Abstract
Mutations in fibrillin-1 or fibrillin-2, the major structural components of extracellular microfibrils, cause pleiotropic manifestations in Marfan syndrome and congenital contractural arachnodactyly, respectively. We recently found that fibrillin-1 and fibrillin-2 control bone formation by regulating osteoblast differentiation through the differential modulation of endogenous TGFβ and bone morphogenetic protein signals. Here, we describe in vivo and ex vivo experiments that implicate the fibrillins as negative regulators of bone resorption. Adult Fbn2−/− mice display a greater than normal osteolytic response to locally implanted lipopolysaccharide-coated titanium particles. Although isolated cultures of Fbn2−/− preosteoclasts exhibited normal differentiation and activity, these features were substantially augmented when mutant or wild-type preosteoclasts were co-cultured with Fbn2−/− but not wild-type osteoblasts. Greater osteoclastogenic potential of Fbn2−/− osteoblasts was largely accounted for by up-regulation of the Rankl gene secondary to heightened TGFβ activity. This conclusion was based on the findings that blockade of TGFβ signaling blunts Rankl up-regulation in Fbn2−/− osteoblasts and bones and that systemic TGFβ antagonism improves locally induced osteolysis in Fbn2−/− mice. Abnormally high Rankl expression secondary to elevated TGFβ activity was also noted in cultured osteoblasts from Fbn1−/− mice. Collectively our data demonstrated that extracellular microfibrils balance local catabolic and anabolic signals during bone remodeling in addition to implying distinct mechanisms of bone loss in Marfan syndrome and congenital contractural arachnodactyly.
Keywords: Bone, Connective Tissue, Extracellular Matrix, Fibrillin, Transforming Growth Factor Beta (TGFbeta), Osteoblasts, Osteolysis, RANKL
Introduction
Fibrillins are ubiquitous extracellular matrix (ECM)2 glycoproteins that impart key physical properties to connective tissues and that control the local bioavailability of transforming growth factor-β (TGFβ) family members (1). The structural role of fibrillins is exerted through the temporal and hierarchical formation of tissue-specific assemblies (microfibrils and elastic fibers), whereas the instructive role reflects the ability of these extracellular molecules to bind TGFβ and bone morphogenetic protein (BMP) complexes (1). Fibrillin monomers self-assemble into microfibrils that associate or interact with several other matrix proteins, including fibulins, microfibril-associated glycoproteins (MAGPs), and latent TGFβ-binding proteins, as well as with elastin in the elastic fibers (2, 3). Binding of latent TGFβ-binding proteins to fibrillin and fibronectin assemblies indirectly targets latent TGFβ complexes to the ECM (4–6). Likewise, sequestration of BMPs in the matrix is mediated in part by direct interaction of their prodomains with the N-terminal regions of fibrillins (7).
Mutations of fibrillin genes in human patients and genetically engineered mice have been correlated with discrete phenotypic outcomes that reflect the dual roles of microfibrils in tissue formation, integrity, and remodeling (8). Like patients afflicted with Marfan syndrome (MFS; OMIM-154700), mice harboring mutations that affect the structure or synthesis of fibrillin-1 display dissecting aortic aneurysm, mitral valve prolapse, muscle hypoplasia, and developmental emphysema (9–11). These manifestations are in part accounted for by promiscuous Smad2/3 signaling secondary to improper ECM sequestration of latent TGFβ complexes (10, 12, 13). Fbn2−/− mice, on the other hand, recapitulate the small and large joint contractures that are the hallmark of individuals affected with congenital contractural arachnodactyly (CCA; OMIM-121050), a disease akin to but clinically distinct from MFS (14, 15). Fbn2−/− mice also display a skeletal patterning defect (syndactyly) that is absent in Fbn1−/− mice and that was genetically linked with decreased BMP7 signaling in the mesenchyme of early autopods (14, 16). Neither the relative patterns of fibrillin gene expression in various tissues, including the forming and mature skeleton, nor the established contributions of fibrillin proteins to microfibril biogenesis can fully account for the discrete phenotypic outcomes in MFS and CCA and their respective mouse models (2, 3). Hence, there is a need to explain how fibrillin-1 and fibrillin-2 contribute differently to the structural properties of extracellular microfibrils and their control of local TGFβ and BMP signals within various developmental and physiological contexts.
We recently began investigating the contribution of fibrillin-1 and fibrillin-2 to bone remodeling because of the following considerations. Microfibrils are abundantly deposited in the non-mineralized bone matrix (osteoid) laid down by differentiating preosteoblasts (17); low bone mass (osteopenia) is one of the few traits in common between MFS and CCA (15); and the bone matrix is the preeminent storage site of TGFβ and BMP complexes (18). Bone remodeling is a complex and tightly regulated process that occurs throughout adult life in response to physiological stimuli and mechanical stresses and according to a locally coupled balance between bone resorption by osteoclasts and bone deposition by osteoblasts (19, 20). Extrinsic regulators of bone remodeling include circulating hormones, which promote bone anabolism and catabolism, signaling molecules, which are synthesized by bone cells or released locally from the ECM during bone resorption, and matrix components, which are directly or indirectly associated with osteoblast and osteoclast differentiation and with ECM mineralization and degradation (19, 20).
