Abstract
The NAD rescue pathway consists of two enzymatic steps operated by nicotinamide phosphoribosyltransferase (Nampt) and nicotinamide mononucleotide adenylyltransferases. Recently, the potent Nampt inhibitor FK866 has been identified and evaluated in clinical trials against cancer. Yet, how Nampt inhibition affects NAD contents and bioenergetics is in part obscure. It is also unknown whether NAD rescue takes place in mitochondria, and FK866 alters NAD homeostasis within the organelle. Here, we show that FK866-dependent reduction of the NAD contents is paralleled by a concomitant increase of ATP in various cell types, in keeping with ATP utilization for NAD resynthesis. We also show that poly- and mono(ADP-ribose) transferases rather than Sirt-1 are responsible for NAD depletion in HeLa cells exposed to FK866. Mass spectrometry reveals that the drug distributes in the cytosolic and mitochondrial compartment. However, the cytoplasmic but not the mitochondrial NAD pool is reduced upon acute or chronic exposure to the drug. Accordingly, Nampt does not localize within the organelles and their bioenergetics is not affected by the drug. In the mouse, FK866-dependent reduction of NAD contents in various organs is prevented by inhibitors of poly(ADP-ribose) polymerases or the NAD precursor kynurenine. For the first time, our data indicate that mitochondria lack the canonical NAD rescue pathway, broadening current understanding of cellular bioenergetics.
Keywords: ATP, Metabolism, Mitochondria, NAD, Nicotinamide
Introduction
Several enzymes that transform NAD, used as a bona fide substrate, into metabolites displaying pleiotypic properties have been recently identified (1–4). Among them, ADP-ribose (ADPR)2 transferases such as poly(ADP-ribose) polymerase (PARP)-1, -2, -3, -10, tankyrases, v-PARP, and others members of the same family are involved in processes such as nuclear homeostasis, cell differentiation, and death (5, 6). Mono(ADP-ribose) transferases (MART) are NAD-consuming enzymes present on cell membranes (but probably also in cytosol and organelles) that are still poorly understood (7). Additional NAD-consuming enzymes are CD-38/CD157, two ectoenzymes synthesizing ADPR, and the intracellular Ca2+-mobilizing compound cyclic ADPR (8), and sirtuins, a family of proteins encompassing seven members (Sirt1–7) involved in numerous processes such as chromatin homeostasis, transcription, cell death, and lifespan extension (9).
Because of the constitutive activity of the NAD-consuming enzymes, eukaryotic cells evolved a rescue pathway leading to NAD resynthesis from nicotinamide (Nam). NAD rescue occurs in parallel to NAD neosynthesis from tryptophan or nicotinic acid (the so-called “kynurenine” and “Priess-Handler” pathways, respectively) (10, 11). Recently, work from the Brenner laboratory (12) identified nicotinamide riboside as an additional NAD precursor. The biochemical route of NAD rescue is composed by two enzymatic steps, the first operated by nicotinamide phosphoribosyltransferase (Nampt), forming nicotinamide mononucleotide from Nam and phosphoribosyl pyrophosphate, and the second driven by nicotinamide mononucleotide adenylyltransferases, leading to NAD formation from nicotinamide mononucleotide and ATP (1).
Despite renewed interest in NAD biology, tools able to specifically modulate the enzymes involved in NAD metabolism are lacking. Indeed, with the exception of PARP inhibitors, which have already reached the clinical arena (13), compounds able to potently and selectively target MART, CD38/CD157, and sirtuins have not been unequivocally identified. Recent work, however, identified FK866 (also known as APO866 or WK175) as an inhibitor of Nampt able to reduce the intracellular NAD pool. Initially identified as a noncompetitive inhibitor (14), further studies indicated that FK866 competes with Nam on binding to the Nampt catalytic pocket (15, 16). For the first time, this drug allowed to pharmacologically modulate the NAD rescue pathway, thereby providing important information about the turnover rate of pyridine nucleotide precursors in different tissues. Importantly, FK866-dependent intracellular NAD depletion proved the efficacy in triggering cell death of both tumor cell lines as well as primary tumor cells (17). Given the safe profile shown by FK866 in phase I clinical trials, the drug is now under scrutiny in phase II trials for the treatment of solid and hematologic malignancies (18, 19). In keeping with the emerging immunomodulatory role of Nampt, further studies also highlighted the potent immunosuppressive properties of FK866 (20–23), pointing to NAD resynthesis as a key regulator of immune cell differentiation and activation.
Despite knowledge on the pharmacodynamic effects of Nampt inhibition by FK866, several questions remain to be answered. For instance, the relative role of the different NAD-consuming enzymes in FK866-induced NAD depletion and tumor cell death is currently unknown. Also, the effect of the drug on NAD homeostasis at the subcellular levels are still not understood. Finally, the impact of Nampt inhibition on cellular bioenergetics waits to be investigated. In the present study we aimed at clarifying these issues by adopting both in vitro and in vivo experimental settings.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions and Evaluation of Viability
HeLa, MT2, fibroblasts, and C6 glioma cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mm glutamine, 10% fetal bovine serum and antibiotics. Cultures were brought to 50–70% confluence and used for the experiments. Pure neuronal cultures were prepared by seeding cortical cells obtained from 16-day-old rat embryos as previously described (24). Neurons were cultured in NeurobasalTM medium with B-27 supplement (Invitrogen) and 0.5 mm glutamine onto poly-d-lysine-coated multiwell plates. Cells were used at 7 days in vitro from preparation. Rat glia cultures were prepared as described (25) and used at 15 days in vitro. Cell cultures were exposed to FK866 (RTI International, Research Triangle Park, NC), N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), phenanthridinone (PHE), N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-N,N-dimethylacetamide (PJ34), meta-iodobenzylguanidine (mIBG, kindly provided by Maria di Girolamo, Istituto Mario Negri Sud, Chieti, Italy), EX-257 (6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide), N-(4-phenylthiazol-2-yl)benzensulphonamide (Ro-618048), or other compounds directly dissolved in the culture medium. Unless otherwise specified, MNNG was 100 μm. Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium assay and phase-contrast microscopy (26). Cell transfection was performed as described (26). Plasmid was kindly provided by Shin-Ichiro Imai (Nampt-GFP) (Washington University School of Medicine, St. Louis, MO) and Rosario Rizzuto (Cytosolic GFP and Mit-GFP)(University of Padua, Italy).
