Abstract
A new isolate, Gordonia sp. strain TY-5, is capable of growth on propane and n-alkanes with C13 to C22 carbon chains as the sole source of carbon. In whole-cell reactions, significant propane oxidation to 2-propanol was detected. A gene cluster designated prmABCD, which encodes the components of a putative dinuclear-iron-containing multicomponent monooxygenase, including the large and small subunits of the hydroxylase, an NADH-dependent acceptor oxidoreductase, and a coupling protein, was cloned and sequenced. A mutant with prmB disrupted (prmB::Kanr) lost the ability to grow on propane, and Northern blot analysis revealed that polycistronic transcription of the prm genes was induced during its growth on propane. These results indicate that the prmABCD gene products play an essential role in propane oxidation by the bacterium. Downstream of the prm genes, an open reading frame (adh1) encoding an NAD+-dependent secondary alcohol dehydrogenase was identified, and the protein was purified and characterized. The Northern blot analysis results and growth properties of a disrupted mutant (adh1::Kanr) indicate that Adh1 plays a major role in propane metabolism. Two additional NAD+-dependent secondary alcohol dehydrogenases (Adh2 and Adh3) were also found to be involved in 2-propanol oxidation. On the basis of these results, we conclude that Gordonia sp. strain TY-5 oxidizes propane by monooxygenase-mediated subterminal oxidation via 2-propanol.
Gaseous n-alkanes ranging from C2 to C5 are recognized as components of nonmethane hydrocarbons, and the increased concentrations of these gases in the atmosphere threaten to destabilize ecosystems through a variety of mechanisms (48). Although these gases are produced as natural intermediates of bacterial, plant, and mammalian metabolism, the main sources of pollution are natural oil seepage and oil spills (42). From a biotechnological perspective, gaseous alkanes are inexpensive carbon sources for microbial cultivation, and the enzymes participating in the oxidation pathway promise to be versatile biocatalysts.
A number of microorganisms have been isolated for their ability to use gaseous n-alkanes as a sole carbon source. In the case of bacteria, these abilities have been found in some Pseudomonas strains (57) and many strains belonging to the order Actinomycetales, such as those of the genera Rhodococcus, Mycobacterium, Corynebacterium, Nocardia, and Pseudonocardia (3, 15). Some of the bacteria are known to degrade various environmental pollutants (trichloroethylene, chloroform, methyl ethers, etc.) through cometabolism with gaseous alkanes (13, 52).
The pathways for the oxidation of gaseous alkanes have received little attention compared with those for the microbial oxidation of methane (34) and liquid n-alkanes (24). Recently, the terminal oxidation pathway of butane (butane → 1-butanol → butyraldehyde → butyrate) by “Pseudomonas butanovora” has been confirmed through enzymological and genetic approaches (2, 14). The first reaction is catalyzed by a soluble butane monooxygenase (sBMO) similar to soluble methane monooxygenase (sMMO) (50). In considering propane oxidation, several possibilities have been proposed (3). Propane could be oxidized by monooxygenase-mediated terminal oxidation via 1-propanol or subterminal oxidation via 2-propanol. As a third possibility, a mixture of 1-propanol and 2-propanol could result from propane oxidation. Since biochemical and genetic findings on propane monooxygenase have been limited, the propane oxidation pathways present in individual isolates have been considered mainly through the properties of alcohol dehydrogenases and other enzymes participating in the metabolism of intermediates such as propanal, acetone, acetol, and so on (3, 28). Published studies indicate that pathways for gaseous alkane oxidation vary depending on the microbial strains and the substrates, but the metabolic reactions in each pathway remain ambiguous. In order to assess each pathway, more extensive enzymological and genetic approaches are required.
We have recently isolated a gram-positive bacterium, Gordonia sp. strain TY-5, that is able to use propane but no other gaseous alkanes. We have cloned the genes for propane monooxygenase and for three types of NAD+-dependent secondary alcohol dehydrogenases. From the results of gene expression and growth characteristics of several mutant strains carrying inactivated genes, we conclude that propane is oxidized through subterminal oxidation in Gordonia sp. strain TY-5. This is the first report confirming that bacterial propane oxidation is catalyzed by an NADH-dependent multicomponent enzyme system belonging to the family of dinuclear-iron oxygenases.
MATERIALS AND METHODS
Enrichment and isolation of a propane-grown bacterium.
A bacterial strain that uses propane as the sole source of carbon and energy was enriched in a 25-ml sealed culture vessel containing 5 ml of AY medium under a gas mixture containing propane, O2, and CO2 (30:65:5, vol/vol/vol). AY medium contained (per liter) 4 g of K2HPO4, 2 g of KH2PO4, 2 g of NH4Cl, 0.2 g of MgSO4 · 7H2O, 4 mg of CaCl2 · 2H2O, 5 mg of H3BO4, 0.4 mg of CuSO4 · 5H2O, 1 mg of KI, 2 mg of FeSO4 · 7H2O, 4 mg of MnSO4 · 4∼7H2O, 4 mg of ZnSO4 · 7H2O, and 1 mg of Na2MoO4 · 2H2O, pH 7.0. The vessel was shaken at 30°C for 4 to 6 days, and then 50 μl of the culture was transferred to another vessel containing fresh medium. After the enrichment culture had been transferred five times, an aliquot of the culture was plated onto AY medium containing 1.5% Bacto Agar, which was placed in a desiccator under the same gas mixture described above at 30°C. Finally, a pure culture was obtained from a single colony, which was grown on yeast-tryptone (2× YT) medium (pH 7.0) containing (per liter) 10 g of Bacto Yeast Extract, 16 g of Bacto Tryptone, and 5 g of NaCl.
Bacterial strains, culture conditions, and vectors.
Strain TY-5, which was isolated from a soil sample as described above, was used in this work. The strain was most closely related to members of the genus Gordonia on the basis of 16S rRNA gene sequencing analysis, which was conducted as described by Hiraishi et al. (17, 18). The morphological and physiological characteristics were obtained from NCIMB Japan (Shimizu, Japan). Gordonia sp. strain TY-5 was grown on AY medium, to which a carbon source (1.0%, wt/vol) was added, at 30°C under shaking conditions. When propane was used as the carbon source, a 500-ml shaking flask containing 100 ml of AY medium under a propane-air mixture (ratio, 3:7) and sealed with a butyl rubber stopper was shaken at 30°C. A large-scale culture (10 liters) was grown in a 15-liter jar fermentor at 30°C with stirring at 300 rpm and aeration at 10 liters/ml.
Escherichia coli DH5α (TaKaRa, Kyoto, Japan) was used for gene cloning and was usually grown on 2×YT medium in the presence of ampicillin (50 mg/liter) or kanamycin (25 μg/liter) when necessary. pBluescript II SK+ (Toyobo, Osaka, Japan), pUC118 (TaKaRa), and pGEM-T Easy (Promega, Madison, Wis.) were used as cloning vectors.
Whole-cell reactions.
Whole-cell reactions with several carbon sources were conducted principally as described by Arp (2). Cells were grown on propane as described above for 3 days, harvested, and suspended in medium with no carbon source. Five milliliters of the cell suspension in a 25-ml sealed culture vessel under a gas mixture containing gaseous alkane and air (3:7, vol/vol) was shaken at 30°C. In order to inhibit further oxidation of the alcohol produced and furnish NADH, excess amounts of 1-butanol and 2-butanol (each at 5 mM) were added to the reaction mixture. A portion of the medium was sampled through a syringe and used for determination of alcohols formed by gas chromatography (45).
Cell extract.
Cells grown on 2-propanol to late exponential phase (optical density at 610 nm of 3.0) were harvested by centrifugation (28,000 × g for 10 min at 4°C) and washed twice with ice-cold AY medium containing no carbon source. The cells were suspended in 20 mM Tris-Cl buffer, pH 7.8; disrupted by six passages through a high pressure laboratory homogenizer (MINI-LAB, type 8.3 H; Rannie a/s, Copenhagen, Denmark) at 80 MPa; and centrifuged at 5,600 × g for 15 min at 4°C and then at 150,000 × g for 1 h at 4°C. The resultant clear supernatant was used as the cell extract.
