Analysis
HEK 293T, NSC-34, and HeLa cell extracts were prepared as described (19). Extracts containing ∼1 mg of protein were immunoprecipitated with 1 μg of antibody for 16 h at 4 °C with gentle inversion mixing, after which protein A-Sepharose was added for 1 h. The beads were collected and washed four times with lysis buffer, and the immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting using the following antibodies for TDP-43 (ProteinTech, 17892-1 (C-terminal) and 10782-2 (N-terminal)), FUS/TLS (Abcam, ab23439 (rabbit); and Santa Cruz, sc-47711 (mouse)), and PABP2 (Abcam, ab81224). A plasmid vector expressing GST-FUS/TLS was generated by insertion of FUS/TLS gene (Open Biosystems, MHS1011–59311) into pGEX-5X-2. HA-TDP-43 expression plasmids were described previously (19). GST fusion proteins were incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4 °C. Binding, washing, and elution of the GST fusion proteins were carried out according to the manufacturer's instructions. For GST pulldown experiments, HEK 293T cells were transfected with plasmid DNA encoding HA-TDP-43 expression plasmids. Twenty-four hours after transfection, the cell pellets were extracted with lysis buffer, and 500 μg of each clarified extract was incubated with GST or GST-FUS/TLS fusion proteins (20 μg) that had been prebound to glutathione-Sepharose 4B beads. After inversion mixing for 4 h at 4 °C, the beads were washed five times with lysis buffer and boiled in 1× SDS-PAGE sample buffer before analysis by SDS-PAGE and immunoblotting. For gel filtration analysis, extracts from 107 HeLa, mouse embryo fibroblasts and NSC-34 cells were filtered through 0.4-μm filter and separated through a Superose 6 column using fast protein liquid chromatography (LLC-501 Plus; GE Healthcare). One-milliliter column fractions were collected, separated using 10% SDS-PAGE, and analyzed by immunoblotting with α-TDP-43, α-FUS/TLS, α-PABP2, α-RNA PolII, and α-MCM3 antibodies. TDP-43 interacting proteins were analyzed using mass spectrometry at ProtTech Inc., and complete protein profiling of TDP-43 subcomplexes was analyzed using a Thermo Scientific LTQ Orbitrap XL mass spectrometer at University of Wisconsin Biotechnology Center.
Quantitative RT-PCR and RNA Immunoprecipitation (RIP)
Total RNA was prepared using RNeasy kit (Qiagen) according to the manufacturer's protocol. The cDNA was generated from 1 μg of total RNA by reverse transcription using an iScript cDNA synthesis kit (Bio-Rad). Two percent of the synthesized cDNA reaction was subjected to reaction in a MyiQ real time PCR (Bio-Rad) with FastStart DNA Master SYBR Green I (Roche Applied Science). The gene-specific primers were: 5′-TTAGGCCTCCTGGACATCAC-3′ (HDAC6-F1), 5′-GCGGTGGATGGAGAAATAGA-3′ (HDAC6-R1), 5′-CGACGTGATCCAAACTCCTC-3′ (HDAC6-F2), and 5′-ATCAGCCATGTCCTGACCTC-3′ (HDAC6-R2) for mRNA of HDAC6. For RIP, HEK 293T cells were washed and harvested in ice-cold PBS and lysed in hypotonic gentle lysis buffer (10 mm Tris-HCl, pH 7.5, 10 mm NaCl, 2 mm EDTA, 0.5% Triton X-100, RNasin, and protease inhibitors). The cells were incubated on ice for 10 min and added with NaCl to 150 mm. After 5 min of incubation on ice, the cells were centrifuged at 14,000 rpm for 15 min. The cell lysate was immunoprecipitated with α-TDP-43, α-FUS/TLS, α-rabbit IgG, and α-mouse IgG antibodies at 4 °C overnight. The RIP fractions were washed five times with wash buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Triton X-100). RNA was eluted with SDS elution buffer (50 mm Tris-HCl, pH 7.0, 10 mm EDTA, and 1.3% SDS) and purified using TRIzol (Invitrogen) according to the manufacturer's instructions. The isolated RNA from RIP was analyzed by quantitative RT-PCR. The results were expressed as the relative fold enrichment of the target precipitation as compared with the normal rabbit IgG control. The HDAC6 primers 5′-ACGGTCCCTCTTCACCTTCT-3′ (3′-UTR-F) and 5′-CTTCTGGGCTGGAGTAGTGG-3′ (3′-UTR-R) were used for quantitative PCR.
RNA Cross-linking Procedures
UV cross-linking of in vitro transcribed biotinylated RNA to protein lysate was performed according to Fiesel et al. with modifications (32). pPD129.36 encoding HDAC6 was obtained from Philipp J. Kahle (Hertie Institute for Clinical Brain Research, Tuebingen, Germany). Biotin-labeled HDAC6 mRNA (1 pmol) was incubated with 250 μl of RNA binding buffer (20 mm Hepes, pH 7.5, 5 mm MgCl2, 50 mm KCl, 150 mm NaCl, 0.5 mm EGTA, 0.5 mm dithiothreitol, 10% glycerol) and 0.1 or 1 μg of HA-TDP-43 or HA-FUS/TLS for 20 min at 30 °C. HA-TDP-43 or HA FUS/TLS were immunopurified from HEK 293T cells transfected with HA-TDP-43 or HA-FUS/TLS using α-HA-agarose-conjugated beads. UV irradiation was performed on ice with 1200 J/m2. Streptavidin beads (Novagen) were added, and precipitations were carried out overnight at 4 °C. The beads were washed with RNA-binding buffer, and elution was performed at 95 °C with SDS loading buffer. Elute and input were separated by 10% SDS-PAGE and analyzed by immunoblotting with α-HA antibodies.