Relevant to the last group of extrinsic factors, gene function studies in mice have shown that collagen I fibrillogenesis is a critical contributor to bone mass and strength (21–26); that osteonectin supports osteoblast formation, maturation, and survival (27); that biglycan is involved in preosteoblast differentiation and osteoblast-supported osteoclastogenesis (28, 29); that osteopontin participates in stimulating osteoclast motility and bone resorption (30); and that MAGP1 restricts osteoclast differentiation and activity (31). Our own genetic studies have recently demonstrated that fibrillin-1 and fibrillin-2 support osteoblast maturation and bone formation by controlling local TGFβ and BMP bioavailability (32). On the one hand, Fbn2−/− mice were found to have one-third less bone mass and half the normal rate of bone formation because augmented TGFβ signaling selectively interferes with osterix-driven collagen I production and osteoblast maturation. On the other hand, accelerated differentiation of Fbn1−/− osteoblasts in the presence of improper latent TGFβ activation was accounted for by slightly higher levels of osterix- and collagen I-coding transcripts and by a greater availability of otherwise matrix-bound BMPs.
The objective of the present study was to assess whether or not fibrillins may also be involved in regulating bone resorption. To this end, osteoclastogenesis was first evaluated in adult Fbn2−/− mice, and these findings were subsequently related to the behavior of mutant osteoclast precursors cultured alone or together with wild-type or mutant osteoblasts. Similar in vitro experiments were performed with bone cells isolated from Fbn1−/− mice. The results of these analyses demonstrated that microfibrils negatively regulate bone resorption by controlling osteoblast-supported osteoclastogenesis through a process that largely involves TGFβ-dependent stimulation of the osteoclastogenic factor receptor activator of nuclear factor-κ B ligand (RANKL). These findings, together with our parallel study of bone formation in fibrillin-deficient mice (32), significantly advance knowledge of the identity and relationship of local extrinsic factors that orchestrate bone formation, mineralization, and resorption during physiological remodeling and fracture healing.
EXPERIMENTAL PROCEDURES
Mice and Bone Resorption Measurements
The present studies employed Fbn1−/− and Fbn2−/− mice that were respectively bred into the C57/Bl6 and 129/SvEv genetic backgrounds (14, 16). Long bones from 3-month-old Fbn2−/− mice and wild-type littermates (n = 5 animals per each genotype) were processed and stained for tartrate-resistant acid phosphatase (TRAP) using the acid phosphatase staining kit (Sigma-Aldrich) according to the manufacturer's instructions. Numbers of surface osteoclasts (multinucleated TRAP-positive cells) in trabecular bone were evaluated with the aid of the OsteoMeasure analysis system (OsteoMetrics, Decatur, GA). Deoxypyridinoline cross-links were measured in morning urine of female mutant and control mice (n = 6 animals per each genotype) using the Pyrilinks-D immunoassay (Metra Biosystems, Mountain View, CA) and normalized to urinary creatinine content. Statistical analyses for these and all the other assays described below were performed using an unpaired t test (Microsoft Excel); significant association was defined when p < 0.05 compared with control.
In Vitro Osteoclastogenic Assays
For osteoclast differentiation assays (n = 5 assays per each genotype and each performed in duplicate), the bone marrow of 6–8-week-old wild-type or Fbn2−/− mice was flushed, and the monocyte fraction (bone marrow monocytes; BMM) was isolated by centrifugation, washed, seeded on 48-well plates at the concentration of 1.75 × 105 cells/well, and cultured for 7 days in differentiation medium, which includes minimum essential medium-α medium containing 15% FBS and 1% penicillin/streptomycin, 30 ng/ml macrophage colony-stimulating factor (M-CSF; R&D Systems, Minneapolis, MN), and 50 ng/ml recombinant RANKL (Sigma-Aldrich) (33). TRAP staining was performed using the acid phosphatase staining kit. For co-culture assays (n = 3 assays per each combination of genotypes and each performed in duplicate), calvarial osteoblasts were isolated from 4-day-old mutant or wild-type mice and plated on 24-well plates at a concentration of 5 × 104 cells/well. Once they reached confluence, BMM were plated at the concentration of 5 × 105 cells/well, and the co-cultures were further grown for 6–10 days in medium supplemented with 10−8 m 1,25-(OH)2 vitamin D3 and 10−6 m prostaglandin E2 (33). Multinucleated TRAP-positive cells were counted, and osteoclast activity was assessed by the pit formation assay using dentine slices (OsteoSite dentine slices, Immunodiagnostic Systems Inc., Fountain Hills, AR) that were preincubated in differentiation medium for 2 h (33). After 7 days of co-cultures, dentine slices were sonicated in 0.5 m ammonium hydroxide stained with toluidine blue for 2 min, washed with water, and photographed under a light microscope (Eclipse TE 200; Nikon, Yokahama, Japan). Resorbed areas were measured using NIH Image analysis software, normalized to the number of osteoclasts in the well, and expressed as the average resorbed area per cell.