Analytical methods.
Gaseous alkanes and alcohols were determined by gas chromatography as previously described (45). Protein was measured with a Bio-Rad protein assay kit (Japan Bio-Rad Laboratories, Tokyo, Japan) with bovine serum albumin as the standard (6). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a 12.5% polyacrylamide gel by the method of Laemmli (29). Prestained protein markers (low range) for SDS-PAGE (Nacalai Tesque, Kyoto, Japan) were used as molecular size standards. The relative molecular mass of a native enzyme was determined by gel filtration with a fast protein liquid chromatography system (Amersham Bioscience, Piscataway, N.J.) with a Superose 12HR 10/30 column equilibrated with 50 mM Tris-Cl buffer (pH 7.5) containing 0.1 M KCl. The standard proteins used were from the Oriental Yeast Co., Ltd. (Tokyo, Japan). In order to determine the N-terminal amino acid sequence, a single band of each purified enzyme on SDS-PAGE was electroblotted onto a PsqPVDF membrane (Millipore Corp. Bedford, Mass.) at 15 V for 40 min with a transfer buffer containing 75 mM Tris base and 580 mM glycine in 20% (vol/vol) methanol. The amino acid sequence was determined by Edman's method with a Perkin-Elmer protein sequencer (model 476A).
Enzyme assays.
NAD+-dependent alcohol dehydrogenase was assayed in a reaction mixture (1 ml) containing 20 mM sodium carbonate buffer (pH 9.5), 1.0 mM NAD+, 200 mM (NH4)2SO4, 100 mM alcohol, and an appropriate amount of enzyme. The reverse reaction was assayed in a reaction mixture (1 ml) containing 20 mM sodium acetate buffer (pH 4.0), 0.25 mM NADH, 200 mM (NH4)2SO4, 100 mM ketone, and an appropriate amount of enzyme. Activities of the forward and reverse reactions were assayed by measuring the increase and decrease in absorbance at 340 nm, respectively, with a Shimadzu spectrophotometer (UV-160) with a cuvette with a 1-cm light path. As a reference, a reaction mixture with no substrate for the forward or reverse reaction was used. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the reduction or oxidation of 1.0 μmol of NAD+ and 1.0 μmol of NADH, respectively, per min at 30°C. Initial velocity studies on the forward reaction of a secondary alcohol dehydrogenase were conducted under standard conditions at 30°C, with respect to NAD+ at fixed concentrations of 2-propanol. The Km for a substrate under an infinite concentration of another substrate was obtained by replotting the slopes and intercepts versus the reciprocal concentration (27).
Cloning and nucleotide sequencing of a gene cluster encoding propane monooxygenase.
The primers used in this work are listed in Table 1. To amplify the DNA fragment carrying the gene cluster encoding propane monooxygenase (prm) from chromosomal DNA of strain TY-5, primers N and C were designed on the basis of the conserved regions between the epoxidase large subunit of alkene monooxygenase (AMO) from Nocardia corallina B-276, which is now commonly referred to as Rhodococcus rhodochrous B-276 (44) (residues 52 to 57 and 467 to 473), and the hydroxylase α subunit of sMMO from Methylococcus capsulatus (56) (residues 70 to 75 and 472 to 478). Chromosomal DNA extracted from Gordonia sp. strain TY-5 with an AquaPure Genomic DNA Isolation Kit (Bio-Rad Laboratories) was used as a template for amplification of a portion of the prm gene cluster by PCR. Ex Taq polymerase (TaKaRa) was used for PCR in accordance with the manufacturer's instructions. The amplified 1.3-kb fragment was gel purified and then ligated with pGEM-T Easy. The propagated recombinant plasmid was digested with EcoRI, and the resulting 1.3-kb insert fragment was gel purified and used as the hybridization probe.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| Primer N | 5′-ATGGAAGCGGAGAAGGA-3′ |
| Primer C | 5′-CTGGCCGATGAGGGTCTTGC-3′ |
| Primer 3′ | 5′-ACTGTTTGCCGTGGATGTTGCGCTGGTTGG-3′ |
| Primer 5′ | 5′-CCTGTACTGGATCGGCGAGGACAAGATCCT-3′ |
| Adh2-N | 5′-ATGAARGCNCTSGTNTTYCAYGGSCC-3′ |
| Adh2-I | 5′-TSCCSACSGCCTCGATSACSACRTCNGCNCC-3′ |
| Adh2-inv-1 | 5′-ATGTCGACGGCGATGACCCG-3′ |
| Adh2-inv-2 | 5′-TGGCGATGACCGACGGACTC-3′ |
| Adh3-N | 5′-ATGACSGTSTACGCSAAGCCSGGNACNGA-3′ |
| Adh3-I | 5′-TASCCSCCGAASGCSGCGTGNGCNGGRTA-3′ |
| Adh3-inv-1 | 5′-AGAGGGTGTCGTTGGCGATGTGGATGGCGT-3′ |
| Adh3-inv-2 | 5′-GGCCGTGTGTGGACCAACACCTACCACGAC-3′ |
| prmA-f | 5′-ATGCGGTCGTACTTCCCGATGGAAGAGGAG-3′ |
| prmA-r | 5′-TCAGACCGTGACGGCCCCGTTCTTGTTGGC-3′ |
| prmB-f | 5′-ATGGCCGACACCCACAAGATCAGCTTCGAA-3′ |
| prmB-r | 5′-TCAGGAGTCGAATGCCGGGCTGGTGAACTT-3′ |
| prmC-f | 5′-ATGTCCGCACCAGCGCAACCCCGGGAACGC-3′ |
| prmC-r | 5′-TCACTTGTTCAACTCCTCTGGGAT ATCGAC-3′ |
| prmD-f | 5′-ATGCAATTCGGAGCCGACACCGAGTTCTCC-3′ |
| prmD-r | 5′-CTAGGAGGCGGTCAGGTCGAAGCCGATGAA-3′ |
| adh1-f | 5′-ATGAGAGCAGTACAGGTGGTCGGATACCAC-3′ |
| adh1-r | 5′-TCAGGGGATCAGGATGGCGCGCCCCCTGAC-3′ |
| adh2-f | 5′-ATGAAGGCACTCGTTTTCCACGGTCCCGGC-3′ |
| adh2-r | 5′-TCAGGCGCTCATGATCACCTTGAGCGCCTT-3′ |
| adh3-f | 5′-ATGACTGTTTACGCGAAGCCGGGTACCGAG-3′ |
| adh3-r | 5′-TTAGAAGAATCCCTTCGCACTCTGAGCGTA-3′ |
| kan-f-Sph | 5′-GCATGCGGACCAGTTGGTGATTTTGAACTT-3′ |
| kan-r-Sph | 5′-GCATGCTTAGAAAAACTCATCGAGCATCAA-3′ |
| kan-f-Bam | 5′-GGATCCGGACCAGTTGGTGATTTTGAACTT-3′ |
| kan-r-Bam | 5′-GGATCCTTAGAAAAACTCATCGAGCATCAA-3′ |
| adh1-500r | 5′-CAATCCGCCTGCGCCGATCACGACGCACGT-3′ |
| adh2-500r | 5′-CCCGATCACCGCGACCACATCACCGGGTTT-3′ |
| adh3-500r | 5′-GAGCGCGGGCGCGAGCTTCCACACGGCCAT-3′ |
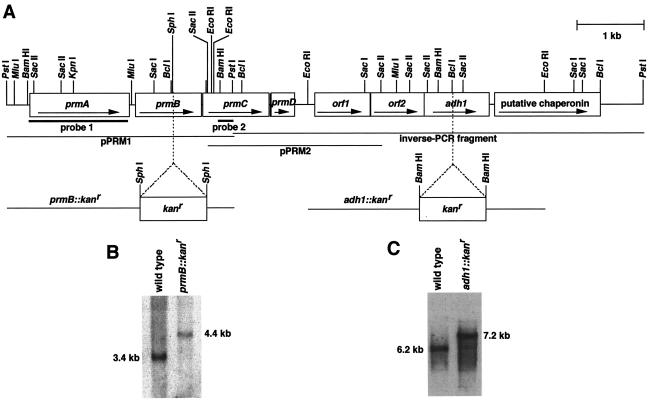
Genomic Southern analysis was done as described previously (58). Southern analysis showed that a 3.4-kb PstI fragment hybridized to the probe. To construct a PstI library, 3.4-kb PstI fragments of strain TY-5 genomic DNA were gel purified and ligated into the PstI site of pBluescript II SK+. E. coli DH5α cells were transformed with the resulting ligation mixtures. The colony hybridization experiment was performed as described previously (58). Clones that showed strong signals were picked from the original plates and used for further studies (pPRM1, Fig. 1). DNA sequencing was performed by the dideoxy chain termination method with the ThermoSequenase fluorescently labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Biosciences K.K., Tokyo, Japan) and a DSQ-1000L DNA sequencer (Shimadzu Co. Ltd., Kyoto, Japan). The 400-bp BamHI-PstI-digested fragment of pPRM1 was used as a probe to clone the region downstream of the prm gene cluster. Genomic Southern analysis showed that a 2.5-kb SacII fragment hybridized to the probe, and this fragment was cloned in a similar colony hybridization experiment (pPRM2, Fig. 1). By an inverse-PCR procedure (38), the region downstream of the prm gene cluster was amplified. Chromosomal DNA from strain TY-5 was digested with PstI and then self-ligated. The ligation mixture was then used as the template for PCR amplification with LA Taq polymerase (TaKaRa) with primer 3′ and primer 5′. The amplified fragment was subcloned into pGEM-T Easy and sequenced (Fig. 1).