NAD, NADH, and ATP Measurement
NAD+ contents were quantified by means of an enzymatic cycling procedure according to Shah et al. (27). Briefly, cells grown in a 48-well plate were killed with 1 n 50 μl of HClO4 and then neutralized with an equal volume of 1 n KOH. After the addition of 100 μl of Bicine (100 mm), pH 8, 50 μl of the cell extract was mixed with an equal volume of the Bicine buffer containing 23 μl/ml of ethanol, 0.17 mg/ml of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium, 0.57 mg/ml of phenazine ethosulfate, and 10 μg of alcohol dehydrogenase. The mixture was kept at room temperature for 20 min and then absorbance at 550 nm was measured. A standard curve allowed quantification of NAD. Intracellular NADH content were measured by quantifying cell autofluorescence as previously reported (26). Briefly, autofluorescence was measured with the a microscopic apparatus (Nikon 2000-TU) equipped with an UV filter and a CCD camera. Five microscopic fields were randomly acquired per each cell culture slide and processed for fluorescence quantitation (Metamorph/Metafluor software). Cellular ATP content and mitochondrial ATP production was measured by means of a commercial kit from PerkinElmer Life Sciences as described (26).
Western Blotting
For Western blotting, cells were scraped, collected in Eppendorf tubes, centrifuged (1500 × g for 5 min at 4 °C) and resuspended in lysis buffer (50 mm Tris, pH 7.4, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (PMSF), 4 μg/ml of aprotinin and leupeptin, 1% SDS). 20–40 μg of protein/lane were loaded. After 4–20% SDS-PAGE and blotting, membranes (Hybond-ECL, Amersham Biosciences) were blocked with phosphate-buffered saline (PBS) containing 0.1% Tween 20 and 5% skim milk (TPBS, 5% milk) and then probed overnight with primary antibodies (1:1000 in TPBS, 5% milk). The anti-poly(ADP-ribose) monoclonal antibody (10H) was from Alexis (Vinci, Italy), the polyclonal anti-Nampt antibody (catalog number AP9010c) was from Abgent (San Diego, CA), and the anti-apoptosis inducing factor was from Santa Cruz (Tamecula, CA). Membranes were then washed with TPBS and incubated 1 h in TPBS, 5% milk containing an anti-mouse peroxidase-conjugated secondary antibody (1:2000). After washing in TPBS, ECL (Amersham Biosciences) was used to visualize the peroxidase-coated bands.
Mitochondrial Isolation and Handling
Mitochondria were isolated from cells using a glass/glass homogenizer in 500 μl of extraction buffer and centrifuged at 600 × g as described (28). Briefly, supernatants were centrifuged at 7000 × g to obtain the mitochondrial pellet that was denatured with perchloric acid for NAD content determination, or resuspended in respiration buffer (10 mm NaCl, 140 mm KCl, 2 mm MgSO4, 1 mm KH2PO4, 100 nm CaCl2, and 20 mm HEPES, pH 7, 2 mm succinate, 1 mm pyruvate, 3 μm ADP) in the presence or absence of different compounds. For subcellular analysis of Nampt distribution, the mitochondrial pellet was washed twice with 0.5 m NaCl and then denatured with lysis buffer. ATP was measured by the ATPlight kit (PerkinElmer Life Sciences).
PARP-1 Activity Assay
For PARP-1 activity evaluation, the enzymatic reaction was carried out in a final volume of 100 μl consisting of: 20 mm Tris-HCl, pH 8.0, 20 mm MgCl2, 5 mm dithiothreitol, 20 μg of sonicated calf thymus DNA, 0.2 μCi of [adenine-2,8-3H]NAD, and 0.13 unit of partially purified bovine PARP-1. Ten microliters of the tested compounds were added to the enzymatic reaction. An equal amount of the vehicle was also added to the control samples. The mixture was incubated at 37 °C for 1 h and the reaction was terminated by adding 500 μl of 50% trichloroacetic acid followed by brief centrifugation. After two gentle washes of the pellet with 1 ml of distilled water, the radioactivity incorporated from [adenine-2,8-3H]NAD into proteins was evaluated by liquid scintillation spectrometry.
Animal Treatment
CD1 male albino mice (20–25 g) (n = 5 per group) were injected intraperitoneally with 100 mg/kg of FK866. Groups of mice also received a subcutaneous injection of kynurenine (100 mg/kg) or PJ34 (10 mg/kg). Animals were sacrificed 8 h later and organs were rapidly collected and stored at −80 °C. From each organ, a few milligrams were collected and processed for NAD content as described above. Procedures involving animals and their care were conducted in compliance with the Italian guidelines for animal care (DL 116/92) in application of the European Communities Council Directive (86/609/EEC) and was formally approved by the Animal Care Committee of the Department of Pharmacology of the University of Florence.
Intracellular FK866 Quantitation by LC-MS/MS
Samples of cytosol or mitochondria extracted from cells exposed or not to 100 μm FK866 for 1 h were extracted with methanol for protein precipitation, centrifuged at 14,000 × g for 15 min, lyophilized, and then resuspended in 100 μl of water, 3% CH3CN. Ten μl of the solution were injected into a HPLC PerkinElmer Series 200 micropump apparatus and chromatographed using a 75 × 2.0-mm inner diameter, 4-μm particle size 80-Å Synergi Fusion (Phenomenex) column. Mobile phase A consisted of water containing 3% CH3CN and 0.1% formic acid, and mobile phase B was CH3CN containing 3% water and 0.15 formic acid. An elution gradient was performed from 5 to 90% B in 10 min; B was held at 90% for 3 min, and then brought back to the starting value in 1 min. The equilibration time before the next injection was 8 min. The eluate, at a flow rate of 200 μl/min, was directly introduced into the LC/MS interface. The LC-MS/MS system consisted of a PerkinElmer Sciex API 365 triple quadrupole mass spectrometry equipped with a Turbo IonSpray Interface. The compounds were ionized by positive ion electrospray and detected using multiple reaction monitoring. The needle and orifice voltage were adjusted to 6.0 kV and 65 V, respectively. The drying gas flow rate and temperature were 7 liters min−1 and 250 °C, respectively. The first and third quadrupole mass analyzers were operated at unit mass resolution. Fragmentation was accomplished with a collision energy of 53 eV; nitrogen was used as the collision gas at a collision gas thickness of 2.6 × 1015 molecules cm−2. The ion transition monitored were m/z 392 → 105, 392 → 132, and 392 → 261. These transitions were selected based on the triple quadrupole collision-induced dissociation product ion spectrum of the analyte.