RESULTS
Interaction between TDP-43 and FUS/TLS in Mammalian Cells
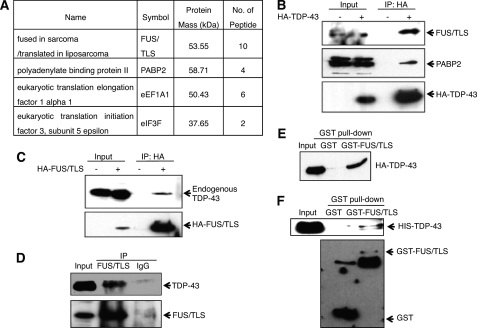
To gain insight into TDP-43 biochemical function, transiently expressed HA-tagged TDP-43 was immunoprecipitated from HeLa cells, and candidate co-precipitating proteins were identified by mass spectrometry (supplemental Fig. S1). A number of candidate TDP-43-interacting proteins were identified, including the nuclear poly(A)-binding protein PABP2, translation initiation factor eIF3f, and translation elongation factor eEF1A1 (Fig. 1A). In addition, a ∼70-kDa protein that co-immunoprecipitated with HA-TDP-43 was identified as FUS/TLS. To confirm the interactions, HA-TDP-43 was immunoprecipitated from transiently transfected HeLa cells, and the immunoprecipitation (IP) fractions were immunoblotted with α-FUS/TLS and α-PABP2 antibodies (Fig. 1B). Both endogenous PABP2 and FUS/TLS were detected in α-HA-TDP-43 IPs, providing preliminary evidence that TDP-43 forms complexes with PABP2 and FUS/TLS in mammalian cells. Additional experiments showed that overexpressed HA-FUS/TLS interacted with endogenous TDP-43 in co-IP assays (Fig. 1C) and that endogenous TDP-43 and FUS/TLS could be co-immunoprecipitated with α-FUS/TLS antibodies (Fig. 1D). In addition, GST pulldown assays demonstrated that purified recombinant GST-FUS/TLS bound to HA-TDP-43 from HEK 293T cell extracts, and this interaction was not abolished upon RNase pretreatment of the cell extract (Fig. 1E and supplemental Fig. S2). Finally, we showed that purified GST-FUS/TLS interacted with purified His-tagged TDP-43 in vitro (Fig. 1F). These findings provide the first direct evidence that TDP-43 and FUS/TLS interact in mammalian cells.
FIGURE 1.
TDP-43 interacts with PABP2 and FUS/TLS. A, identification of TDP-43 interacting proteins by mass spectrometry. HeLa cells transfected with HA-TDP-43 were immunoprecipitated with α-HA-conjugated agarose, and the immunoprecipitated proteins were separated by SDS-PAGE. The gel was stained with Colloidal Blue. Candidate TDP-43-associated proteins were analyzed by mass spectrometry. B, co-IP of HA-TDP-43 with endogenous FUS/TLS and PABP2. HA-TDP-43 was immunoprecipitated with α-HA, and the immunoprecipitated fractions were analyzed by immunoblotting with α-HA, α-FUS/TLS, and α-PABP2 antibodies. C, co-IP of HA-FUS/TLS and endogenous TDP-43. HeLa cells transfected with HA-FUS/TLS were immunoprecipitated with α-HA, and the immunoprecipitated fractions were analyzed by immunoblotting with α-HA and α-TDP-43 antibodies. D, interaction of endogenous TDP-43 and FUS/TLS. Exponentially growing HeLa cells were immunoprecipitated with α-FUS/TLS antibodies, and the immunoprecipitated fraction was analyzed by immunoblotting with α-FUS/TLS and α-TDP-43 antibodies. E, HA-TDP-43 interacts with GST-FUS/TLS in vitro. GST-FUS/TLS fusion proteins conjugated to glutathione-Sepharose 4B beads were incubated with HEK 293T cell extract containing HA-TDP-43, and bound proteins were analyzed by immunoblotting with α-GST and α-HA antibodies. F, interaction of purified proteins. Purified GST and GST-FUS/TLS fusion proteins conjugated to glutathione-Sepharose 4B beads were incubated with purified HIS-TDP-43, and bound proteins were analyzed by immunoblotting with α-GST and α-HIS antibodies.
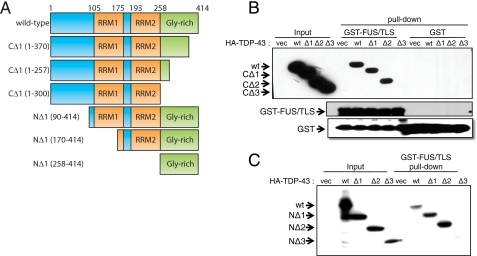
The TDP-43 Gly-rich and RRM2 Domains Contribute to FUS/TLS Binding
A panel of HA-tagged TDP-43 C- and N-terminal deletion mutants were constructed to determine which domain(s) were required for binding to FUS/TLS in GST-FUS/TLS pulldown assays (Fig. 2A). Deletion of the C-terminal 114 amino acids of TDP-43 had no effect on GST-FUS/TLS binding, whereas deletion of the C-terminal 157 amino acids, which includes the entire Gly-rich domain, ablated binding to GST-FUS/TLS in vitro (Fig. 2B). With respect to N-terminal TDP-43 deletion mutants, we found that TDP-43NΔ2, which lacks amino acids 1–169 but retains RRM2 and the Gly-rich domain, retained association with GST-FUS/TLS (Fig. 2C). On the other hand, a TDP-43NΔ3 mutant lacking RRM2 failed to interact with GST-FUS/TLS in vitro (Fig. 2C). These results suggest that the C-terminal Gly-rich and RRM2 domains are important for interaction with FUS/TLS.