FIG. 1.
The 9.2-kb gene region of Gordonia sp. strain TY-5. (A) Genetic organization of the gene cluster and restriction map. The orientation of the identified genes is indicated by arrows. A kan gene (1,033 bp) was inserted into the SphI site within the prmB gene and into the BclI site within the adh1 gene. The overlapping inserts of the two plasmids, pPRM1 and pPRM2, and the inverse-PCR product representing the gene region analyzed are indicated by lines. (B) Genomic Southern analysis of PstI-digested total DNAs (5.0 μg of each) from the wild-type strain and the mutant strain with prmB disrupted, with the 32P-labeled prmB fragment as a probe. (C) Genomic Southern analysis of PstI-digested total DNAs (5.0 μg of each) from the wild-type strain and the mutant strain with prmB disrupted, with the 32P-labeled adh1 fragment as a probe.
Cloning of Adh2- and Adh3-encoding genes.
To amplify DNA fragments encoding Adh2 and Adh3 from strain TY-5 chromosomal DNA by PCR, the upstream primers for adh2 and adh3 were designed from the N termini of the purified enzymes (Adh2-N and Adh-3, respectively), and the downstream primers were designed on the basis of multiple aligned sequences of NAD+-dependent alcohol and aldehyde dehydrogenases. Cloning of the adh2 and adh3 genes was conducted by essentially the same procedures. Chromosomal DNA extracted from strain TY-5 as mentioned above was used as the template for amplification. To clone the full-length adh2 gene, strain TY-5 chromosomal DNA was digested with SacII, religated, and used as a template for an inverse PCR with primers Adh2-inv-1 and Adh2-inv-2. This inverse-PCR amplification product was cloned and sequenced. In the case of adh3, the chromosomal DNA was digested with BglII, religated, and used as a template for an inverse PCR with primers Adh3-inv-1 and Adh3-inv-2.
Northern blot analysis.
Gordonia sp. strain TY-5 was cultured at 30°C on AY medium containing a carbon source to exponential phase (optical density at 610 nm of 0.8 to 0.9) and harvested. In order to examine the transcriptional induction by gaseous alkanes that did not support growth, succinate-grown cells were harvested and washed with AY medium containing no carbon source. The resultant cells were shaken in AY medium containing a gaseous alkane for 4 h at 30°C and harvested. Total cellular RNA was extracted with an RNeasy Mini Kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer's protocol, electrophoresed on a 0.8% agarose gel in 20 mM morpholinepropanesulfonic acid (MOPS) buffer containing 1.0 mM EDTA and 2.2 M formaldehyde, and then transferred to a nylon membrane filter (GeneScreen Plus; NEN Life Science Products, Boston, Mass.) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization was carried out as described previously (33). DNA probes specific for individual genes were generated by PCR with the following primer combinations: prmA-f and prmA-r for prmA, prmB-f and prmB-r for prmB, prmC-f and prmC-r for prmC, and prmD-f and prmD-r for prmD.
Construction of mutants with disrupted genes.
The kanamycin resistance gene (kan), including its own promoter, was amplified by PCR with pKT231 as the template. For inactivation of prmB, primers flanking the SphI site, kan-f-Sph and kan-r-Sph, were used. The amplified kan gene was inserted into the SphI site within the prmB gene of pPRM1 that had been linearized and dephosphorylated. The resulting plasmid, pDisPrmB, was digested with PstI and then introduced into Gordonia sp. strain TY-5 by double-crossover homologous recombination. For inactivation of adh1, primers flanking the BamHI site, kan-f-Bam and kan-r-Bam, were used. The amplified kan gene was inserted into the BclI site within the adh1 gene in the EcoRI fragment (Fig. 1), yielding pDisAdh1. The plasmid was digested with EcoRI and then introduced into Gordonia sp. strain TY-5 as described above. Kanamycin-resistant transformants were selected, and each gene disruption was confirmed by Southern analysis (Fig. 1).
Purification of enzymes catalyzing NAD+-dependent 2-propanol dehydrogenation.
Purification was performed at 4°C. The cell extract, which contained 1,650 mg of protein, was used for enzyme purification. A precipitate obtained by ammonium sulfate fractionation (1.6 to 3.2 M) was dialyzed against 20 mM Tris-Cl buffer, pH 8.5 (buffer A), containing 1.2 M ammonium sulfate. The dialyzed solution was applied to a Butyl-Toyopearl 650 M column (2.3 by 20 cm; Tosoh, Tokyo, Japan) that was preequilibrated with the dialyzing buffer and then eluted with a linear gradient containing decreasing ammonium sulfate concentrations (1.2 to 0 M) in buffer A. The active fractions were collected, dialyzed against buffer A, and then applied to a DEAE-Toyopearl 650 column (2.2 by 20 cm; Tosoh) preequilibrated with buffer A. Two active fractions, I and II, were separated by elution with a linear gradient containing increasing KCl concentrations (0.2 to 0.5 M). The activity peaks of fractions I and II were eluted with buffers containing approximately 0.33 and 0.42 M KCl, respectively. After dialysis against buffer A, each fraction was chromatographed on a MonoQ HR 5/5 column (0.5 by 5 cm; Amersham Biosciences) preequilibrated with buffer A with a linear gradient containing increasing concentrations of KCl (0.1 to 0.5 M). During the chromatography process, fraction I was divided into two active fractions, Ia and Ib, whose activity peaks were found in the eluates containing 0.18 and 0.3 M KCl. Fractions Ia and Ib were heated to 60 and 70°C, respectively, for 20 min and centrifuged at 15,000 × g for 15 min. The heat-treated preparations of fractions Ia and Ib and the MonoQ preparation of fraction II were homogeneous on SDS-PAGE and were stored at 0°C until use. Fractions Ia, Ib, and II are designated Adh2, Adh3, and Adh1, respectively, for consistency with the gene-cloning experiments described above.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the GenBank database and assigned the following accession numbers: 16S rRNA gene sequence of strain TY-5, AB112923; 9.2-kb PstI fragment, AB112920; adh2, AB112921; adh3, AB112922.
RESULTS
Properties of newly isolated strain TY-5.