In Vivo Osteolysis Assay
Implementation and analysis of experimentally induced osteolysis was carried out according to Bi et al. (29). Briefly, titanium particles with adherent lipopolysaccharide (LPS) endotoxin (8 × 106 particles/μl; Johnson Matthey, London, UK) or vehicle (25 μl of PBS) were implanted in the parietal bones of isoflurane-anesthetized 1-month-old wild-type or Fbn2−/− mice (n = 3 animals per each genotype), which were sacrificed 1 week later to harvest and process the calvarias for x-ray imaging and histology. In parallel experiments, mutant and wild-type mice (n = 3 animals per each genotype and treatment) were systemically treated with the TGFβ type I receptor (ALK5) inhibitor SD-208 (60 mg/kg; Tocris Biosciences, Ellisville, MO) or vehicle (1% methylcellulose, Sigma-Aldrich) twice daily for 7 days following LPS-induced osteolysis. The extent of osteolysis and the number of osteoclasts were evaluated by computer-aided histomorphometry of x-ray and light microscopy images using NIH Image J analysis software.
RNA Analyses
Real-time quantitative PCR (qPCR) employed total cellular RNA isolated using the RNeasy mini kit (Qiagen) and whose concentration and purity were determined spectrophotometrically (NanoDrop, Thermo Scientific, Waltham, MA). Reverse transcription assays for individual transcripts were each performed in triplicate on at least 3 independent RNA samples per genotype. Amplification assays were carried out with random hexamer primers (Invitrogen) and AffinityScript multiple temperature reverse transcriptase (Stratagene, La Jolla, CA) using 1 μg of total RNA according to manufacturer's instructions. The cDNAs were amplified using SYBR Green Supermix with 6-carboxy-X-rhodamine (Fermentas, Glen Burnie, MD) on a Mastercycler ep realplex instrument (Eppendorf, Westbury, NY) using β-actin as an internal control. Amplification primers were purchased from SABiosciences, a Qiagen company (Frederick, MD) and included those specific for Rankl (PPM03047E), Opg (PPM03404E), M-csf (PPM02990E), and β-actin (PPM0245A). Thermal cycling conditions were incubation at 95 °C for 10 min followed by 40 cycles each consisting of 95 °C for 15 s (denaturation), 60 °C for 30 s (annealing), and 72 °C for 30 s (extension). In some experiments, qPCR assays were performed on total RNA purified from wild-type and Fbn2−/− osteoblasts (n = 5 independent samples per each genotype and treatment and each assayed in triplicate) plated and cultured for 4 days with 1 μm of the ALK5 inhibitor SB431542 (Sigma-Aldrich). For RNA interference (RNAi) experiments (34), wild-type and Fbn2−/− osteoblasts (n = 3 independent experiments per each genotype and treatment and each performed in duplicate) were transfected with 50 μm small interfering RNA (siRNA) specific for Alk5 (10620312; Invitrogen) or non-targeting siRNA (J-040125; Dharmacon, Thermo Scientific). Likewise, wild-type calvarial osteoblasts (n = 3 independent experiments per each genotype and treatment and each performed in duplicate) were transfected with 50 μm siRNA specific for Fbn1 or Fbn2 (D-011034–03 and D-045311–03, respectively; Dharmacon, Thermo Scientific) along with the same non-targeting siRNA as control. Two days after siRNA transfection, total RNA was collected, and qPCR was employed to estimate the extent of gene silencing and the levels of Rankl transcripts. All qPCR data represent multiple biological replicates each analyzed in triplicate and expressed relative to the indicated controls arbitrarily averaged as 1 unit.