FIGURE 2.
The Gly-rich and RRM2 domains of TDP-43 contribute to FUS/TLS binding. A, stick diagrams of TDP-43 deletion mutants used in the GST-FUS/TLS pulldown assays. B, GST-FUS/TLS pulldown assay using C-terminal deletion mutants of TDP-43. HA-tagged wild-type TDP-43 or deletion mutants of TDP-43 were expressed in HEK 293T cells, and the cell lysates were incubated with GST or GST-FUS/TLS proteins conjugated to glutathione-Sepharose 4B beads. Bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with α-GST and α-HA antibodies. C, interaction of FUS/TLS with N-terminal TDP-43 truncation mutants. The indicated TDP-43 N-terminal truncation mutants were expressed in HEK 293T cell and tested for interaction with GST-FUS/TLS in GST pulldown assays. These findings demonstrate that a region spanning amino acids 170–414 of TDP-43 is sufficient for binding to GST-FUS/TLS in vitro. vec, vector; wt, wild type.
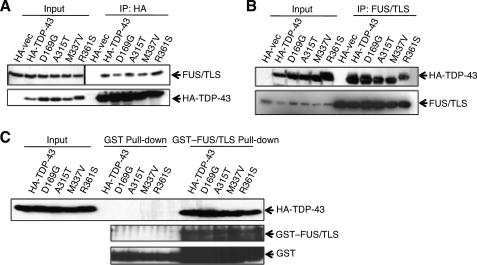
We also compared the FUS/TLS binding profiles of HA-tagged wild-type TDP-43 and four ALS-associated TDP-43 mutants (D169G, A315T, M337V, and R361S). Wild-type and mutant TDP-43 proteins interacted with FUS/TLS to a similar extent in reciprocal co-immunoprecipitation experiments (Fig. 3, A and B) and GST-FUS/TLS pulldown assays (Fig. 3C), suggesting that ALS mutations do not grossly alter the TDP-43-FUS/TLS interaction.
FIGURE 3.
Wild-type and ALS-associated TDP-43 mutants interact with FUS/TLS comparably. A, co-immunoprecipitation assay. HeLa cells were transfected with HA-tagged wild-type TDP-43 or ALS-associated TDP-43 mutants. Wild-type and mutant HA-TDP-43 proteins were immunoprecipitated with α-HA. The IP fractions were analyzed by immunoblotting with α-HA and α-FUS/TLS antibodies. B, reciprocal co-immunoprecipitation assay. HeLa cells were transfected with plasmids encoding HA-tagged wild-type TDP-43 or ALS-associated TDP-43 mutants, and cell extracts were immunoprecipitated with α-FUS/TLS antibodies. The IP fractions were analyzed by immunoblotting with α-HA and α-FUS/TLS antibodies. C, GST-FUS/TLS pulldown of ALS-associated TDP-43 mutants. GST and GST-FUS/TLS were induced in BL 21 cells and purified using glutathione-Sepharose 4B beads. HA-tagged wild-type TDP-43 or ALS-associated mutants of TDP-43 were expressed in HEK 293T cells, and corresponding cell extracts were incubated with GST or GST-FUS/TLS. The affinity-purified proteins were separated by 10% SDS-PAGE and analyzed by Western blotting with α-GST and α-HA antibodies. vec, vector.
Gel Filtration Reveals Two TDP-43 Complexes
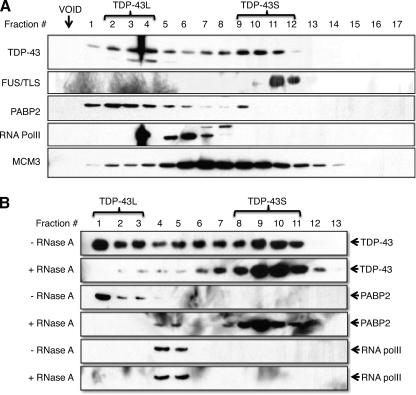
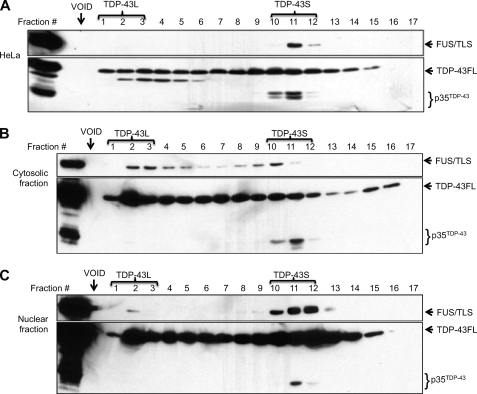
Based on the co-immunoprecipitation findings, we wished to determine whether PABP2 and FUS/TLS interacted with TDP-43 within a common complex. TDP-43 exhibited a heterogeneous migration pattern upon gel filtration chromatography of HeLa cell extracts; however, two major peaks of TDP-43 elution were observed (Fig. 4A). The first peak of TDP-43 immunoreactivity, denoted TDP-43-Large (TDP-43L), was observed immediately after the column void volume and, using the 500-kDa MCM (minichromosome maintenance) complex as a reference, was estimated to be ∼1–3 MDa in size. A second TDP-43 elution peak, with an estimated size of ∼300–400 kDa, was also observed and designated TDP-43-Small (TDP-43S).