The new isolate, strain TY-5, was able to grow on propane and n-alkanes with C13 to C22 carbon chains as the sole source of carbon but was incapable of growth on methane, ethane, n-alkanes with C4 to C12 carbon chains, and n-alkenes with C2 to C5 carbon chains. Strain TY-5 was a high-G+C-containing and mycolic-acid-containing, gram-positive bacterium and was most closely related to members of the genus Gordonia on the basis of 16S rRNA gene sequencing analysis, the highest similarities (99.4% identity) being found for the sequence of G. polyisoprenivorans.
Whole-cell reactions with propane.
Significant propane oxidation activity (157 nmol of 2-propanol formed · mg of protein−1 · h−1) was detected in reaction mixtures with whole cells of Gordonia sp. strain TY-5 when 1-butanol and 2-butanol (each at 5 mM) were added to the reaction mixture to prevent further oxidation of any alcohols formed as reaction products. No production of the terminal oxidation product, 1-propanol, was detected under the same conditions. The reaction rate was comparable to that of butane oxidation to 1-butanol by whole cells of P. butanovora (2). These results imply that propane metabolism in Gordonia sp. strain TY-5 is initiated by a monooxygenase that catalyzes subterminal oxidation.
Sequence analysis of the genes encoding Prm and Adh1.
The complete nucleotide sequence of a 9.2-kb chromosomal DNA fragment from Gordonia sp. strain TY-5 represented by the overlapping inserts of recombinant plasmids pPRM1 and pPRM2 and inverse-PCR fragments was determined on both strands (Fig. 1). Analysis of the sequence revealed eight putative open reading frames (ORFs) encoded on the same strand in the same direction. The upstream four complete ORFs in this region were closely spaced with respect to each other and designated (in the order of transcription) prmA, prmB, prmC, and prmD. Each ORF has its own putative ribosomal binding site, but there were no significant similarities to conserved sequences of bacterial promoters in the upstream region. In the sequence downstream of prmD, a G+C-rich region of dyad symmetry not followed by a series of thymidine residues was identified, which corresponds to a rho-dependent transcription terminator. These observations suggest the possibility that the gene cluster prmABCD functions as a polycistronic transcriptional unit.
A BLAST search against the available sequence databases suggested that the prmABCD gene products comprise an enzyme belonging to a family of enzymes containing nonheme carboxylate-bridged dinuclear-iron sites (61). The enzyme encoded by prmABCD has significant sequence similarity to several three- or four-component monooxygenases (Table 2). The highest overall similarities were found for the proteins of tetrahydrofuran monooxygenase of Pseudonocardia sp. strain K1 (59), which are encoded in the same order as those of the prm gene cluster. The second highest sequence similarities were found with the AMO of R. rhodochrous B-276 (44). PrmA was similar to the hydroxylase large subunits of tetrahydrofuran monooxygenase, the epoxidase of AMO, and the α subunit of sMMO. The existence of a pair of conserved amino acid sequences (Glu-X-X-His) in the putative sequence of prmA is consistent with the assignment of several monooxygenases in the family of dinuclear-iron oxygenases (12, 30, 43, 53), suggesting that this protein could catalyze the hydroxylation of propane. From the similarity analysis, the prm gene cluster most likely encodes the propane monooxygenase (Prm), of which prmA, prmB, prmC, and prmD encode the large subunit of the hydroxylase, the NADH-dependent acceptor oxidoreductase (reductase), the small subunit of hydroxylase, and the coupling protein, respectively.
TABLE 2.
Sequence similarity between propane monooxygenase and multicomponent monooxygenase proteins
| Gene | Size (aa)a | Identity (%) | Similarity (%) | Organism | Accession no. | Reference |
|---|---|---|---|---|---|---|
| prmA | 498 | |||||
| thmA | 545 | 39.16 | 68.47 | Pseudonocardia sp. strain K1 | AJ296087 | 59 |
| amoC | 501 | 34.74 | 63.25 | Rhodococcus rhodochrous B-276 | D37875 | 44 |
| bmoX | 530 | 29.12 | 59.04 | Pseudomonas butanovora | AY093933 | 50 |
| mmoX | 526 | 27.71 | 58.23 | Methylocystis sp. strain M | U81594 | 31 |
| mmoX | 527 | 27.11 | 58.43 | Methylococcus capsulatus (Bath) | JQ0702 | 56 |
| mmoX | 527 | 27.11 | 58.23 | Methylomonas sp. strain KSP III | AB025021 | 49 |
| prmB | 346 | |||||
| thmD | 360 | 42.20 | 75.14 | Pseudonocardia sp. strain K1 | AJ296087 | 59 |
| amoD | 342 | 34.68 | 70.76 | Rhodococcus rhodochrous B-276 | D37875 | 44 |
| tomA5 | 354 | 31.21 | 69.08 | Burkholderia cepacia G4 | AF319657 | |
| phkF | 358 | 30.92 | 70.23 | Burkholderia kururiensis | AB063252 | |
| tbc1F | 354 | 30.92 | 69.94 | Burkholderia cepacia JS150 | AF282897 | 26 |
| mmoC | 348 | 30.35 | 67.63 | Methylococcus capsulatus (Bath) | P2868 | 56 |
| prmC | 368 | |||||
| thmB | 346 | 27.72 | 65.22 | Pseudonocardia sp. strain K1 | AJ296087 | 59 |
| amoA | 343 | 24.46 | 61.41 | Rhodococcus rhodochrous B-276 | D37875 | 44 |
| mmoY | 389 | 22.83 | 59.24 | Methylococcus capsulatus (Bath) | M90050 | 56 |
| bmoY | 391 | 21.74 | 58.97 | Pseudomonas butanovora | AY093933 | 50 |
| mmoY | 395 | 19.29 | 58.15 | Methylocystis sp. strain M | U81594 | 31 |
| mmoY | 394 | 19.02 | 57.61 | Methylosinus trichosporium OB3b | P27354 | 9 |
| prmD | 111 | |||||
| thmC | 117 | 30.63 | 65.77 | Pseudonocardia sp. strain K1 | AJ296087 | 59 |
| amoB | 117 | 23.42 | 57.66 | Rhodococcus rhodochrous B-276 | D37875 | 44 |
aa, amino acids.
Four ORFs were identified downstream of the prm gene cluster (Fig. 1). From the results of BLAST searches, ORF1 and ORF2 encode hypothetical proteins and ORF4 showed high similarity to GroEL from Bacillus subtilis (47). The putative polypeptide predicted from ORF3 (adh1) was similar to NAD+-dependent and zinc-containing alcohol dehydrogenases from several organisms (Table 3). The ORF for adh1 consists of 1,026 bp, and the deduced amino acid sequence includes 341 amino acid residues with a theoretical molecular mass of 35,532 Da. In the putative amino acid sequence, an alcohol dehydrogenase motif, GHENAGWVEAIGDAV (residues 62 to 77), an NAD+-binding motif, GLGHIG (residues 80 to 85), and ligands of a catalytic zinc atom (Cys39 and His64) (41) were identified. As described below, the theoretical molecular mass was close to the relative molecular mass of purified Adh1 and the deduced N-terminal amino acid sequence was obtained from the purified enzyme.
Transcript analysis.
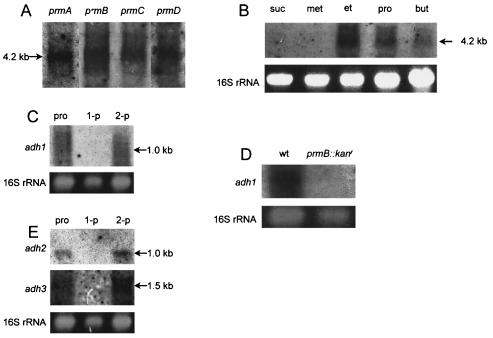
To determine the transcriptional regulation of the prm gene cluster, Northern blot analysis of total RNA from propane-grown Gordonia sp. strain TY-5 cells was conducted with probes for prmA, prmB, prmC, and prmD. The sizes of the hybridizing bands obtained with all four probes were identical and corresponded to the entire length of the transcription product of prmABCD (4.2 kb) (Fig. 2A). Next, Northern blot analysis was conducted on total RNA from cells that had been grown on succinate and then incubated with a carbon source (methane, ethane, propane, butane, or succinate). The probe for prmC hybridized against the total RNAs of cells induced weakly with ethane and butane, with the band sizes corresponding to the entire length of the operon product (Fig. 2B). Judging from the results and the gene organization described above, expression of the prm gene cluster is regulated at the mRNA level and the prm gene cluster is transcribed as a polycistronic operon that is induced by propane during growth. The organism did not grow on ethane and butane but was able to grow on the corresponding alcohols, ethanol and 1- or 2-butanol, respectively (data not shown). These results imply that the prm gene product is only active in response to propane, although the prm operon is induced by ethane, propane, and butane. The enzymological properties of Prm have yet to be elucidated.