RESULTS
Fbn2−/− Mice Display Increased Osteolytic Activity
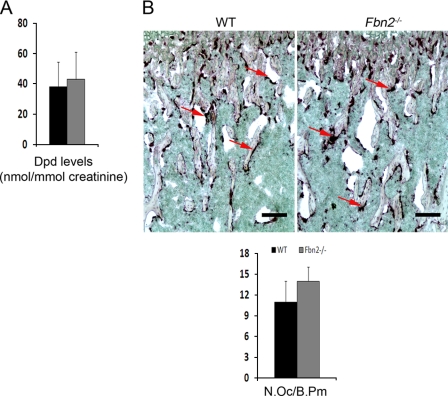
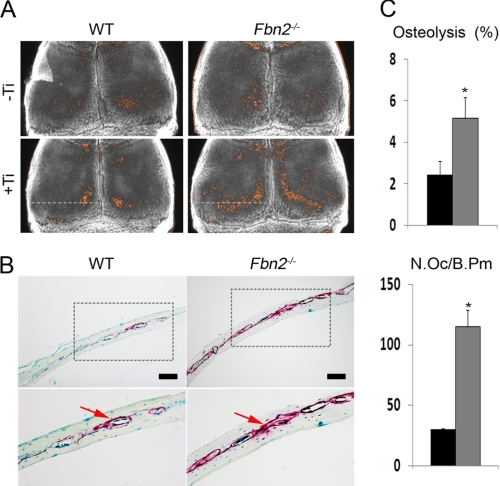
Fbn2−/− mice are viable and fertile, and by 3 months of age, they display 27% less bone mass and a 55% reduction in bone formation rate, which cell culture experiments have correlated with severely impaired osteoblast maturation secondary to greater latent TGFβ activation (14, 32). Examination of two static parameters of bone resorption, urinary excretion of deoxypyridinoline cross-links and the number of TRAP-positive cells, failed to identify significant changes between 3-month-old Fbn2−/− mice and wild-type littermates (Fig. 1, A and B). An acute bone resorption response (osteolysis) was therefore elicited locally in Fbn2−/− mice to uncover potential changes in osteoclast activity (29). Specifically, LPS-coated titanium particles were implanted on the surface of parietal bones of 2-month-old Fbn2−/− and wild-type mice. A week later, the calvarias were harvested and examined by contact x-ray to visualize sites of LPS-induced osteolysis (Fig. 2A). Computer-aided quantitative resolution of the x-ray images revealed that osteolysis occurred more extensively in Fbn2−/− than wild-type calvarias (Fig. 2B). This conclusion was further corroborated by histological analyses of parietal bone sections that documented a statistically significant increase of TRAP-positive osteoclasts in mutant when compared with control samples (Fig. 2C). Taken at face value, these results suggested that heightened osteoclastogenesis is the major determinant of experimentally induced local osteolysis in Fbn2−/− mice.
FIGURE 1.
Bone resorption in Fbn2−/− mice. A, urinary concentration of deoxypyridinoline (Dpd) cross-links normalized to creatinine levels in wild-type (black) and mutant (dark gray) mice (n = 5 per each genotype). B, representative growth plate sections of TRAP-stained wild-type (WT) and Fbn2−/− tibias (scale bar = 200 μm) with arrows pointing to TRAP-positive cells; bar graphs below summarize the estimated number of osteoclasts per bone perimeter (N.Oc/B.Pm) in wild-type (black) and mutant (gray) bones (n = 5 per each genotype). Error bars represent ± S.D.
FIGURE 2.
Experimental osteolysis in Fbn2−/− mice. A, illustrative x-ray radiographs of parietal bones from WT and Fbn2−/− mice that were sham-operated (top; −Ti) or implanted with LPS-titanium beads (bottom; +Ti) (n = 3 per each genotype and treatment). Resorption areas (orange) in each parietal bone were delineated in a blinded fashion from the x-ray radiographs using NIH Image-based histomorphometry. B, representative images of TRAP-stained parietal bones (scale bar = 100 μm) that were sectioned along the white dotted line shown in the x-ray images of panel A; below are magnified images (20×) of the boxed areas above with arrows pointing to TRAP-positive cells. C, bar graphs summarize the percentage of osteolysis (resorbed bone over total bone area; top) and the number of osteoclasts per bone perimeter (N.Oc/B.Pm, bottom) in wild-type (black) and mutant (dark gray) mice. Error bars represent ± S.D., and asterisks signify statistically significant differences (p < 0.05).
Fibrillin Deficiency Stimulates Osteoblast-supported Osteoclastogenesis
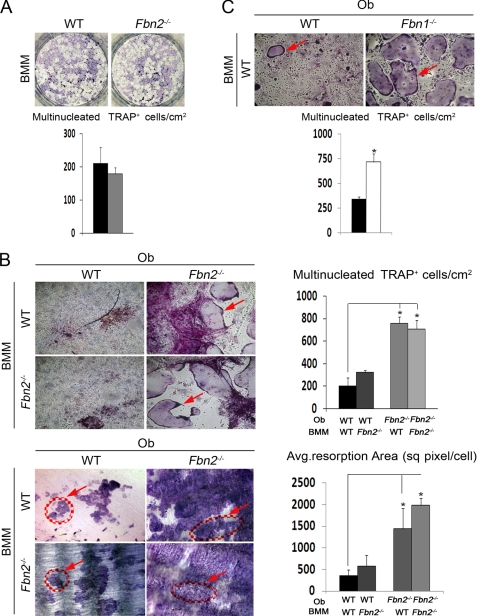
To decipher the cellular mechanism underlying the above observations, osteoclast precursors (BMM) were first isolated from the bone marrow of 2-month-old Fbn2−/− mice and cultured in vitro either in the presence of M-CSF and RANKL, which stimulate osteoclast proliferation and differentiation, or together with mutant or wild-type calvarial osteoblasts, which synthesize both osteoclastogenic factors as well as the RANKL decoy osteoprotegerin (19, 20). Although no differences were noted in the number of TRAP-positive multinucleated cells between isolated wild-type and mutant BMM cultures, significantly more TRAP-positive multinucleated cells and larger resorption pits were observed when either wild-type or mutant BMM were co-cultured with mutant but not with wild-type osteoblasts (Fig. 3, A and B). The finding that BMM co-cultured with mutant osteoblasts are significantly more active than BMM in the presence of stimulatory factors implied that osteoblasts deficient for fibrillin-2 have greater than normal potential to support osteoclastogenesis. The results also suggested that this gain-of-function phenotype of Fbn2−/− osteoblasts is likely to translate in the elevated osteolytic response of Fbn2−/− mice.