RESULTS
Effect of FK866 on NAD and ATP Contents in Different Cell Cultures
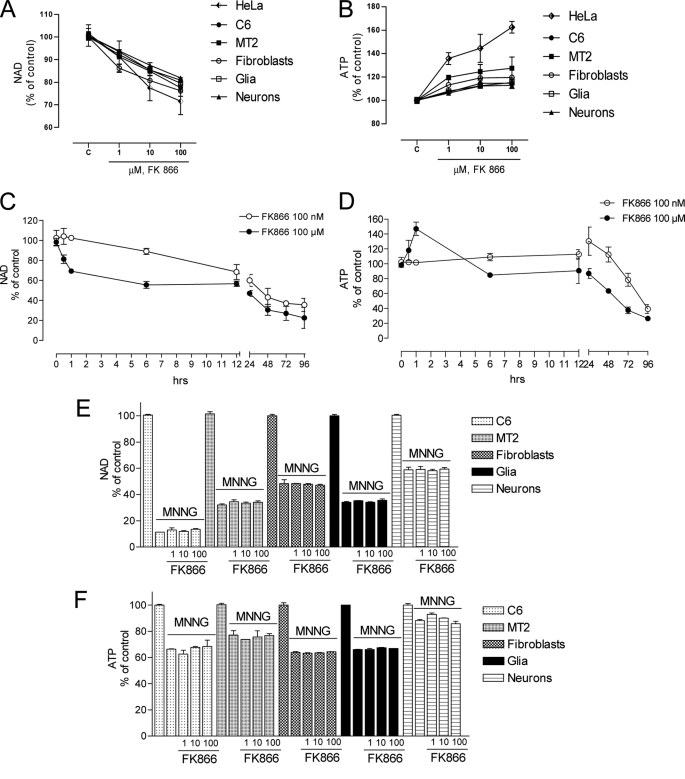
Reportedly, the effect of FK866 on intracellular NAD content has been examined after several hours (6–72 h) of exposure to the drug. Thus, to gather information on the basal turnover rate of NAD rescue we exposed different culture cell types to a range of FK866 concentrations, and measured NAD contents over time. Given that the bioenergetic cost of the resynthesis of NAD has not yet been quantified, we also measured ATP contents in cells challenged with FK866. These measurements were done in different neoplastic lines (HeLa (cervix), MT2 (macrophages), C6 (astrocytes), and 3T3 (fibroblasts)), and primary neural cultures (mixed glia or pure neurons). As shown in Fig. 1A, a 1-h exposure to the drug prompted concentration-dependent NAD depletion in all cell types under resting conditions, indicating significant ongoing activity of Nampt as well as NAD consumption. In keeping with utilization of ATP by Nampt, a concomitant concentration-dependent increase of ATP contents was also found in all cultured cell types exposed to FK866 (Fig. 1B). Given that HeLa cells appeared the most sensitive to the effect of FK866 on NAD and ATP, this cell line was used for further experiments. A time course analysis showed that cells exposed to 100 μm FK866 underwent ∼20% NAD depletion after 30 min exposure. Pyridine nucleotide content was further diminished at 6 h, remained constant between 6 and 12 h, and underwent further depletion between 24 and 96 h, reaching ∼20% of the control levels. FK866 at 100 nm was less efficient in reducing NAD content in the early 6 h of exposure. At this concentration a pattern of NAD depletion similar to that prompted by 100 μm was obtained between 12 and 96 h (Fig. 1C). The time course analysis on the effects of 100 μm FK866 on ATP levels showed that the increase of ATP vanished after 6 h when ATP depletion similar to those of NAD occurred between 12 and 96 h. FK866 at 100 nm prompted no changes of ATP up to 12 h, and a linear depletion between 24 and 72 h. At 96 h ATP depletion induced by the two concentrations was similar (Fig. 1D).
FIGURE 1.
Effect of FK866 on NAD and ATP contents in various cell cultures under basal and PARP-1-activating conditions. Concentration-dependent effects of FK866 on NAD (A) and ATP (B) contents in various cultured cell types. Basal contents of NAD and ATP contents were comparable among different cell types and comprised between 10.4 ± 4.4 and 23.5 ± 7.8 nmol/mg of protein, respectively. Time course of the effects of FK866 at 100 μm and 100 nm on NAD (C) and ATP (D) contents in HeLa cells. Effects of FK866 (100 μm, 30 min preincubation) on NAD (E) and ATP (F) contents of cultured cells exposed 1 h to the PARP-1 activating compound MNNG (100 μm). Each point/column represents the mean ± S.E. of at least three experiments conducted in duplicate.
A large body of information indicates that PARP-1 hyperactivation is a potent inducer of cell death (29). Cellular energy failure due to excessive ATP consumption for NAD resynthesis has been postulated to underlie PARP-1-dependent cell death (30, 31). Although NAD depletion seems to be the cause of death of neurons undergoing PARP-1 hyperactivation (32), whether modulation of NAD resynthesis affects PARP-1-dependent cell death is unclear (28). Hence, we analyzed the effect of FK866 on PARP-1 activation-dependent NAD and ATP depletion in the above mentioned cell cultures. We reasoned that the drug should prevent ATP utilization for NAD resynthesis, thereby unmasking the contribution of the NAD rescue pathway to PARP-1 hyperactivity-dependent energy failure. However, neither NAD nor ATP depletion prompted by PARP-1 hyperactivation with MNNG was affected by FK866 exposure in the various cell cultures (Fig. 1, E and F).