FIG. 2.
(A) Northern blot analysis of the prmABCD gene cluster. A 20-μg portion of total RNA was loaded into each lane, and prmABCD transcription was detected by hybridization with 32P-labeled prmA, prmB, prmC, and prmD fragments as probes. Total RNA was prepared from Gordonia sp. strain TY-5 cells grown on propane. (B) prmABCD transcription was detected by hybridization with a 32P-labeled prmA fragment as the probe. Total RNAs were prepared from cells induced with methane (met), ethane (et), propane (pro), and butane (but) as described in Materials and Methods and from cells grown on succinate (suc). (C) adh1 transcription was detected by hybridization with a 32P-labeled adh1 fragment as the probe. Total RNAs were prepared from cells grown on propane, 1-propanol (1-p), and 2-propanol (2-p). (D) adh1 transcription was detected by hybridization with a 32P-labeled adh1 fragment as the probe. Total RNA was prepared from cells with prmB disrupted that were induced with 2-propanol. wt, wild type. (E) Transcription of adh2 and adh3 was detected by hybridization with 32P-labeled adh2 and adh3 fragments, respectively, as probes. Total RNAs were prepared from wild-type cells grown on propane, 1-propanol, and 2-propanol.
To examine the expression and regulation of adh1, Northern blot analysis was carried out with total RNA from cells grown on propane, 1-propanol, and 2-propanol (Fig. 2C). Each prm gene and the adh1 gene were used as probes. One intensively hybridizing band was detected with the probe for adh1 with the mRNA from propane- and 2-propanol-grown cells but not 1-propanol- or succinate-grown cells. The sizes of all hybridizing bands were identical and corresponded to the entire length of the transcription product of adh1 (1.0 kb). No hybridization band appeared when the probes for the prm genes were used, suggesting that the prm gene cluster and the adh1 gene are in distinct transcriptional units. These results suggest that adh1 is inducibly expressed in response to propane and 2-propanol and that the enzyme plays a significant role in propane oxidation via 2-propanol but does not participate in growth on 1-propanol.
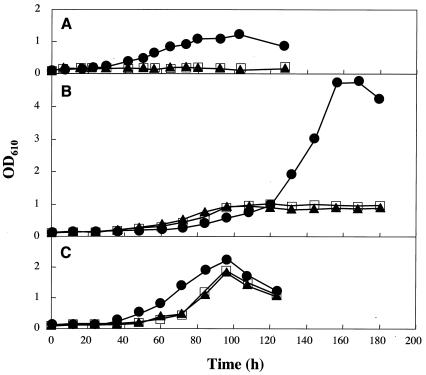
Inactivation of the prmB and adh1 genes.
In order to determine the physiological roles of the products of the prm and adh1 genes, prmB and adh1 were inactivated by homologous recombination with gene disruption plasmids as described in Materials and Methods. Results of studies of the growth of the wild-type strain and the mutants on propane, 2-propanol, and 1-propanol are shown in Fig. 3. Neither the prmB (prmB::Kanr) mutant nor the adh1 mutant (adh1::Kanr) was able to grow on propane (Fig. 3A), indicating that the products of the prmB operon and the adh1 gene are required for propane metabolism. Curiously, growth of the prmB mutant on 2-propanol was depressed similarly to that of the adh1 disruption mutant (Fig. 3B), despite the fact that prmB and adh1 are distinct transcriptional units. In order to clarify this phenomenon, the expression of the adh1 gene in the prmB mutant was investigated through Northern blot analysis (Fig. 2D). The results indicated that adh1 was not transcribed in the 2-propanol-grown mutant with prmB disrupted. Therefore, the insertion of Kanr into prmB inhibited transcription of the downstream gene, adh1.
FIG. 3.
Growth of the wild-type strain (circles), a mutant strain with prmB disrupted (prmB::Kanr) (triangles), and a mutant strain with adh1 disrupted (adh1::Kanr) (squares) on propane (A), 2-propanol (B), and 1-propanol (C). OD610, optical density at 610 nm.
Notably, both mutant strains still grew to some extent on 2-propanol (Fig. 3B). This suggested that another enzyme(s) might participate in 2-propanol oxidation. Indeed, the other two genes for NAD+-dependent secondary alcohol dehydrogenases, adh2 and adh3, were found on the chromosomal DNA as described below and were transcribed in the cells grown on propane and 2-propanol (Fig. 2E). Inducible activities of the three genes were compared by real-time quantitative PCR with the same total RNA as used for Northern blot analysis with the following primers: adh1-f and adh1-500r for adh1, adh2-f and adh2-500r for adh2, and adh3-f and adh3-500r for adh3 (data not shown). When total RNA preparations from propane- and 2-propanol-grown cells were used, the three genes were transcribed, consistent with the results of Northern blot analyses. Under these conditions, transcription of adh1 was eight and four times higher than that of adh2 and adh3, respectively. This quantitative gene transcription analysis suggests that Adh1 is the primary dehydrogenase involved in 2-propanol oxidation.
Both the prmB and adh1 mutants grew to the same maximum level as the wild-type strain on 1-propanol, although the growth of the mutant strains was somewhat delayed compared with that of the wild-type strain (Fig. 3C). The NAD+-dependent alcohol dehydrogenases Adh1, Adh2, and Adh3 were not induced with 1-propanol, indicating that another enzyme participates in 1-propanol oxidation.
Comparison of properties of three NAD+-dependent alcohol dehydrogenases.
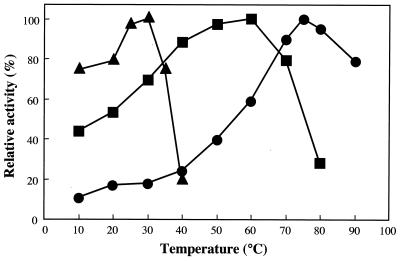
As described in Materials and Methods, three NAD+-dependent alcohol dehydrogenases were found in cells of 2-propanol-grown Gordonia sp. strain TY-5. Three enzymes, Adh1, Adh2, and Adh3, were purified 19.9-, 145-, and 17.2-fold, respectively, from the extract of 2-propane-grown cells. The relative molecular masses of Adh1, Adh2, and Adh3 were estimated to be 38, 42, and 50 kDa, respectively, by SDS-PAGE and 67, 72, and 100 kDa, respectively, by gel filtration, indicating that these enzymes are dimeric. Maximum activities of all three enzymes were found at pH 10 for the forward reactions. For the reverse reaction (acetone reduction to 2-propanol), Adh1, Adh2, and Adh3 were most active at pHs 6.0, 4.0, and 5.5, respectively. The three enzymes had different temperature profiles. The optimum temperatures for Adh1, Adh2, and Adh3 were 30, 60 and 75°C, respectively (Fig. 4). The Km and kcat values of the three enzymes for 2-propanol were as follows: Adh1, 4.4 mM and 2.7 s−1; Adh2, 0.024 mM and 21.6 s−1; Adh3, 4.3 mM and 2.6 s−1. The Kms for NAD+ were 0.071 mM for Adh1, 0.088 mM for Adh2, and 0.14 mM for Adh3. NAD+ could not be replaced by NADP+ (at concentrations of up to 5 mM) for 2-propanol oxidation by the three enzymes. The activities of the three enzymes with a variety of alcohols and ketones are listed in Table 4. Adh1 was active toward primary alcohols with C2 to C5 carbon chains and secondary alcohols with C3 to C6 carbon chains. Adh2 was active toward only ethanol and 1-propanol among the primary alcohols tested. Adh3 was specific for the secondary alcohols, with only negligible activity for primary alcohols. On the basis of their substrate specificities, the three enzymes were classified as NAD+-dependent secondary alcohol dehydrogenases.