FIGURE 3.
Osteoclastogenesis in co-cultures with fibrillin-deficient osteoblasts. A, osteoclast differentiation of BMM from WT and Fbn2−/− mice with bar graphs below summarizing the number of TRAP-positive multinucleated cells per cm2 in control (black) and mutant (gray) samples (n = 5 per each genotype). B, osteoclast differentiation of WT or Fbn2−/− BMM co-cultured with WT or Fbn2−/− osteoblasts (Ob) showing TRAP-positive cells (red arrow; top) and toluidine blue-stained resorption pits (red circles with arrows; bottom); bar graphs on the right of the respective panels summarize the osteoclast number and the average resorbed area per cell (n = 3 per each genotype and experimental combination). sq pixel, square pixel. C, osteoclast differentiation of WT BMM co-cultured with WT or Fbn1−/− osteoblasts with red arrow pointing to TRAP-positive cells; bar graphs below summarize the number of TRAP-positive multinucleated cells per cm2 in WT (black) and Fbn1−/− (light gray) samples (n = 3 per each genotype and experimental combination). Error bars represent ± S.D., and asterisks signify statistically significant differences (p < 0.05).
Next, the above conclusion was evaluated in Fbn1−/− mice to establish whether or not both fibrillins play a negative role in controlling the osteoclastogenic potential of osteoblasts. In this case, however, the neonatal demise of Fbn1−/− mice limited our analysis to performing differentiation assays of wild-type BMM cultured together with mutant neonatal osteoblasts. Within this experimental limitation, the co-culture assays showed that osteoblasts deficient for fibrillin-1 stimulate osteoclastogenesis virtually to the same extent as those deficient for fibrillin-2 (Fig. 3C). Collectively these in vitro experiments suggested that a common molecular mechanism may be responsible for the enhanced osteoclastogenic potential of Fbn1−/− and Fbn2−/− osteoblasts.
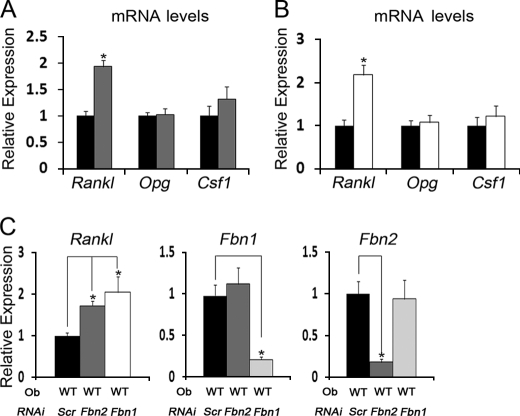
Increased RANKL Expression Characterizes Fibrillin-deficient Osteoblasts
Local control of osteoclast activity by osteoblasts is exerted through the balanced synthesis of factors that promote (RANKL and M-CSF) or inhibit (osteoprotegerin) osteoclastogenesis (19, 20). Accordingly, we investigated whether changes in the relative expression of the Rankl, Csf1, and Opg genes could account for the ability of Fbn1−/− or Fbn2−/− calvarial osteoblasts to stimulate osteoclastogenesis more than the wild-type counterparts. In contrast to Csf1 and Opg, we found that mutant osteoblasts produce significantly more Rankl mRNA than control cells (Fig. 4, A and B). We therefore concluded that loss of either fibrillin-1 or fibrillin-2 similarly stimulates RANKL production, and consequently, enhances the osteoclastogenic potential of mutant osteoblasts.
FIGURE 4.
RANKL expression in fibrillin-deficient osteoblasts. A and B, steady-state levels of indicated transcripts in WT (black) and Fbn2−/− (gray) osteoblasts (n = 6 per each genotype; A) and in WT (black) and Fbn1−/− (white) osteoblasts (n = 4 per each genotype; B). Control WT values are arbitrarily averaged as 1 unit. C, steady-state levels of Fbn and Rankl transcripts in WT osteoblasts (Ob) transfected with non-targeting siRNA (Scr) or siRNA targeting Fbn1 or Fbn2 (n = 3 per each genotype and assay); control Scr values are arbitrarily averaged as 1 unit. Error bars represent ± S.D., and asterisks signify statistically significant differences (p < 0.05).