Effects of Modulation of NAD Consumption and Neosynthesis on NAD and ATP Changes Induced by FK866
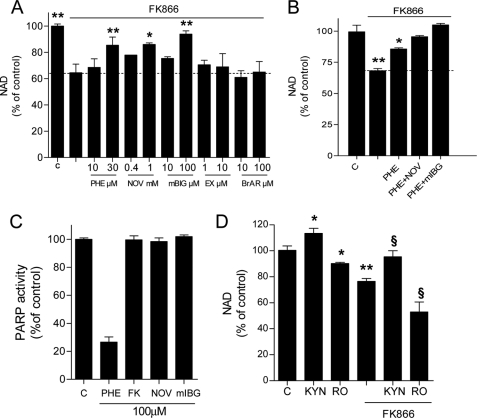
Data shown in Fig. 1 unmask ongoing NAD consumption and ATP utilization for NAD resynthesis in different cell types. To understand the relative contribution of poly- and mono(ADP-ribose) transferases as well as sirtuins and CD38 to basal NAD catabolism, we measured the content of NAD in HeLa cells acutely exposed to FK866 in the presence or absence of inhibitors of the above mentioned enzymes. The PARP inhibitors PHE and PJ34 partially reduced FK866-induced NAD depletion (Fig. 2A and not shown). Interestingly, similar effects were obtained with MART inhibitors novobiocin (33) and mIBG (34) (Fig. 2A). On the contrary, the two structurally unrelated sirtuin inhibitors EX-257 (35) and sirtinol (36) had no effect on NAD content reduction triggered by FK866 (Fig. 2A and not shown). When cells were exposed to PHE and novobiocin an additive effect on reduction of FK866-triggered NAD depletion was obtained, whereas PHE plus mIBG led to a complete inhibition of the effects of FK866 (Fig. 2B). Of note, the increase of ATP in cells exposed to FK866 was not affected by any of the above mentioned compounds (not shown). Also, PHE, novobiocin, and mIBG, as well as FK866 did not inhibit pure human PARP-1 activity when tested in an in vitro assay at concentrations up to 100 μm (Fig. 1C). We also wondered whether an increased metabolic flux through the kynurenine pathway could reduce FK866-induced NAD depletion. As shown in Fig. 2D, kynurenine increased basal NAD contents and counteracted the effects of FK866. Conversely, exposure of cells to the inhibitor of kynurenine monooxygenase Ro-618048 (37) reduced basal NAD content and increased NAD depletion prompted by FK866 (Fig. 2D).
FIGURE 2.
Effects of various compounds on FK866-induced NAD depletion in HeLa cells. A, effects of the PARP-1 inhibitor PHE, the MART inhibitors novobiocin (NOV) and mIBG, and the Sirt-1 inhibitor EX-257 (EX) on NAD depletion prompted by FK866 (100 μm/1 h). Each drug was preincubated 30 min at the indicated concentrations. B, additive effects of the PARP-1 inhibitor PHE (30 μm) and MART inhibitors novobiocin (NOV, 1 mm) and mIBG (100 μm) on reduction of NAD depletion prompted by FK866 (100 μm/1 h). C, effects of FK866, the PARP-1 inhibitor PHE and MART inhibitors novobiocin (NOV) and mIBG (all at 100 μm) on in vitro activity of purified PARP-1. D, effects of kynurenine (KYN, 200 μm) and the kynurenine monooxygenase inhibitor Ro-618048 (RO, 30 μm) on NAD contents in cells under control conditions or exposed to FK866 (100 μm/1 h). KYN and RO have been preincubated 60 min. Each column represents the mean ± S.E. of three experiments conducted in duplicate. A, *, p < 0.05; **, p < 0.01 versus FK866. B, *, p < 0.05; **, p < 0.01 versus control. D, *, p < 0.05; **, p < 0.01 versus control; §, p < 0.05, versus FK866. Analysis of variance and Tukey's post hoc test were used.
NAD Depletion by FK866 Is Cell Compartment Specific
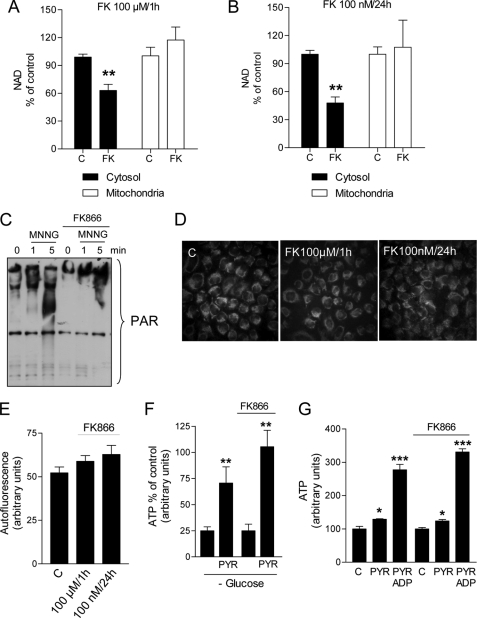
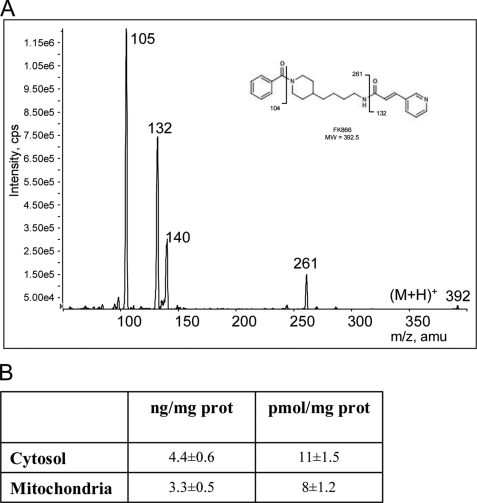
Intracellular NAD is present in specific subcellular compartments apparently not freely interchangeable. The main NAD pools are the cytoplasmic and mitochondrial ones (38). Although cytoplasmic NAD moves freely through nuclear pores, thereby tending to equilibrate with this compartment, mitochondrial membranes are supposed to be impermeable to pyridine nucleotides leading to a nonexchangeable mitochondrial NAD pool (39). We therefore attempted to determine the effects of FK866 on nucleocytoplasmic and mitochondrial NAD contents. Data shown in Fig. 3A indicate that Nampt inhibition by 100 μm FK866 led to a ∼40% reduction of the cytosolic NAD pool 1 h after the challenge. Remarkably, the mitochondrial NAD content was not affected by FK866. A similar pattern of compartment-specific NAD depletion was obtained in cells exposed to 100 nm FK866 for 24 h (Fig. 3B). In keeping with the nucleo/cytoplasmic drop of NAD contents, PARP-1-dependent poly(ADP-ribose) formation was significantly reduced in cells exposed to FK866 compared with controls (Fig. 3C). Given the lack of NAD depletion in mitochondria of cells exposed to FK866, we analyzed the effects of the compound on cell autofluorescence (a classic index of mitochondrial NADH content (40)). We found that autofluorescence was not affected in cells exposed to the drug (Fig. 3, D and E). Furthermore, the ATP content of cells exposed or not to 100 nm FK866 for 24 h and cultured overnight in the absence of glucose was similarly increased by a 1-h exposure to 1 mm pyruvate (Fig. 3F), suggesting that FK866 did not impair mitochondrial ATP production. In keeping with this assumption, ATP production was similarly triggered by pyruvate or pyruvate plus ADP in mitochondria isolated from control or FK866-challenged cells (Fig. 3G). To rule out the possibility that mitochondria are not permeable to FK866, by means of a LC-MS/MS apparatus we measured the intracellular distribution of the drug in cells exposed to 100 μm for 1 h. As shown in Fig. 4A, FK866 fragmentation gave a specific transition spectrum of m/z 392 → 105, 392 → 132, and 392 → 261. We found that FK866 was distributed both in the cytosolic and mitochondrial compartments reaching amounts of 4.4 ± 0.6 and 3.3 ± 0.5 ng/mg of protein, respectively (Fig. 4B).