FIGURE 4.
Gel filtration analysis of TDP-43 and FUS/TLS complexes. A, gel filtration analysis of TDP-43, FUS/TLS, PABP2, RNA PolII, and MCM3 from HeLa whole cell extracts. HeLa extracts were size-fractionated using Superose 6 and immunoblotted with the indicated antibodies. The approximate locations of TDP-43L and TDP-43S complexes are shown. B, TDP-43L complexes are RNA-dependent. HeLa extracts were treated with RNase A or vehicle, resolved by gel filtration, and analyzed with α-TDP-43, α-PABP2, and α-RNA PolII antibodies.
The gel filtration profile of FUS/TLS in HeLa cells was much more homogeneous than that of TDP-43, with virtually all FUS/TLS eluting in several fractions that overlapped extensively with TDP-43S (Fig. 4A). On the other hand, PABP2 co-fractionated with TDP-43L but not TDP-43S, suggesting that its interaction with TDP-43 occurs within the context of the larger complex. Finally, we found that RNA PolII eluted between TDP-43L and TDP-43S, suggesting that it does not form stable complexes with TDP-43 or FUS/TLS. These findings suggested that TDP-43 exists in two complexes in HeLa cells: a large TDP-43L complex that contains PABP2 and a smaller TDP-43S complex that overlaps extensively with FUS/TLS.
The large size of TDP-43L and its co-fractionation with PABP suggested that it may represent an RNP particle. To test this idea we compared gel filtration profiles of TDP-43 from RNase-treated versus untreated cells. RNase treatment strongly right-shifted the TDP-43 and PABP2 elution profiles such that virtually the entire cellular pool of TDP-43 was present in fractions corresponding to TDP-43S (Fig. 4B). By contrast, the RNA PolII elution profile was not changed after RNase treatment (Fig. 4B). This finding implied that TDP-43L is a stable RNP complex and that TDP-43S is not.
To further substantiate that TDP-43L is an RNP, we immunoprecipitated pooled TDP-43L fractions with α-TDP-43 antibodies and performed protein profiling using mass spectrometry. Numerous ribosomal proteins, RNA-binding proteins, and translational factors were identified as TDP-43 interacting proteins in the TDP-43L complex (supplemental Table S1). These results strongly support the idea that TDP-43L complexes are RNP in nature. Finally, to test the complex forming property of ALS-associated TDP-43 mutants, we compared the gel filtration profiles of HA-tagged wild-type TDP-43 and four ALS-associated TDP-43 mutants (D169G, A315T, M337V, and R361S). The column elution profiles of wild-type and mutant TDP-43 proteins were not discernibly different, suggesting that the complex forming potential of mutant TDP-43 proteins is largely intact (supplemental Fig. S3).
TDP-43 Fragments Are Excluded from TDP-43L Complexes
TDP-43 is cleaved into 25- and 35-kDa C-terminal fragments in cell culture and degenerating ALS spinal cord motor neurons (4, 33, 34). It has been hypothesized that these aggregation-prone TDP-43 cleavage fragments are an important determinant of TDP-43-dependent neuropathology (34). To ascertain the complex-forming properties of TDP-43 fragments, we performed additional gel filtration runs using longer autoradiographic exposures to visualize scarce TDP-43 species. This analysis showed that a 35-kDa TDP-43-immunoreactive doublet, designated p35TDP-43, co-eluted with TDP-43S but not TDP-43L (Fig. 5A). To validate this finding and to determine whether TDP-43-immunoreactive fragments corresponded to N-terminal or C-terminal regions of TDP-43, HeLa gel filtration fractions were immunoblotted with antibodies generated against the C-terminal 154 amino acids of TDP-43. These antibodies also recognized the p35TDP-43 doublet that co-fractionated with FUS/TLS (supplemental Fig. S4). Additionally, transfection of HeLa cells or HEK 293T cells with TDP-43 siRNA abolished the 35-kDa immunoreactive band. This finding strongly suggests it to be a bona fide TDP-43 fragment (supplemental Fig. S5).
FIGURE 5.
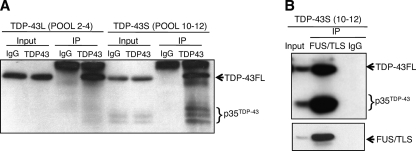
35-kDa TDP-43 fragments are excluded from TDP-43L complexes. A, immunoprecipitation of TDP-43 and p35TDP-43 from pooled gel filtration fractions. TDP-43L (fractions 2–4) and TDP-43S (fractions 10–12) were immunoprecipitated with IgG or α-TDP-43 antibodies and immunoblotted with α-TDP-43 antibodies. B, FUS/TLS co-immunoprecipitates with full-length TDP-43 and p35TDP-43. Gel filtration fractions 10–12 (TDP-43S) were immunoprecipitated with α-FUS/TLS and α-mouse IgG antibodies, and the immunoprecipitated fractions were analyzed by immunoblotting with α-TDP-43 and α-FUS/TLS antibodies.
To ascertain the generality of findings obtained using HeLa cell extracts, we examined gel filtration profiles of TDP-43 and FUS/TLS in mouse embryo fibroblasts and NSC-34 cells derived from fusion of mouse motor neurons with neuroblastoma cells. In both cell lines p35TDP-43 fragments were found to co-fractionate with TDP-43S complexes and were excluded from TDP-43L complexes (supplemental Fig. S6). Although results from mouse embryo fibroblasts and NSC-34 cells largely recapitulated findings in HeLa cells, we observed a greater proportion of FUS/TLS in high molecular mass complexes (supplemental Fig. S6). This latter finding indicates that there may be cell type-dependent differences in FUS/TLS complexes; however, TDP-43 fragments are consistently excluded from TDP-43L RNP complexes in all of the cell lines tested.