We have recently shown that primary calvarial osteoblasts produce and assemble both fibrillin-1 and fibrillin-2 microfibrils and that loss of either protein has virtually no effect on the expression levels of the other fibrillin gene (32). In contrast to the impaired maturation of Fbn2−/− osteoblasts, however, Fbn1−/− osteoblasts mature more rapidly than wild-type cells, and these opposite phenotypes can be replicated in wild-type osteoblasts in which either Fbn1 or Fbn2 expression is silenced by siRNA (32). Increased levels of Rankl mRNA in wild-type osteoblasts subject to Fbn1 or Fbn2 silencing further corroborated the phenotypic equivalence between siRNA-induced and germ line loss-of-function mutations of microfibrils, in addition to emphasizing the cell-autonomous nature of the Rankl up-regulation in fibrillin-deficient osteoblast cultures (Fig. 4C).
Heightened TGFβ Signaling Stimulates Experimental Osteolysis in Fbn2−/− Bones
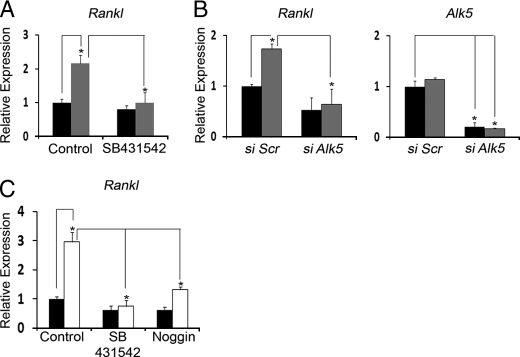
Parallel work has equated the distinct maturation potential of Fbn1−/− and Fbn2−/− osteoblasts with heightened TGFβ and BMP signaling in the former cells and with greater TGFβ signaling in the latter cells (32). The finding that Fbn1−/− and Fbn2−/− osteoblasts share higher TGFβ activity and Rankl expression prompted us to investigate the possibility of a causal connection between these two cellular abnormalities. To this end, Rankl expression was first compared in Fbn2−/− and wild-type osteoblast cultures in which TGFβ signaling was inhibited by either chemical or genetic means. The results documented an appreciable normalization of Rankl expression in mutant osteoblasts that were either treated with the ALK5 kinase inhibitor SB431542 or transfected with Alk5 siRNA (Fig. 5, A and B).
FIGURE 5.
TGFβ signaling regulates RANKL production. A and B, steady-state levels of Rankl in WT (black) and Fbn2−/− (gray) osteoblasts cultured with or without the ALK5 kinase inhibitor SB431542 (n = 5 per each genotype; A) or transfected with non-targeting (Scr) or Alk5-targeting siRNA (n = 5 per each genotype; B). Histograms on the right of the latter panel indicate Alk5 levels in osteoblasts transfected with non-targeting or Alk5-targeting siRNA. C, steady-state levels of Rankl in WT (black) and Fbn1−/− (white) osteoblast cultures treated with SB431542 (SB) or noggin (n = 3 per each genotype and treatment). WT values in control and non-targeting samples are each arbitrarily averaged as 1 unit. Error bars represent ± S.D., and asterisks signify statistically significant differences (p < 0.05).
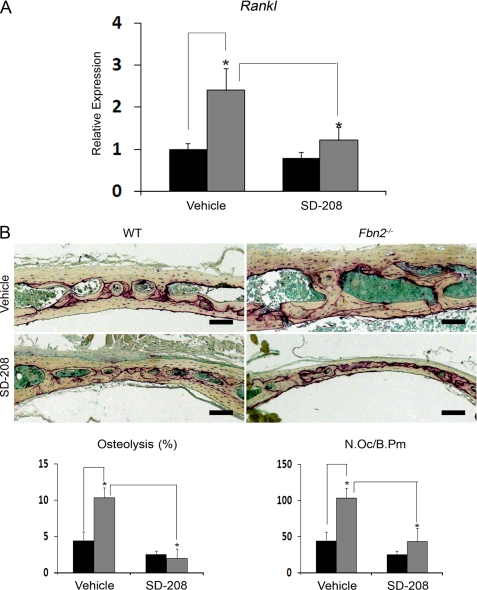
Next, two independent lines of evidence correlated the in vitro data to the in vivo condition. First, qPCR analyses showed that systemic administration of the ALK5 kinase inhibitor SD-208 decreases considerably the abnormally high levels of Rankl transcripts noted in the bones of adult Fbn2−/− mice (Fig. 6A). Second, light microscopy documented that greater LPS-induced osteolysis was blunted in 2-month-old Fbn2-null mice that were systemically treated with SD-208 for 7 days following the implantation of titanium beads (Fig. 6B). Together the ex vivo and the in vivo experiments concurred in supporting the notion that loss of fibrillin-2 deposition in the forming osteoid augments the osteoclastogenic potential of mutant osteoblasts by increasing Rankl expression, in part, through heightened TGFβ signaling.
FIGURE 6.