FIGURE 3.
Effects of FK866 on subcellular NAD contents, poly(ADP-ribose) formation, and ATP production. Quantitation of cytosolic and mitochondrial NAD contents in HeLa cells exposed to FK866 at 100 μm for 1 h (A) or 100 nm for 24 h (B). C, Western blotting evaluation of the effects of FK866 (100 nm/24) on poly(ADP-ribose) (PAR) formation prompted by the PARP-1 activator MNNG (100 μm). Note that poly(ADP-ribosyl)ated proteins appear as a smear due to their highly increased molecular weight. D, visualization of autofluorescence in cells under control conditions or exposed to FK866 at 100 μm for 1 h or 100 nm for 24 h. E, quantitation of cell autofluorescence of cells shown in D. F, ATP content of cells exposed or not to 100 nm FK866 for 24 h, cultured overnight in the absence of glucose, and pulsed for 1 h with 1 mm pyruvate (PYR). Values represent the % of ATP of cells cultured in the presence of glucose (control). G, ATP production by mitochondria isolated from control or FK866-challenged cells and exposed 5 min to 1 mm pyruvate (PYR) or 1 mm pyruvate plus 3 μm ADP. In A, B, F, and G, each column represent the mean ± S.E. of 7 (A), 5 (B), or 3 (F and G) experiments. In C the blot is representative of three different experiments. In D an experiment representative of 4 is shown. In E each column represents the mean ± S.E. of the fluorescence present in each microscopic field (3 fields per slide have been acquired). A and B, *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control. Analysis of variance and Tukey's post hoc test were used.
FIGURE 4.
LC-MS/MS evaluation of cytosolic and mitochondrial concentrations of FK866 in HeLa cells. A, ion transition spectrum of FK866 fragmentation. B, cytosolic and mitochondrial concentrations of FK866 in cells exposed to 100 μm for 1 h. Values are the mean of three independent experiments.
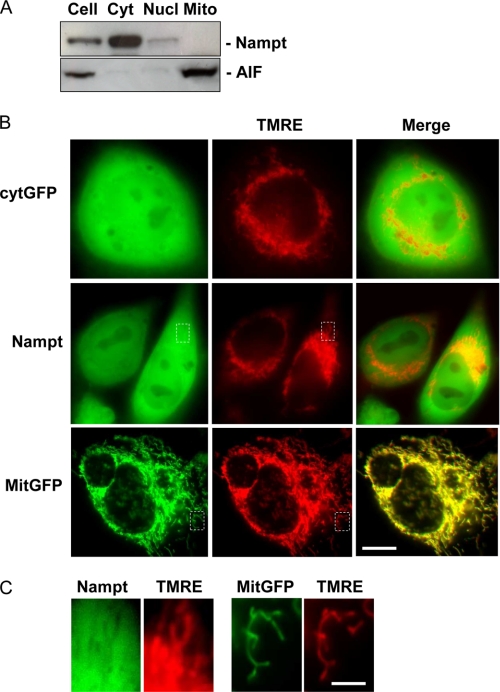
Evidence that the mitochondrial NAD pool was not affected by FK866 suggested that mitochondrial and cytoplasmic NAD pools are not freely exchangeable, and that mitochondria do not contain Nampt. To investigate the latter hypothesis, we analyzed the mitochondrial presence of Nampt by Western blotting. Fig. 5A shows that the enzyme was present in the cytosol, slightly localized in the nuclear fraction, and absent in mitochondria. The nuclear presence of Nampt, although consistent with a previous report (41), might also be due to the contamination of the nuclear fraction by unbroken cells. In keeping with the absence of Nampt in mitochondria, MitoProtII software found no evidence for a mitochondrial targeting sequence in human Nampt, and a probability of mitochondrial import of 0.044 (Mn-superoxide dismutase and Cu/Zn-superoxide dismutase, chosen, respectively, as both positive and negative controls, gave a probability of 0.96 and 0.01). To corroborate this finding, we then transfected cells with different plasmids encoding cytosolic GFP, Nampt-GFP, or mitochondrial GFP. After 24 h, cells were treated with the mitochondrial dye TMRE. As shown in Fig. 5B, distribution of Nampt-GFP was identical to that of the cytosolic GFP. Colocalization analysis showed that Nampt-GFP or cytosolic GFP fluorescence did not merge with TMRE fluorescence. Conversely, complete colocalization was obtained between mitochondrial GFP and TMRE staining. Further confirming that Nampt does not localize within mitochondria, higher magnifications allowed visualization of the negative mitochondrial profile in the cytoplasm of Nampt-GFP-transfected cells (Fig. 5C).
FIGURE 5.
Subcellular localization of Nampt in HeLa cells. A, Western blot analysis of Nampt in whole extract, or cytoplasmic, nuclear, and mitochondrial fractions. Apoptosis inducing factor (AIF) is shown as a mitochondrial marker. B, fluorescence of cells transfected with cytosolic GFP (Cyt-GFP), Nampt-GFP, or mit-GFP together with that of their mitochondria stained with TMRE is shown. The merging of green and red fluorescence is also shown. Note that TMRE fluorescence becomes yellow after merging with that originating from mit-GFP, whereas remains red or appears orange after merging with that of Cyt-GFP or Nampt-GFP. C, magnifications of areas delimited by dotted lines in B are shown. Note the negative image of the TMRE-positive mitochondria in the cytoplasmic area of cells expressing Nampt-GFP. In A, a representative blot of two is shown. In B and C, representative images of 3 experiments are shown. Bars = 4 μm (A) and 0.2 μm (B).