To more definitively show that p35TDP-43 co-fractionated with TDP-43S and not TDP-43L, we pooled HeLa gel filtration fractions containing TDP-43L (2–4) or TDP-43S (10–12) and performed co-immunoprecipitation experiments with α-TDP-43 antibodies. Both full-length TDP-43 and p35TDP-43 were immunoprecipitated with α-TDP-43 antibodies from TDP-43S-containing fractions, whereas only full-length TDP-43 was immunoprecipitated from TDP-43L-containing fractions (Fig. 5A). This finding is consistent with the notion that p35TDP-43 is largely excluded from TDP-43L complexes.
To determine whether endogenously produced TDP-43 fragments retain the capacity to associate with FUS/TLS, we immunoprecipitated pooled TDP-43S fractions with α-FUS/TLS antibodies and tested for the presence of TDP-43 and p35TDP-43. We found that both full-length TDP-43 and p35TDP-43 were detected in α-FUS/TLS IPs from TDP-43S-containing fractions (Fig. 5B). These findings demonstrate that endogenous TDP-43 and FUS/TLS interact in the context of TDP-43S complexes and that endogenously expressed fragments retain the capacity to bind to FUS/TLS.
Analysis of Cytosolic and Nuclear TDP-43 Complexes
TDP-43 and FUS/TLS are predominantly nuclear proteins; however, cytosolic localization patterns have also been reported. We therefore sought to determine whether TDP-43L, TDP-43S, and FUS/TLS complexes were enriched in nuclear or cytosolic compartments. Gel filtration revealed a general trend that HeLa nuclear fractions were enriched in TDP-43S-containing complexes, whereas TDP-43L complexes were enriched in the cytosol (Fig. 6, B and C). p35TDP-43 eluted with TDP-43S in both cytosolic and nuclear fractions. Interestingly, the majority of FUS/TLS co-fractionated with nuclear TDP-43S complexes; however, FUS/TLS was also observed in high molecular mass cytosolic complexes that overlapped with TDP-43L (Fig. 6, B and C). These findings suggest that TDP-43S and TDP-43L complexes are differentially localized between nucleus and cytoplasm, respectively.
FIGURE 6.
TDP-43S complexes are enriched in the nucleus. A, gel filtration analysis of full-length TDP-43, p35TDP-43, and FUS/TLS from HeLa whole cell extracts. HeLa extracts were size-fractionated using Superose 6 and immunoblotted with α-TDP-43 and α-FUS/TLS antibodies. The approximate locations of TDP-43L and TDP-43S complexes are shown. B, gel filtration analysis of TDP-43, p35TDP-43, and FUS/TLS from HeLa cell cytosolic fractions. C, gel filtration analysis of TDP-43, p35TDP-43, and FUS/TLS from HeLa cell nuclear fractions.
TDP-43 and FUS/TLS Co-regulate HDAC6 Expression
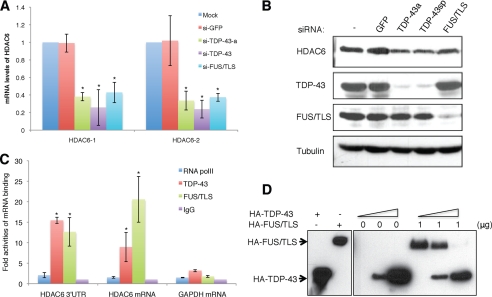
The above findings established the existence of distinct TDP-43 complexes and potential direct interaction between TDP-43 and FUS/TLS. To probe the functional significance of the TDP-43-FUS/TLS interaction, we tested whether the proteins interacted in the context of a defined RNA substrate. Recent studies demonstrated that TDP-43 enhanced the mRNA stabilities of light neurofilament and HDAC6 mRNAs through binding to 3′-UTR sequences (32, 35). We found that knockdown of TDP-43 in HEK 293T cells reduced steady-state HDAC6 mRNA expression relative to cells transfected with control siRNA, which supports the findings of Fiesel et al. (32) (Fig. 7A). In addition, knockdown of FUS/TLS also reduced steady-state HDAC6 expression, suggesting that FUS/TLS regulates HDAC6. Correspondingly, HDAC6 protein was reduced in either TDP-43 or FUS/TLS depleted cells, and the simultaneous knockdown of TDP-43 and FUS/TLS resulted in a slight additional reduction in HDAC6 expression (Fig. 7B and supplemental Fig. S7). These findings reveal a co-requirement of TDP-43 and FUS/TLS for optimal HDAC6 expression in HeLa cells.
FIGURE 7.
TDP-43 and FUS/TLS regulate HDAC6 mRNA expression. A, HEK 293T cells were transfected with siRNAs for TDP-43-a (single siRNA), TDP-43 (SMARTpool), FUS/TLS, or GFP. Forty-eight h later total RNA was prepared from the cells, and cDNA was analyzed by quantitative RT-PCR using two different primer sets for HDAC6 and normalized to GAPDH. *, p < 0.05. B, HEK 293T extracts transiently transfected with indicated siRNAs were separated by 10% SDS-PAGE and analyzed by immunoblotting with α-TDP-43, α-FUS/TLS, α-HDAC6, and α-β-tubulin antibodies. C, HEK 293T extracts were immunoprecipitated with α-TDP-43, α-FUS/TLS, and α-rabbit IgG antibodies, and the co-immunoprecipitated RNA was analyzed with quantitative RT-PCR using primer sets for 3′-UTR and mRNA of HDAC6, and GAPDH. *, p < 0.05. D, HA-TDP-43 and HA-FUS/TLS were expressed in HEK 293T cells and purified using α-HA-conjugated agarose beads. Biotin-labeled HDAC6 mRNA conjugated to streptavidin beads was incubated with different amounts of purified HA-TDP-43 and/or HA-FUS/TLS, and bound proteins were eluted and analyzed by SDS-PAGE and immunoblotting with α-HA antibodies.