FIG. 4.
Effect of temperature on the 2-propanol-oxidizing activities of Adh1 (triangles), Adh2 (circles), and Adh3 (squares).
TABLE 4.
Substrate specificities of purified Adh1, Adh2, and Adh3
| Activity and substrate | Relative activitya (%)
|
||
|---|---|---|---|
| Adh1 | Adh2 | Adh3 | |
| Primary alcohol oxidation | |||
| Methanol | NDb | ND | ND |
| Ethanol | 25.6 | 22.9 | ND |
| 1-Propanol | 21.4 | 32.6 | ND |
| 1-Butanol | 50.3 | ND | ND |
| 1-Pentanol | 14.8 | ND | ND |
| 1-Hexanol | ND | ND | ND |
| 1-Heptanol | ND | ND | ND |
| 1-Octanol | ND | ND | ND |
| Secondary alcohol oxidation | |||
| 2-Propanol | 100 | 100 | 100 |
| 2-Butanol | 119 | 82.8 | 62.9 |
| (R)-(−)-2-Butanol | 13.7 | 165 | 44.6 |
| (S)-(+)-2-Butanol | 178 | 125 | 78.7 |
| 2-Pentanol | 129 | 40.0 | 63.5 |
| 2-Hexanol | 19.2 | 12.7 | 78.1 |
| 2-Heptanol | ND | ND | 109 |
| 2-Octanol | ND | ND | 112 |
| Ketone reduction | |||
| Acetone | 36.9 | 54.9 | 34.9 |
| 2-Butanone | 65.1 | 64.9 | 30.6 |
100% relative activity was defined as the activity with 2-propanol as the substrate.
ND, activity of <5%.
Cloning and sequence comparison with Adh2 and Adh3.
Two genes for alcohol dehydrogenases, adh2 and adh3, which were located at different loci on the chromosomal DNA, were cloned and sequenced. The calculated molecular masses and deduced N-terminal amino acid sequences were in good agreement with those obtained from the purified enzymes. A BLAST search against the available sequence databases suggested that the deduced amino acid sequences of Adh2 and Adh3 are homologous to those of several NAD+-dependent dehydrogenases (Table 3). Adh2 has an NAD+-binding motif, GTGPVG (between residues 180 and 185), an alcohol dehydrogenase motif, GHEGVGTITEVGDAV (between residues 63 and 77), and ligands typical of a reactive zinc atom, Cys38 and His60, indicating that Adh2 is a typical zinc-containing, NAD+-dependent alcohol dehydrogenase. Adh3 has the alternative NAD+-binding domain GFGVEAG (between residues 219 and 225), which has been found in several aldehyde dehydrogenases capable of catalyzing irreversible reactions (16, 35). The catalytic-site sequences containing Glu and Cys residues of the S-ethyl dipropylcarbamothionate-inducible aldehyde dehydrogenase (thcA) from Rhodococcus sp. strain NI86/21 (35) were completely conserved in Adh3: LELGGKSP (between residues 261 to 268) for Glu and FALNQGEVCTAPS (between residues 293 to 305) for Cys. Judging from the deduced sequence, Adh3 is an unusual alcohol dehydrogenase that is highly homologous in primary structure to aldehyde dehydrogenases, although it was not active with any of the aldehydes tested.
TABLE 3.
Sequence similarity between Adh1, Adh2, and Adh3 and NAD+-dependent dehydrogenases
| Proteina | Size (aa)b | Identity (%) | Similarity (%) | Organism | Accession no. | Reference |
|---|---|---|---|---|---|---|
| Adh1 | 341 | |||||
| ADH | 347 | 35.48 | 69.21 | Sulfolobus solfataricus | P39462 | 1 |
| ADH | 347 | 34.90 | 69.21 | Sulfolobus sp. strain RC3 | P50381 | 7 |
| ADHHT | 339 | 32.55 | 68.14 | Bacillus stearothermophilus | P42328 | 8 |
| SADH | 346 | 32.26 | 67.74 | Rhodococcus ruber | Q8KLT9 | |
| ADHT | 337 | 31.38 | 68.62 | Bacillus stearothermophilus | P12311 | 46 |
| Adh2 | 351 | |||||
| ADH | 366 | 35.04 | 69.52 | Alcaligenes eutrophus | P14940 | 25 |
| FDH | 424 | 33.05 | 66.38 | Methylobacter marinus | P47734 | 55 |
| ADH | 362 | 29.91 | 66.10 | Pseudomonas putida | L35343 | 22 |
| AACR | 349 | 29.63 | 63.82 | Bacillus cereus | Q93R65 | 20 |
| XDH | 363 | 27.64 | 62.96 | Trichoderma reesei | AAO42466 | |
| Adh3 | 506 | |||||
| THCA | 506 | 79.45 | 96.05 | Rhodococcus erythropolis | P46369 | 35 |
| HDALD | 506 | 71.94 | 90.12 | Oleomonas sagaranensis | Q9Z918 | |
| ACOD | 506 | 71.74 | 89.72 | Alcaligenes eutrophus | P46368 | 40 |
| ALD1 | 503 | 71.34 | 90.51 | Acinetobacter sp. M-1 | Q9FDS1 | 23 |
| ALDB | 506 | 70.75 | 89.13 | Xanthobacter autotrophicus | O50206 | 5 |
Abbreviations: ADH, alcohol dehydrogenase; ADHHT, higher thermostable alcohol dehydrogenase; SADH, secondary alcohol dehydrogenase; ADHT, thermostable alcohol dehydrogenase; FDH, formaldehyde dehydrogenase; AACR, acetylacetoin reductase; XDH, xylitol dehydrogenase; THCA, thiocarbamate-inducible aldehyde dehydrogenase; HDALD, aldehyde dehydrogenase from a petroleum-degrading bacterium, strain HD-1; ACOD, acetaldehyde dehydrogenase; ALD1, aldehyde dehydrogenase 1; ALDB, chloroacetaldehyde dehydrogenase.
aa, amino acids.
DISCUSSION
Gordonia sp. strain TY-5 was able to grow with only propane among the gaseous alkanes (C2 to C5) as its sole source of carbon and energy, and it was not capable of growth on methane and alkenes. Propane oxidation to 2-propanol was detected in whole-cell reactions. The prmABCD gene cluster located on the chromosome of TY-5 was cloned and sequenced. Significant function of the prm genes in n-propane metabolism has been demonstrated by the following results: (i) the mutant with prmB disrupted lost the ability to grow on propane, (ii) the prmABCD genes were transcribed polycistronically and transcribed in response to propane, and (iii) the deduced proteins from the prmABCD gene cluster were characteristic of a dinuclear-iron-containing multicomponent monooxygenase.
Soluble dinuclear-iron-containing monooxygenases are classified into two groups based on the subunit structures (59, 65). In one group are the monooxygenases composed of three components, a hydroxylase, a reductase, and a coupling protein (11, 19, 34, 36, 37, 44, 50, 59). Representatives include AMO and tetrahydrofuran monooxygenase (Thm), which have hydroxylases composed of two subunits and other hydroxylases consisting of three subunits. The second group consists of monooxygenases composed of four components, including an additional ferredoxin (51, 60, 64, 65). The Prm of Gordonia sp. strain TY-5 belongs to the former class, and its hydroxylase is composed of two subunits. Detailed analysis of prmABCD based on the functional studies of AMO (44), sMMO (61), and Thm (59) predicts the following model for Prm. Hydroxylation of propane occurs at a dinuclear iron of the large subunit (PrmA) accompanied by the small subunit (PrmC), and electrons needed for O2 activation are provided by an NADH reductase (PrmB). The third component, the regulatory protein PrmD, may influence the reaction rate or product distribution.