Effect of Acute FK866 Treatment on Mouse Tissue NAD Contents
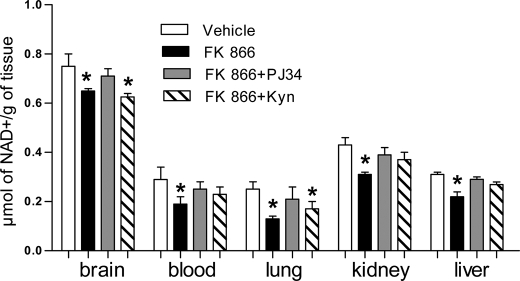
A clear picture of the impact of acute Nampt inhibition on the content of NAD in vivo is not available. This information would be of relevance to understand the rate of NAD resynthesis in vivo. We therefore measured NAD content in different mouse organs after a single intraperitoneal injection of FK866. Eight hours after intraperitoneal drug injection, NAD content was reduced in all the organs analyzed such as brain, lung, blood, kidney, and liver (Fig. 6). Nicotinamide injection as been proposed as an antidote to counteract the NAD depleting effects of FK866 (17). We therefore asked whether the same effects could be obtained by boosting the de novo NAD synthesis through the kynurenine pathway or by inhibiting enzymatic NAD consumption. Interestingly, FK866-dependent NAD depletion was reduced in all the organs analyzed when mice were treated with a concomitant subcutaneous injection of the PARP-1 inhibitor PJ34. Conversely, the NAD precursor kynurenine reduced FK866-dependent NAD depletion in the blood, liver, and kidney but not brain and lung (Fig. 6).
FIGURE 6.
Effects of PARP-1 inhibition of boosting of the kynurenine pathway on the effects of FK866 on NAD contents in various mouse organs. Mice were injected intraperitoneally with FK866 (100 mg/kg) and subcutaneously with PARP-1 inhibitors PJ34 (10 mg/kg) or kynurenine (100 mg/kg) and NAD contents were measured in different tissues 8 h later. Each column represents the mean ± S.E. of 5 mice. *, p < 0.05 versus vehicle. Analysis of variance and Tukey's post hoc test was used.
DISCUSSION
In the present study, by means of a recently identified Nampt inhibitor, we gathered information on the biochemistry, bioenergetic relevance, and subcellular compartmentation of the NAD rescue pathway. We report that acute exposure to the Nampt inhibitor FK866 prompts intracellular NAD depletion paralleled by a concomitant increase of ATP in both transformed and primary cell cultures of different species and tissues. Evidence that depletion of NAD and increments of ATP were comparable among the different cells we have studied suggests that Nampt activity and basal NAD consumption does not differ substantially among cells of different origin. Considering the ongoing trial with FK866 in cancer patients (18, 19), these data suggest that the impact of the drug on NAD contents is comparable among different tissues.
An important finding of the present study is that, upon FK866 exposure, the initial drop of NAD contents was followed by a plateau phase between 6 and 12 h and a subsequent drop at later time points (Fig. 1C). Theoretically, the plateau is in contrast with evidence for Nampt inhibition concomitant to ongoing NAD consumption. Yet, the present finding that NAD content drops in the nucleo/cytoplasmic compartment and not in mitochondria of FK866-treated cells is in keeping with the plateau. We reason that the secondary drop of NAD content is due to cell stress/death, a well known event in cells exposed to FK866 (14, 17, 42, 43).
Although our findings underscore the significance of the NAD rescue pathway to cellular bioenergetics, they challenge the relevance of this pathway to PARP-1-dependent energy failure. Indeed, ATP depletion prompted by PARP-1 hyperactivation was not affected by FK866, despite the biochemical evidence of Nampt inhibition. This is in agreement with work suggesting that PARP-1 hyperactivation triggers cell death by mechanisms not directly related to NAD resynthesis (28, 32, 44).
Several studies reported the cytotoxic properties of FK866 on neoplastic cells, an event ascribed to NAD depletion and ensuing bioenergetic failure (14, 17, 18, 42). Yet, it remains to be understood whether NAD depletion was due to basal enzymatic consumption, and the relative contribution of the various NAD-consuming enzymes. In this study, we gather information on the relative contribution of the different NAD-hydrolyzing enzymes to FK866-dependent NAD depletion. Interestingly, poly- and mono(ADP-ribosylation) appears to consume the same amount of NAD under resting conditions. Although we used two structurally unrelated MART inhibitors (unable to affect PARP-1 activity, Fig. 2C), given their unknown degree of specificity over enzymes regulating NAD contents, caution must be exercised when drawing conclusions from the present findings. Indeed, equivalence of PARPs and MARTs to basal NAD consumption is in contrast with the general view that poly(ADP-ribosyl)ation is more represented than mono-ADPR transfer within the cell. However, emerging evidence that several PARPs are bona fide MARTs (6) might in part explain our finding. Furthermore, the additive effect of the two MART inhibitors on rescue of the NAD contents prompted by a PARP inhibitor in cells exposed to FK866 (Fig. 2B) suggests that the compounds were targeting different enzymes. It is also worth noting that two structurally unrelated sirtuin inhibitors (sirtinol and EX-257) did not modify FK866-induced NAD depletion, thereby suggesting negligible formation of acetyl-ADPR in resting HeLa cells. These findings are in keeping with the ability of a mixture of PARP and MART inhibitors (PHE plus mIBG) to completely prevent the effect of FK866 on the NAD pool (Fig. 2B). On the contrary, the kynurenine pathway downstream of indoleamine dioxygenase appears functional in these cells under control conditions, suggesting that kynurenines can be used as rescue agents in cancer patients over FK866.