TGFβ signaling stimulates osteolysis in Fbn2−/− mice. A, steady-state levels of Rankl transcripts in calvarial bones from adult wild-type (WT, black) and Fbn2−/− (gray) mice systemically treated for 2 months with either vehicle or the ALK5 kinase inhibitor SD-208 (n = 3 per each genotype and treatment). WT values in vehicle-treated samples are arbitrarily averaged as 1 unit. B, illustrative images of LPS-titanium-induced osteolysis in parietal bone sections of wild-type and Fbn2−/− mice systemically treated with vehicle or SD-208 (scale bar = 100 μm). Bar graphs below summarize the amount of absorbed tissue per bone surface and the number of osteoclasts per bone perimeter (N.Oc/B.Pm) in vehicle- and SD-208-treated wild-type (black) and mutant (gray) mice (n = 3 per each genotype and treatment). Error bars represent ± S.D., and asterisks signify statistically significant differences (p < 0.05).
The above conclusion was indirectly extended to mice deficient for fibrillin-1 by documenting that the ALK5 kinase inhibitor SB431542 normalizes the abnormal up-regulation of Rankl expression in Fbn1−/− osteoblasts (Fig. 5C). In line with published evidence from mice with disrupted BMP signaling (35, 36), Rankl expression was also down-regulated in Fbn1−/− osteoblasts treated with the BMP antagonist noggin, but to a lesser extent than the TGFβ blockade (Fig. 5C). Taken at face value, this last finding suggested a probable cooperation between local TGFβ and BMP signals in regulating RANKL production by osteoblasts.
DISCUSSION
We have recently implicated fibrillin-1 and fibrillin-2 in the regulation of bone formation by respectively balancing the local release of TGFβ and BMP molecules and by restricting TGFβ bioavailability during osteogenic differentiation (32). This conclusion was based on the following findings. First, loss of fibrillin-2 impairs osteoblast maturation by increasing latent TGFβ activation, which in turn interferes with osterix-stimulated production of collagen I; second, loss of fibrillin-1 accelerates osteoblast differentiation by elevating TGFβ and BMP signaling to a degree that apparently restores the osterix-driven maturation process. Here, we showed that both fibrillins act as negative regulators of bone resorption and that both proteins exert most of their action through the modulation of TGFβ-dependent production of RANKL by osteoblasts. Together our studies therefore demonstrate that extracellular microfibrils are intimately involved in bone anabolism and catabolism by providing the structural scaffold that modulates local TGFβ and BMP bioavailability.
Extracellular regulation of TGFβ and BMP bioavailability is a critical but poorly characterized aspect of organ development and tissue remodeling (1). Current evidence indicates that latent TGFβ-binding proteins mediate targeting of latent TGFβ complexes to ECM components, including nascent fibronectin and fibrillin assemblies, whereas BMPs are directly sequestered in the matrix through high affinity interactions between their prodomains and the N termini of fibrillins (4–7). Mutations that affect the structure or expression of fibrillins are therefore expected to dysregulate TGFβ and BMP signaling by interfering with the ECM-mediated processes of localizing and concentrating the ligands during organ development and releasing them in a timely fashion during tissue remodeling. This prediction is based on the characterization of fibrillin mutant mice, which has also documented the contextual specificity of microfibril regulation of TGFβ and BMP bioavailability (9–14, 33). Relevant to the latter point, mouse studies have implied that the phenotypic diversity of Fbn1 and Fbn2 mutant mice depends on yet to be defined mechanisms that specify which of all possible interactions between the growth factors and fibrillins may prevail in an individual tissue and at a given developmental stage or physiological process. Earlier examples of highly contextual interactions between extracellular microfibrils and growth factors include the unique promotion by fibrillin-2 of BMP signaling in the emerging autopods and the selective restriction by fibrillin-1 of TGFβ activity in the postnatal aortas, regenerating muscles, and developing lungs and mitral valves (10–14). The present investigations, together with our parallel study of bone formation in fibrillin-deficient mice (32), add another layer of complexity to the contextual specificity of microfibril control of TGFβ and BMP bioavailability by demonstrating that fibrillins have both unique and overlapping functions in modulating growth factor signaling during bone remodeling.
The influence of locally released TGFβ and BMP signals on the activity of and cross-talk between osteoblasts and osteoclasts remains largely undefined mostly because cell culture experiments have often yielded contradictory results, depending on the cell type and experimental conditions employed (18, 37). For example, osteoclasts co-cultured with osteoblasts have been reported to have opposite responses to high (inhibition) and low (stimulation) concentrations of TGFβ as a result of parallel changes in RANKL and osteoprotegerin production by osteoblasts (38–40). Our co-culture experiments adhere to a model in which differentiating preosteoclasts are continuously exposed to higher than normal amounts of RANKL and active TGFβ. Importantly, this in vitro evidence was corroborated in vivo by documenting the efficacy of systemic TGFβ antagonism to reduce both Rankl up-regulation and experimentally induced osteolysis in Fbn2−/− mice. Our results are also consistent with previous reports indicating that dysregulated TGFβ signaling in osteoblasts perturbs osteoclast activity and bone resorption in mice and that germ line ablation of a TGFβ-inducible transcription factor impairs RANKL production and osteoblast-supported osteoclastogenesis (41–43).