Several polypeptides participating in the oxidation of gaseous n-alkanes have been described (15, 39, 63). Since very little is known about the genetics of these systems, we were unable to determine whether any of the reported polypeptides are related to the gene products of prmABCD. Recently, the genes encoding the sBMO from P. butanovora were cloned. The enzyme is composed of three components, a dinuclear-iron-site-containing hydroxylase, a reductase, and a third component (an effector or regulator) like sMMO (50). Among the components of Prm, the putative large and small subunits of the hydroxylase show a relatively higher sequence similarity to those of sBMO, while sequences of the other components show lower similarities. There have been no reports that described propane-oxidizing activity in Thm and sBMO (50, 59), and AMO is incapable of hydroxylation of any n-alkanes (32). The monooxygenase encoded by the prm operon of Gordonia sp. strain TY-5 is the first bacterial enzyme that has been genetically confirmed to participate in propane oxidation.
Some strains belonging to the order Actinomycetales, such as strains of the genera Rhodococcus (54, 62) and Nocardioides (15), are known to degrade n-alkanes with a wide range of carbon chain lengths. Gordonia sp. strain TY-5 can utilize liquid alkanes with longer carbon chains (C13 to C22), as well as propane. Since the prmB mutant strain could still grow on the liquid alkanes, the organism should possess another monooxygenase(s) for the oxidation of liquid alkanes.
We have concluded that propane is oxidized by monooxygenase-mediated subterminal oxidation via 2-propanol from the following results: (i) whole cells of Gordonia sp. strain TY-5 produced 2-propanol and not 1-propanol from propane, (ii) adh1 was transcribed in response to propane and 2-propanol and not in response to 1-propanol, (iii) disruption of adh1 prevented the organism from growing on propane and 2-propanol but did not affect its ability to utilize 1-propanol, and (iv) strain TY-5 could grow on acetone, acetol, and methyl acetate, which are intermediates in the subterminal oxidation pathway (data not shown).
There are several possible pathways for the oxidation of propane (3). Among them, the terminal oxidation pathway of Mycobacterium vaccae JOB-5 (10), both the terminal and subterminal oxidation pathways of P. fluorescens NRRL-B-1244 (21), and the subterminal oxidation pathway of R. rhodochrous PNKb1 (4) have been proposed mainly on the basis of the properties of the alcohol dehydrogenases that participate in 1- or 2-propanol oxidation. Three different secondary alcohol dehydrogenases, Adh1, Adh2, and Adh3, were found in Gordonia sp. strain TY-5, purified, and characterized. All three genes were transcribed in response to propane and 2-propanol but not in response to 1-propanol. Among them, Adh1 appears to be an important dehydrogenase in the propane oxidation pathway. Interestingly, Adh2 and Adh3 had somewhat higher activity for 2-propanol and showed higher specificity for secondary alcohols than did Adh1, but the mutant with adh1 disrupted, in which adh2 and adh3 were expressed, was able to grow only partially on 2-propanol and was incapable of growth with propane. This implies that the data on catalytic properties and induction profiles are not enough to confirm the physiological role of an enzyme.
REFERENCES
- 1.Ammendola, S., C. A. Raia, C. Caruso, L. Camardella, S. D'Auria, M. de Rosa, and M. Rossi. 1992. Thermostable NAD+-dependent alcohol dehydrogenase from Sulfolobus solfataricus: gene and protein sequence determination and relationship to other alcohol dehydrogenases. Biochemistry 31:12514-12523. [DOI] [PubMed] [Google Scholar]
- 2.Arp, D. J. 1999. Butane metabolism by butane-grown ‘Pseudomonas butanovora’. Microbiology 145:1173-1180. [DOI] [PubMed] [Google Scholar]
- 3.Ashraf, W., A. Mihdhir, and J. C. Murrell. 1994. Bacterial oxidation of propane. FEMS Microbiol. Lett. 122:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Ashraf, W., and J. C. Murrell. 1990. Purification and characterization of a NAD+-dependent secondary alcohol dehydrogenase from propane-grown Rhodococcus rhodochrous PNKb1. Arch. Microbiol. 153:163-168. [Google Scholar]
- 5.Bergeron, H., D. Labbe, C. Turmel, and P. C. K. Lau. 1998. Cloning, sequence and expression of a linear plasmid-based and a chromosomal homolog of chloroacetaldehyde dehydrogenase-encoding genes in Xanthobacter autotrophicus GJ10. Gene 207:9-18. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cannio, R., G. Fiorentino, P. Carpinelli, M. Rossi, and S. Bartolucci. 1996. Cloning and overexpression in Escherichia coli of the genes encoding NAD-dependent alcohol dehydrogenase from two Sulfolobus species. J. Bacteriol. 178:301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannio, R., M. Rossi, and S. Bartolucci. 1994. A few amino acid substitutions are responsible for the higher thermostability of a novel NAD+-dependent bacillar alcohol dehydrogenase. Eur. J. Biochem. 222:345-352. [DOI] [PubMed] [Google Scholar]
- 9.Cardy, D. L. N., V. Laidler, G. P. C. Salmond, and J. C. Murrell. 1991. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol. Microbiol. 5:335-342. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, J. P., and J. J. Perry. 1985. Purification and characterization of the secondary alcohol dehydrogenase from propane-utilizing Mycobacterium vaccae JOB5. J. Gen. Microbiol. 131:2901-2907. [DOI] [PubMed] [Google Scholar]
- 11.Ehrt, S., F. Schirmer, and W. Hillen. 1995. Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIB8250. Mol. Microbiol. 18:13-20. [DOI] [PubMed] [Google Scholar]
- 12.Elango, N., R. Radhakrishnan, W. A. Froland, B. J. Wallar, C. A. Earhart, J. D. Lipscomb, and D. H. Ohlendorf. 1997. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 6:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamamura, N., C. Page, T. Long, L. Semprini, and D. J. Arp. 1997. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 63:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamamura, N., R. T. Storfa, L. Semprini, and D. J. Arp. 1999. Diversity in butane monooxygenases among butane-grown bacteria. Appl. Environ. Microbiol. 65:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamamura, N., C. M. Yeager, and D. J. Arp. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 67:4992-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidalgo, E., Y. M. Chen, E. C. C. Lin, and J. Aguilar. 1991. Molecular cloning and DNA sequencing of the Escherichia coli K-12 ald gene encoding aldehyde dehydrogenase. J. Bacteriol. 173:6118-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiraishi, A. 1992. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:210-213. [DOI] [PubMed] [Google Scholar]
- 18.Hiraishi, A., Y. K. Shin, Y. Ueda, and J. Sugiyama. 1994. Automated sequencing of PCR-amplified 16S rDNA on ‘Hydrolink’ gels. J. Microbiol. Methods 19:145-154. [Google Scholar]
- 19.Horinouchi, M., K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1997. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol. Lett. 155:99-105. [DOI] [PubMed] [Google Scholar]
- 20.Hosaka, T., S. Ui, T. Ohtsuki, A. Mimura, M. Ohkuma, and T. Kudo. 2001. Characterization of the NADH-linked acetylacetoin reductase/2,3-butanediol dehydrogenase gene from Bacillus cereus YUF-4. J. Biosci. Bioeng. 91:539-544. [DOI] [PubMed] [Google Scholar]
- 21.Hou, C. T., R. N. Patel, A. I. Laskin, I. Barist, and N. Barnabe. 1983. Thermostable NAD-linked secondary alcohol dehydrogenase from propane-grown Pseudomonas fluorescens NRRL B1244. Appl. Environ. Microbiol. 46:98-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, M., F. B. Oppermann, and A. Steinbuechel. 1994. Molecular characterization of the Pseudomonas putida 2,3-butanediol catabolic pathway. FEMS Microbiol. Lett. 124:141-150. [DOI] [PubMed] [Google Scholar]
- 23.Ishige, T., A. Tani, Y. Sakai, and N. Kato. 2000. Long-chain aldehyde dehydrogenase that participates in n-alkane utilization and wax ester synthesis in Acinetobacter sp. strain M-1. Appl. Environ. Microbiol. 66:3481-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishige, T., A. Tani, Y. Sakai, and N. Kato. Wax ester production by bacteria. Curr. Opin. Microbiol., in press. [DOI] [PubMed]