NAD contents within intracellular organelles and fluxes through their membranes are only in part understood (38). Although mitochondria harbor a significant portion of cellular NAD (45, 46), they are reported to be impermeable to pyridine nucleotides (39). It is still unknown, therefore, whether NAD reaches the mitochondrial matrix from outside or is it synthesized internally, as well as whether it exits the organelle through the flickering of the mitochondrial transition pore. In this regard, NAD fluxes through mitochondrial membranes have been reported, and found dependent on cell culture conditions (47). Also, a specific carrier is able to transport NAD inside yeast mitochondria (48), but the mammalian counterpart has not been identified yet. In this light, our findings that FK866 reduces cytosolic NAD and PARP-1-dependent poly(ADP-ribose) formation, but does not affect mitochondrial NAD(H) pool and energy production are of particular significance. Indeed, on the one hand they are in keeping with mitochondrial impermeability to NAD (45), and inaccessibility of mitochondrial NAD to PARP-1 (46). On the other, they suggest that Nampt activity is not relevant to regulate mitochondrial NAD content. Accordingly, we report that human Nampt does not bear a mitochondria localization sequence and is not present in the organelles. Furthermore, GFP-fused Nampt does not localize within mitochondria. It is worth noting that these results are in apparent contrast with Western blotting experiments showing the mitochondrial presence of Nampt (49). At present, we cannot explain this inconsistency. We reason, however, that localization of Nampt within mitochondria along with knowledge that several NAD-consuming enzymes such as sirtuins (9), mono-ADPR transferases (7) and, possibly, poly(ADP-ribose) forming proteins (50, 51) are present in the organelles should render the mitochondrial NAD pool sensitive to FK866. Different kinetics of basal NAD degradation between the nucleocytoplasmic and mitochondrial compartment should also be considered. Yet, evidence that FK866 is present in mitochondria at concentrations similar to those reached in the cytoplasm rules out the possibility that the drug does not reduce mitochondrial NAD because it cannot reach the matrix.
In keeping with in vitro data, results showing that NAD decreases in numerous organs of mice acutely injected with FK866 point to a significant rate of NAD resynthesis in mammalian organs under basal conditions. The fact that PARP-1 inhibition partially prevents FK866-dependent NAD depletion in vivo is in keeping with in vitro data and unmasks constitutive PARP-1 activity in mouse organs and its impact on NAD homeostasis. This is consistent with the notion that PARP-1 activity is not invariantly related to genotoxicity but occurs in the presence of complex DNA structure or activation of specific signaling pathways (52–55). The ability of kynurenine to diminish NAD depletion in organs of mice treated with FK866 strengthens the functional relevance of this metabolic route to NAD formation in vivo, and corroborates the hypothesis that kynurenine treatment may be exploited as a rescue strategy for patients receiving FK866.
Acknowledgments
We are extremely grateful to Shin-Ichiro Imai and Rosario Rizzuto for providing plasmids used in the present work. The kind gift of mIBG by Mariella Di Girolamo is also gratefully acknowledged.
This work was supported by grants from the Fondazione Giuseppe Tomasello ONLUS, Italian Ministry of Research and Technology (Prin 2007), Associazione Italiana Sclerosi Multipla (AISM), University of Florence, and Ente Cassa di Risparmio di Firenze.
- ADPR
- ADP-ribose
- FK866
- (E)-N-[4-(1-benzoylpiperidin-4-yl)butyl]-3-(pyridin-3-yl)acrylamide
- MART
- mono(ADP-ribose) transferase
- PARP
- poly(ADP-ribose) polymerase
- MNNG
- N-methyl-N′-nitro-N-nitrosoguanidine
- Nampt
- nicotinamide phosphoribosyltransferase
- TMRE
- tetramethylrhodamine methyl ester
- mIBG
- meta-iodobenzylguanidine
- PHE
- phenanthridinone
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1.Berger F., Ramírez-Hernández M. H., Ziegler M. (2004) Trends Biochem. Sci. 29, 111–118 [DOI] [PubMed] [Google Scholar]
- 2.Sauve A. A. (2008) J. Pharmacol. Exp. Ther. 324, 883–893 [DOI] [PubMed] [Google Scholar]
- 3.Imai S. (2009) Cell Biochem. Biophys. 53, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch-Nolte F., Haag F., Guse A. H., Lund F., Ziegler M. (2009) Sci. Signal. 2, mr1. [DOI] [PubMed] [Google Scholar]
- 5.Hassa P. O., Haenni S. S., Elser M., Hottiger M. O. (2006) Microbiol. Mol. Biol. Rev. 70, 789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hottiger M. O., Hassa P. O., Luscher B., Schuler H., Koch-Nolte F. (2010) Trends Biochem. Sci. 35, 208–219 [DOI] [PubMed] [Google Scholar]
- 7.Di Girolamo M., Dani N., Stilla A., Corda D. (2005) FEBS J. 272, 4565–4575 [DOI] [PubMed] [Google Scholar]
- 8.Malavasi F., Deaglio S., Funaro A., Ferrero E., Horenstein A. L., Ortolan E., Vaisitti T., Aydin S. (2008) Physiol. Rev. 88, 841–886 [DOI] [PubMed] [Google Scholar]
- 9.Finkel T., Deng C. X., Mostoslavsky R. (2009) Nature 460, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner C. (2005) Structure 13, 1239–1240 [DOI] [PubMed] [Google Scholar]
- 11.Bogan K. L., Brenner C. (2008) Annu. Rev. Nutr. 28, 115–130 [DOI] [PubMed] [Google Scholar]
- 12.Bieganowski P., Brenner C. (2004) Cell 117, 495–502 [DOI] [PubMed] [Google Scholar]
- 13.Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M. J., Ashworth A., Carmichael J., Kaye S. B., Schellens J. H., de Bono J. S. (2009) N. Engl. J. Med. 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 14.Hasmann M., Schemainda I. (2003) Cancer Res. 63, 7436–7442 [PubMed] [Google Scholar]