The quantitative PCR results above suggest that TDP-43 and FUS/TLS are both recruited to HDAC6 mRNA. To test this possibility, we performed RIP analysis of HDAC6 mRNA occupancy using α-TDP-43 and α-FUS/TLS antibodies. These experiments revealed that HDAC6 mRNA was immunoprecipitated with α-TDP-43 and α-FUS/TLS but not RNA PolII or control IgG antibodies (Fig. 7C). Thus, both TDP-43 and FUS/TLS interact with HDAC6 mRNA. To test for specificity of RIP using α-TDP-43 and α-FUS/TLS antibodies, we performed RIP analysis in cells that had been depleted for TDP-43 or FUS/TLS using RNAi. Knockdown of TDP-43 reduced TDP-43 recruitment to HDAC6 mRNA, and knockdown of FUS/TLS reduced FUS/TLS recruitment to HDAC6 mRNA, which strongly supports the specificity of the observed RIP signals (supplemental Fig. S8). These results support a model in which TDP-43 and FUS/TLS specifically bind to HDAC6 mRNA to regulate its expression.
The RIP analysis suggests that both TDP-43 and FUS/TLS occupy HDAC6 mRNA in intact cells, but the experiments could not address whether the proteins bind simultaneously or competitively. To test for direct interaction of TDP-43 or FUS/TLS with HDAC6 mRNA, we generated biotin-labeled HDAC6 mRNA through in vitro transcription. Biotin-labeled mRNA was incubated with immunopurified HA-TDP-43 and/or HA-FUS/TLS from transiently transfected HEK 293T cells. After UV cross-linking, the protein-biotin-HDAC6 complexes were purified using streptavidin-conjugated beads and analyzed by immunoblotting with α-HA antibodies. HA-FUS/TLS as well as HA-TDP-43 interacted with HDAC6 mRNA (Fig. 7D). To test for cooperative or competitive binding, we incubated biotin-HDAC6 mRNA and HA-FUS/TLS with increasing amounts of HA-TDP-43. As shown in Fig. 7D, the addition of increasing amounts of purified HA-TDP-43 caused a decrease in FUS/TLS binding to HDAC6 mRNA in vitro, suggesting that TDP-43 and FUS/TLS occupy a common binding site (Fig. 7D).
TDP-43 participates in pre-mRNA splicing but has also been implicated in mRNA stabilization (36). To test whether decreased mRNA stability accounted for reduced HDAC6 mRNA expression in TDP-43 or FUS/TLS knockdown cells, we measured decay of steady-state HDAC6 mRNA expression following treatment of HEK 293T cells with the RNA PolII inhibitor, actinomycin D. As expected, steady-state levels of HDAC6 were strongly reduced in TDP-43 and FUS/TLS-depleted cells; however, knockdown of the factors did not accelerate the disappearance of HDAC6 mRNA following actinomycin D treatment (supplemental Fig. S9). This finding suggests that TDP-43 and FUS/TLS are not required for HDAC6 mRNA stabilization and may instead regulate HDAC6 processing and/or nuclear export.
DISCUSSION
TDP-43 and FUS/TLS are structurally related RNA-binding proteins whose respective gene mutations cause convergent ALS phenotypes in humans. The underlying basis for phenotypic convergence is not known. The structural relatedness of TDP-43 and FUS/TLS suggests that misfolded variants of either protein may acquire a related toxic gain of function. Nonexclusively, TDP-43 and FUS/TLS may function in common biochemical pathways sensitive to dominant mutations and/or dosage of either protein. The present findings showing that TDP-43 and FUS/TLS directly interact to regulate HDAC6 mRNA expression provide evidence for a common pathway model.
A recent proteomic analysis of FLAG-TDP-43-interacting proteins identified factors involved in RNA splicing and translation, and FUS/TLS peptides were identified in FLAG-TDP-43 immunoprecipitates in that study (37). Here we showed that TDP-43 and FUS/TLS interacted in intact cells at endogenous expression levels (Figs. 1D and 5B), directly interacted as purified proteins in vitro in an RNA-independent manner (Fig. 1F and supplemental Fig. S2), and associated via the C-terminal region of TDP-43 harboring the RRM2 and Gly-rich domains (Fig. 2, B and C). The combined binding studies provide definitive evidence that fractions of TDP-43 and FUS/TLS exist in complex in mammalian cells.
The finding that cellular TDP-43 partitions as two separable complexes on size exclusion media suggests that disparate functions of TDP-43 may be carried out by molecularly distinct complexes. The larger of the two TDP-43 complexes, TDP-43L, is several MDa in size, RNase A-sensitive, and enriched in ribosomal proteins and translation factors (supplemental Table S1), which were also identified as FLAG-TDP-43-binding proteins from unfractionated cell extracts by Freibaum et al. (37). Thus, we propose that TDP-43L complexes represent TDP-43 associated with RNPs. On the other hand, the TDP-43S complex was ∼300–400 kDa in size, was RNase-resistant, and overlapped extensively with FUS/TLS (Fig. 4A). Subcellular fractionation further revealed that TDP-43S complexes were enriched in the nucleus (Fig. 6). Although the molecular constituents of TDP-43S have not been unambiguously identified, it is plausible that such nuclear complexes are involved in transcriptional control or RNA splicing functions attributed to TDP-43 and FUS/TLS (38, 39).