- 25.Jendrossek, D., A. Steinbuechel, and H. G. Schlegel. 1988. Alcohol dehydrogenase gene from Alcaligenes eutrophus: subcloning, heterologous expression in Escherichia coli, sequencing, and location of Tn5 insertions. J. Bacteriol. 170:5248-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahng, H. Y., J. C. Malinverni, M. M. Majoko, and J. Kukor. 2001. Genetic and functional analysis of the tbc operons for catabolism of alkyl- and chloroaromatic compounds in Burkholderia sp. strain JS150. Appl. Environ. Microbiol. 67:4805-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, N., H. Sahm, and F. Wagner. 1979. Steady-state kinetics of formaldehyde dehydrogenase and formate dehydrogenase from a methanol-utilizing yeast, Candida boidinii. Biochim. Biophys. Acta 566:12-20. [DOI] [PubMed] [Google Scholar]
- 28.Kulikova, A. K., and A. M. Bexborodov. 2001. Assimilation of propane and characterization of propane monooxygenase from Rhodococcus erythropolis 3/89. Appl. Biochem. Microbiol. 37:164-167. [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophages T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lipscomb, J. D. 1994. Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 48:371-399. [DOI] [PubMed] [Google Scholar]
- 31.McDonald, I. R., H. Uchiyama, S. Kambe, O. Yagi, and J. C. Murrell. 1997. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl. Environ. Microbiol. 63:1898-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirua, A., and H. Dalton. 1995. Purification and characterization of the alkene monooxygenase from Nocardia corallina B-276. Biosci. Biotech. Biochem. 59:853-859. [Google Scholar]
- 33.Mitsui, R., Y. Sakai, H. Yasueda, and N. Kato. 2000. A novel operon encoding formaldehyde fixation: the ribulose monophosphate pathway in the gram-positive facultative methylotrophic bacterium Mycobacterium gastri MB19. J. Bacteriol. 182:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murrell, J. C., B. Gilbert, and I. R. McDonald. 2000. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173:325-332. [DOI] [PubMed] [Google Scholar]
- 35.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothionate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman, L. M., and L. P. Wackett. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066-14076. [DOI] [PubMed] [Google Scholar]
- 37.Nordlund, P., J. Powlowski, and V. Shingler. 1990. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J. Bacteriol. 172:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padda, R. S., K. K. Pandey, S. Kaul, V. D. Nair, R. K. Jain, S. K. Basu, and T. Chakrabarti. 2001. A novel gene encoding a 54 kDa polypeptide is essential for butane utilization by Pseudomonas sp. IMT37. Microbiology 147:2472-2491. [DOI] [PubMed] [Google Scholar]
- 40.Priefert, H., N. Krueger, D. Jendrossek, B. Schmidt, and A. Stainthorpe. 1992. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J. Bacteriol. 174:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid, M. R., and C. A. Fewson. 1994. Molecular characterization of microbial alcohol dehydrogenase. Crit. Rev. Microbiol. 20:13-56. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg, E., and E. Z. Ron. 1996. Bioremediation of petroleum contamination, vol. 6. Cambridge University Press, Cambridge, England.
- 43.Rosenzweig, A. C., C. A. Frederick, S. J. Lippard, and P. Nordlund. 1993. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 366:537-543. [DOI] [PubMed] [Google Scholar]
- 44.Saeki, H., and K. Furuhashi. 1994. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene monooxygenase. J. Ferment. Bioeng. 78:399-406. [Google Scholar]
- 45.Sakai, Y., J. H. Maeng, S. Kubota, A. Tani, Y. Tani, and N. Kato. 1996. A non-conventional dissimilation pathway for long chain n-alkanes in Acinetobacter sp. M-1 that start with a dioxygenase reaction. J. Ferment. Bioeng. 81:286-291. [Google Scholar]
- 46.Sakoda, H., and T. Imanaka. 1992. Cloning and sequencing of the gene coding for alcohol dehydrogenase of Bacillus stearothermophilus and rational shift of the optimum pH. J. Bacteriol. 174:1397-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt, A., M. Schiesswohl, U. Volker, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J. Bacteriol. 174:3993-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seila, R. L., H. H. Main, J. L. Arriaga, G. Martines, and A. Ramadan. 2001. Atmospheric volatile organic compound measurements during the 1996 Paso del Norte ozone study. Sci. Total Environ. 276:153-169. [DOI] [PubMed] [Google Scholar]
- 49.Shigematsu, T., S. Hanada, M. Eguchi, Y. Kamagata, T. Kanagawa, and R. Kurane. 1999. Soluble methane monooxygenase gene clusters from trichloroethylene-degrading Methylomonas sp. strains and detection of methanotrophs during in situ bioremediation. Appl. Environ. Microbiol. 65:5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sluis, M. K., L. A. Sayavedra-Soto, and D. J. Arp. 2002. Molecular analysis of the soluble butane monooxygenase from ‘Pseudomonas butanovora’. Microbiology 148:3617-3629. [DOI] [PubMed] [Google Scholar]
- 51.Small, F. J., and S. A. Ensign. 1997. Alkene monooxygenase from Xanthobacter strain Py2. J. Biol. Chem. 272:24913-24920. [DOI] [PubMed] [Google Scholar]
- 52.Smith, C. A., K. T. O'Reilly, and M. R. Hyman. 2003. Characterization of the initial reactions during the cometabolic oxidation of methyl tert-butyl ether by propane-grown Mycobacterium vaccae JOB5. Appl. Environ. Microbiol. 69:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, T. J., S. E. Slade, N. P. Burton, J. C. Murrell, and H. Dalton. 2002. Improved system for protein engineering of the hydroxylase component of soluble methane monooxygenase. Appl. Environ. Microbiol. 68:5265-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smits, T. H., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speer, B. S., L. Chistoserdova, and M. E. Lidstrom. 1994. Sequence of the gene for a NAD(P)-dependent formaldehyde dehydrogenase (class III alcohol dehydrogenase) from a marine methylotroph, Methylobacter marinus A45. FEMS Microbiol. Lett. 121:349-355. [DOI] [PubMed] [Google Scholar]
- 56.Stainthorpe, A. C., V. Lees, G. P. C. Salmond, H. Dalton, and J. C. Murrell. 1990. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath). Gene 91:27-34. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi, J., Y. Ichikawa, H. Sagae, I. Komura, H. Kanou, and K. Yamada. 1980. Isolation and identification of n-butane-assimilating bacterium. Agric. Biol. Chem. 44:1835-1840. [Google Scholar]
- 58.Tani, A., T. Ishige, Y. Sakai, and N. Kato. 2001. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 183:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiemer, B., J. R. Andreesen, and T. Schrader. 2003. Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Arch. Microbiol. 179:266-277. [DOI] [PubMed] [Google Scholar]
- 60.van Hylckama Vlieg, J. E. T., H. Leemhuis, J. H. L. Spelberg, and D. B. Janssen. 2000. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J. Bacteriol. 182:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallar, B. J., and J. D. Lipscomb. 1996. Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev. 96:2625-2658. [DOI] [PubMed] [Google Scholar]
- 62.Whyte, L. G., T. H. Smits, D. Labbe, B. Witholt, C. W. Greer, and J. B. van Beilen. 2002. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRLB-16531. Appl. Environ. Microbiol. 68:5933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woods, N. R., and J. C. Murrell. 1989. The metabolism of propane in Rhodococcus rhodochrous PNKb1. J. Gen. Microbiol. 135:2335-2344. [Google Scholar]
- 64.Yen, K. M., M. R. Karl, L. M. Blatt, M. J. Simon, R. B. Winter, P. R. Fausset, H. S. Lu, A. A. Harcourt, and K. K. Chen. 1991. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 173:5315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou, N.-Y., A. Jenkins, C. K. Chion, and D. J. Leak. 1999. The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenase and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol. Appl. Environ. Microbiol. 65:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]