- 15.Khan J. A., Tao X., Tong L. (2006) Nat. Struct. Mol. Biol. 13, 582–588 [DOI] [PubMed] [Google Scholar]
- 16.Kang G. B., Bae M. H., Kim M. K., Im I., Kim Y. C., Eom S. H. (2009) Mol. Cells 27, 667–671 [DOI] [PubMed] [Google Scholar]
- 17.Nahimana A., Attinger A., Aubry D., Greaney P., Ireson C., Thougaard A. V., Tjørnelund J., Dawson K. M., Dupuis M., Duchosal M. A. (2009) Blood 113, 3276–3286 [DOI] [PubMed] [Google Scholar]
- 18.Pogrebniak A., Schemainda I., Azzam K., Pelka-Fleischer R., Nüssler V., Hasmann M. (2006) Eur. J. Med. Res. 11, 313–321 [PubMed] [Google Scholar]
- 19.Holen K., Saltz L. B., Hollywood E., Burk K., Hanauske A. R. (2008) Invest. New Drugs 26, 45–51 [DOI] [PubMed] [Google Scholar]
- 20.Rongvaux A., Galli M., Denanglaire S., Van Gool F., Drèze P. L., Szpirer C., Bureau F., Andris F., Leo O. (2008) J. Immunol. 181, 4685–4695 [DOI] [PubMed] [Google Scholar]
- 21.Van Gool F., Galli M., Rongvaux A., Andris F., Leo O. (2009) Med. Sci. 25, 551–553 [DOI] [PubMed] [Google Scholar]
- 22.Bruzzone S., Fruscione F., Morando S., Ferrando T., Poggi A., Garuti A., D'Urso A., Selmo M., Benvenuto F., Cea M., Zoppoli G., Moran E., Soncini D., Ballestrero A., Sordat B., Patrone F., Mostoslavsky R., Uccelli A., Nencioni A. (2009) PLoS ONE 4, e7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallí M., Van Gool F., Rongvaux A., Andris F., Leo O. (2010) Cancer Res. 70, 8–11 [DOI] [PubMed] [Google Scholar]
- 24.Chiarugi A. (2002) Brain Res. Mol. Brain Res. 109, 179–188 [DOI] [PubMed] [Google Scholar]
- 25.Chiarugi A., Moskowitz M. A. (2003) J. Neurochem. 85, 306–317 [DOI] [PubMed] [Google Scholar]
- 26.Cipriani G., Rapizzi E., Vannacci A., Rizzuto R., Moroni F., Chiarugi A. (2005) J. Biol. Chem. 280, 17227–17234 [DOI] [PubMed] [Google Scholar]
- 27.Shah G. M., Poirier D., Duchaine C., Brochu G., Desnoyers S., Lageaux J., Verreault A., Hoflack J. C., Kirkland J. B., Poirier G. G. (1995) Anal. Biochem. 227, 1–13 [DOI] [PubMed] [Google Scholar]
- 28.Formentini L., Macchiarulo A., Cipriani G., Camaioni E., Rapizzi E., Pellicciari R., Moroni F., Chiarugi A. (2009) J. Biol. Chem. 284, 17668–17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jagtap P., Szabó C. (2005) Nat. Rev. Drug Discov. 4, 421–440 [DOI] [PubMed] [Google Scholar]
- 30.Berger N. A. (1985) Radiat. Res. 101, 4–15 [PubMed] [Google Scholar]
- 31.Chiarugi A. (2002) Trends Pharmacol. Sci. 23, 122–129 [DOI] [PubMed] [Google Scholar]
- 32.Alano C. C., Garnier P., Ying W., Higashi Y., Kauppinen T. M., Swanson R. A. (2010) J. Neurosci. 30, 2967–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banasik M., Komura H., Shimoyama M., Ueda K. (1992) J. Biol. Chem. 267, 1569–1575 [PubMed] [Google Scholar]
- 34.Smets L. A., Loesberg C., Janssen M., van Rooij H. (1990) Biochim. Biophys. Acta 1054, 49–55 [DOI] [PubMed] [Google Scholar]
- 35.Napper A. D., Hixon J., McDonagh T., Keavey K., Pons J. F., Barker J., Yau W. T., Amouzegh P., Flegg A., Hamelin E., Thomas R. J., Kates M., Jones S., Navia M. A., Saunders J. O., DiStefano P. S., Curtis R. (2005) J. Med. Chem. 48, 8045–8054 [DOI] [PubMed] [Google Scholar]
- 36.Grozinger C. M., Chao E. D., Blackwell H. E., Moazed D., Schreiber S. L. (2001) J. Biol. Chem. 276, 38837–38843 [DOI] [PubMed] [Google Scholar]
- 37.Röver S., Cesura A. M., Huguenin P., Kettler R., Szente A. (1997) J. Med. Chem. 40, 4378–4385 [DOI] [PubMed] [Google Scholar]
- 38.Dölle C., Niere M., Lohndal E., Ziegler M. (2010) Cell Mol. Life Sci. 67, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Lisa F., Ziegler M. (2001) FEBS Lett. 492, 4–8 [DOI] [PubMed] [Google Scholar]
- 40.Chance B. (2004) Methods Enzymol. 385, 361–370 [DOI] [PubMed] [Google Scholar]
- 41.Kitani T., Okuno S., Fujisawa H. (2003) FEBS Lett. 544, 74–78 [DOI] [PubMed] [Google Scholar]
- 42.Wosikowski K., Mattern K., Schemainda I., Hasmann M., Rattel B., Löser R. (2002) Cancer Res. 62, 1057–1062 [PubMed] [Google Scholar]
- 43.Billington R. A., Genazzani A. A., Travelli C., Condorelli F. (2008) Autophagy 4, 385–387 [DOI] [PubMed] [Google Scholar]
- 44.Yu S. W., Andrabi S. A., Wang H., Kim N. S., Poirier G. G., Dawson T. M., Dawson V. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18314–18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Lisa F., Menabò R., Canton M., Barile M., Bernardi P. (2001) J. Biol. Chem. 276, 2571–2575 [DOI] [PubMed] [Google Scholar]
- 46.Alano C. C., Tran A., Tao R., Ying W., Karliner J. S., Swanson R. A. (2007) J. Neurosci. Res. 85, 3378–3385 [DOI] [PubMed] [Google Scholar]
- 47.Rustin P., Parfait B., Chretien D., Bourgeron T., Djouadi F., Bastin J., Rötig A., Munnich A. (1996) J. Biol. Chem. 271, 14785–14790 [DOI] [PubMed] [Google Scholar]
- 48.Todisco S., Agrimi G., Castegna A., Palmieri F. (2006) J. Biol. Chem. 281, 1524–1531 [DOI] [PubMed] [Google Scholar]
- 49.Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du L., Zhang X., Han Y. Y., Burke N. A., Kochanek P. M., Watkins S. C., Graham S. H., Carcillo J. A., Szabó C., Clark R. S. (2003) J. Biol. Chem. 278, 18426–18433 [DOI] [PubMed] [Google Scholar]
- 51.Rossi M. N., Carbone M., Mostocotto C., Mancone C., Tripodi M., Maione R., Amati P. (2009) J. Biol. Chem. 284, 31616–31624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Homburg S., Visochek L., Moran N., Dantzer F., Priel E., Asculai E., Schwartz D., Rotter V., Dekel N., Cohen-Armon M. (2000) J. Cell Biol. 150, 293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lonskaya I., Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Biol. Chem. 280, 17076–17083 [DOI] [PubMed] [Google Scholar]
- 54.Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Mol. Biol. 348, 609–615 [DOI] [PubMed] [Google Scholar]
- 55.Szabo C., Pacher P., Swanson R. A. (2006) Trends Pharmacol. Sci. 27, 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]