Most cellular FUS/TLS co-fractionated with TDP-43S complexes during gel filtration chromatography of whole cell extracts (Figs. 4A and 6A). Although their elution profiles were not identical, the proteins co-immunoprecipitated in the presence of RNase from pooled TDP-43S fractions (Fig. 1F and supplemental Fig. S2C). These findings suggest that TDP-43 and FUS/TLS interact in the context of the smaller TDP-43S complex but do not rule out the possibility that TDP-43 and FUS/TLS also interact within the context of larger TDP-43L-RNP complexes. In fact, high molecular mass complexes of FUS/TLS were observed in the cytoplasm, and they overlapped with TDP-43L (Fig. 6B). We speculate that such cytosolic complexes are involved in the transport and/or localized translation of mRNA, functions previously ascribed to FUS/TLS (31).
While our paper was under review Ling et al. (40) reported that ALS-associated mutations in TDP-43 increased its binding affinity for FUS/TLS. However, we saw no evidence for increased binding of mutant TDP-43 proteins to FUS/TLS in reciprocal co-IP experiments in which either HA-TDP-43 or endogenous FUS/TLS was the immunoprecipitated target (Fig. 3, A and B). Wild-type and mutant HA-TDP-43 proteins also showed comparable binding to GST-FUS/TLS in vitro (Fig. 3C). The reason for the discrepant findings is not entirely clear given that the TDP-43M337V mutant was examined in both studies. However, the Ling et al. study employed stably expressed GFP-TDP-43 fusion proteins for co-immunoprecipitation studies, whereas we investigated transiently expressed TDP-43 proteins harboring a smaller HA epitope. We also found that ALS-associated mutations did not grossly affect TDP-43 gel filtration elution profiles, which would be predicted if mutation altered TDP-43 complexes (supplemental Fig. S3). Thus, although we cannot rule out subtle binding differences in these assays, our findings do not support increased binding of ALS-associated TDP-43 mutant proteins to FUS/TLS. This important question clearly requires additional study.
Although TDP-43 mutants appeared grossly normal in gel filtration and FUS/TLS interaction assays, C-terminal fragments of TDP-43, which have received attention as possible ALS disease determinants, showed altered complex forming potential in all of the cell lines tested (15–17, 34). Specifically, the major 35-kDa TDP-43 fragment, p35TDP-43, was excluded from the RNA-dependent TDP-43L complexes but retained its ability to interact with FUS/TLS (Fig. 5). Although the disease relevance of TDP-43 fragments is still uncertain, we speculate that C-terminal fragments of TDP-43 produced during the course of disease form soluble complexes with FUS/TLS that possess altered functional properties. A comprehensive understanding of FUS/TLS-dependent gene expression will facilitate the testing of this hypothesis.
The finding that TDP-43 and FUS/TLS are jointly required for HDAC6 mRNA expression provides strong evidence that these proteins regulate overlapping downstream pathways including pre-mRNA splicing and/or mRNA export. Our findings are compatible with the results of Fiesel et al. (32) showing that TDP-43 regulates expression of HDAC6, which plays key roles in autophagic clearance of protein aggregates and is linked to suppression of neurodegeneration (41–43). Interestingly, although knockdown of either TDP-43 or FUS/TLS caused strong down-regulation of HDAC6, simultaneous knockdown of both factors was less than additive (supplemental Fig. S7). This finding suggests that the proteins work in complex or possibly sequentially in HDAC6 mRNA processing. Based on the results of the actinomycin D experiments, HDAC6 mRNA stabilization appears not to be regulated by TDP-43 and FUS/TLS (supplemental Fig. 9). However, because TDP-43 or FUS/TLS knockdown caused dramatic reductions in steady-state HDAC6 mRNA expression prior to treatment of actinomycin D, we cannot exclude that TDP-43 and FUS/TLS make a minor contribution to HDAC6 mRNA stabilization.
Purified FUS/TLS also bound to HDAC mRNA in vitro, and titration experiments suggest that TDP-43 and FUS/TLS compete for common binding sites (Fig. 7D). One model that can accommodate these findings posits that TDP-43 and FUS/TLS occupy HDAC6 mRNA as homodimers or heterodimers. In the presence of high in vitro concentrations of TDP-43, HDAC6 mRNA is occupied primarily by TDP-43 homodimers, which form spontaneously (44). Understanding the stoichiometry, dynamics, and functional consequences of TDP-43 and FUS/TLS interactions at the level of individual RNA substrates should yield important insights into the biological interaction of these proteins in health and neurodegenerative disease.
In conclusion, we have shown that TDP-43 exists in distinct biochemical complexes in mammalian cells and interacts with FUS/TLS. Although much remains to be learned about the TDP-43-FUS/TLS functional relationship in the context of HDAC6 and other mRNA substrates, participation in a common biochemical complex may partially explain the convergent ALS phenotypes associated with TARDBP or FUS/TLS gene mutations in humans.
Supplementary Material
Acknowledgments
We thank Dr. Shigeki Miyamoto and Dr. Richard Anderson for access to chromatography instrumentation and Grzegorz Sabat for mass spectrometric analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA124722 and NS059001. This work was also supported by the American Cancer Society and a Shaw Scientist Award (to R. S. T.) from the Greater Milwaukee Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S9.
- ALS
- amyotrophic lateral sclerosis
- HDAC
- histone deacetylase
- RNP
- ribonucleoprotein
- PolII
- polymerase II
- RIP
- RNA immunoprecipitation
- IP
- immunoprecipitation.
REFERENCES
- 1.Nelson L. M. (1995) Clin. Neurosci. 3, 327–331 [PubMed] [Google Scholar]
- 2.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X. (1993) Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 3.Ilieva H., Polymenidou M., Cleveland D. W. (2009) J. Cell Biol. 187, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 5.Buratti E., Baralle F. E. (2008) Front. Biosci. 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 6.Sephton C. F., Good S. K., Atkin S., Dewey C. M., Mayer P., 3rd, Herz J., Yu G. (2010) J. Biol. Chem. 285, 6826–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 10.Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., Onodera O. (2008) Ann. Neurol. 63, 538–542 [DOI] [PubMed] [Google Scholar]
- 12.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 13.Pesiridis G. S., Lee V. M., Trojanowski J. Q. (2009) Hum Mol. Genet. 18, R156–R162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson B. S., McCaffery J. M., Lindquist S., Gitler A. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igaz L. M., Kwong L. K., Chen-Plotkin A., Winton M. J., Unger T. L., Xu Y., Neumann M., Trojanowski J. Q., Lee V. M. (2009) J. Biol. Chem. 284, 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009) J. Biol. Chem. 284, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann M., Kwong L. K., Lee E. B., Kremmer E., Flatley A., Xu Y., Forman M. S., Troost D., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2009) Acta Neuropathol. 117, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S. H., Shi Y., Hanson K. A., Williams L. M., Sakasai R., Bowler M. J., Tibbetts R. S. (2009) J. Biol. Chem. 284, 8083–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caccamo A., Majumder S., Deng J. J., Bai Y., Thornton F. B., Oddo S. (2009) J. Biol. Chem. 284, 27416–27424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urushitani M., Sato T., Bamba H., Hisa Y., Tooyama I. (2010) J. Neurosci. Res. 88, 784–797 [DOI] [PubMed] [Google Scholar]
- 22.Hanson K. A., Kim S. H., Wassarman D. A., Tibbetts R. S. (2010) J. Biol. Chem. 285, 11068–11072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabashi E., Lin L., Tradewell M. L., Dion P. A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H. D., Vande Velde C., Rouleau G. A., Drapeau P. (2010) Hum. Mol. Genet. 19, 671–683 [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Ray P., Rao E. J., Shi C., Guo W., Chen X., Woodruff E. A., 3rd, Fushimi K., Wu J. Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3169–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Ceuterick-de Groote C., Van Broeckhoven C., Kumar-Singh S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feiguin F., Godena V. K., Romano G., D'Ambrogio A., Klima R., Baralle F. E. (2009) FEBS Lett. 583, 1586–1592 [DOI] [PubMed] [Google Scholar]
- 28.Kwiatkowski T. J., Jr., Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T., Valdmanis P., Rouleau G. A., Hosler B. A., Cortelli P., de Jong P. J., Yoshinaga Y., Haines J. L., Pericak-Vance M. A., Yan J., Ticozzi N., Siddique T., McKenna-Yasek D., Sapp P. C., Horvitz H. R., Landers J. E., Brown R. H., Jr. (2009) Science 323, 1205–1208 [DOI] [PubMed] [Google Scholar]
- 29.Vance C., Rogelj B., Hortobágyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K. L., Tripathi V., Al-Saraj S., Al-Chalabi A., Leigh P. N., Blair I. P., Nicholson G., de Belleroche J., Gallo J. M., Miller C. C., Shaw C. E. (2009) Science 323, 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. (1997) J. Cell Sci. 110, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 31.Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G. G., Takumi T. (2005) Curr. Biol. 15, 587–593 [DOI] [PubMed] [Google Scholar]
- 32.Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., Walter M., Anderson M. L., Kern J. V., Rasse T. M., Schmidt T., Springer W., Kirchner R., Bonin M., Neumann M., Baekelandt V., Alunni-Fabbroni M., Schulz J. B., Kahle P. J. (2010) EMBO J. 29, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimoto Y., Ito D., Yagi T., Nihei Y., Tsunoda Y., Suzuki N. (2010) J. Biol. Chem. 285, 608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igaz L. M., Kwong L. K., Xu Y., Truax A. C., Uryu K., Neumann M., Clark C. M., Elman L. B., Miller B. L., Grossman M., McCluskey L. F., Trojanowski J. Q., Lee V. M. (2008) Am. J. Pathol. 173, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strong M. J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. (2007) Mol. Cell Neurosci. 35, 320–327 [DOI] [PubMed] [Google Scholar]
- 36.Volkening K., Leystra-Lantz C., Yang W., Jaffee H., Strong M. J. (2009) Brain Res. 1305, 168–182 [DOI] [PubMed] [Google Scholar]
- 37.Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagier-Tourenne C., Polymenidou M., Cleveland D. W. (2010) Hum Mol. Genet. 19, R46–R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strong M. J. (2010) J. Neurol. Sci. 288, 1–12 [DOI] [PubMed] [Google Scholar]
- 40.Ling S. C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S. H., Garrido C., Yao T. P., Vourc'h C., Matthias P., Khochbin S. (2007) Genes Dev. 21, 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J. Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y. S., Pandey U. B., Kaushik S., Tresse E., Lu J., Taylor J. P., Cuervo A. M., Yao T. P. (2010) EMBO J. 29, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P. (2007) Nature 447, 859–863 [DOI] [PubMed] [Google Scholar]
- 44.Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K., Yuan H. S. (2009) Nucleic Acids Res. 37, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.