In addition to TGFβ, osteoclast precursors can also respond to BMP directly in isolated cultures or indirectly through BMP action on co-cultured osteoblasts (18). The latter mechanism was correlated with increased RANKL production in vitro and independently validated in mice in which BMP signaling is selectively inactivated in maturing osteoblasts (35, 44). Subsequent mouse studies suggested that BMP signaling in osteoblasts targets osteoclastogenesis directly by elevating Rankl expression and indirectly by stimulating sclerostin production through inhibition of Wnt signaling (36). Our finding that noggin can partially inhibit Rankl transcription in Fbn1−/− osteoblasts is in agreement with these previous reports. Additionally, inhibition of Rankl up-regulation in Fbn1−/− osteoblasts by both SB431542 and noggin suggests that TGFβ and BMP signals cooperate in stimulating RANKL production, and this may conceivably translate into greater osteoclastogenic activity in Fbn1−/− than in Fbn2−/− mice. Although we are unable to evaluate osteolysis in Fbn1−/− mice, we can reasonably assume that the same abnormality may apply to Fbn1 mutant mice that model adult MFS (9, 45). Along these same lines, one would expect that MFS or CCA patients may be at a greater risk than healthy individuals for implant wear-induced osteolysis and that TGFβ antagonism may be an effective means to manage this and related orthopedic problems. Indeed proof-of-concept experiments in mice have recently shown that systemic ALK5 inhibition has both anabolic and anti-catabolic effects on bone with the net effect of improving bone fracture resistance (46).
In summary, the present study identifies the fibrillin microfibrils as the first extrinsic factor that links together targeting of TGFβ and BMP complexes to the bone matrix with contextual specification of their respective signals and with osteoblast-promoted formation and degradation of the bone matrix (Fig. 6). This finding sheds new light on the extracellular mechanisms that coordinate the timely release and calibrate the threshold levels of TGFβ and BMP signals during bone remodeling. As such, this information has great potential to improve the design of therapeutic interventions to ameliorate bone loss and of bioengineered formulations to improve bone repair.
Acknowledgments
We thank Catherine Liu and Maria del Solar for technical support and Karen Johnson for organizing the manuscript. We are indebted to Drs. Hal Dietz, Patricia Ducy, Gerard Karsenty, Dan Rifkin, and Lynn Sakai for many helpful discussions and to Drs. Theresa Guise, Marian Young, and Mone Zaidi for helpful experimental advice.
This work was supported, in whole or in part, by National Institutes of Health Grant AR42044 (to F. R.). This work was also supported by the National Marfan Foundation.
- ECM
- extracellular matrix
- ALK5
- activin-like kinase-5 or TGFβ receptor I
- BMM
- bone marrow monocytes
- BMP
- bone morphogenetic protein
- CCA
- congenital contractural arachnodactyly
- M-CSF
- macrophage colony stimulating factor
- MFS
- Marfan syndrome
- RANKL
- receptor activator of nuclear factor-κ B ligand
- TRAP
- tartrate-resistant acid phosphatase
- qPCR
- quantitative PCR.
REFERENCES
- 1.Ramirez F., Rifkin D. B. (2009) Curr. Opin. Cell Biol. 21, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubmacher D., Tiedemann K., Reinhardt D. P. (2006) Curr. Top. Dev. Biol. 75, 93–123 [DOI] [PubMed] [Google Scholar]
- 3.Ramirez F., Sakai L. Y. (2010) Cell Tissue Res. 339, 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taipale J., Saharinen J., Hedman K., Keski-Oja J. (1996) J. Histochem. Cytochem. 44, 875–889 [DOI] [PubMed] [Google Scholar]
- 5.Isogai Z., Ono R. N., Ushiro S., Keene D. R., Chen Y., Mazzieri R., Charbonneau N. L., Reinhardt D. P., Rifkin D. B., Sakai L. Y. (2003) J. Biol. Chem. 278, 2750–2757 [DOI] [PubMed] [Google Scholar]
- 6.Dallas S. L., Sivakumar P., Jones C. J., Chen Q., Peters D. M., Mosher D. F., Humphries M. J., Kielty C. M. (2005) J. Biol. Chem. 280, 18871–18880 [DOI] [PubMed] [Google Scholar]
- 7.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez F., Dietz H. C. (2007) Curr. Opin. Genet. Dev. 17, 252–258 [DOI] [PubMed] [Google Scholar]
- 9.Pereira L., Lee S. Y., Gayraud B., Andrikopoulos K., Shapiro S. D., Bunton T., Biery N. J., Dietz H. C., Sakai L. Y., Ramirez F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3819–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neptune E. R., Frischmeyer P. A., Arking D. E., Myers L., Bunton T. E., Gayraud B., Ramirez F., Sakai L. Y., Dietz H. C. (2003) Nat. Genet. 33, 407–411 [DOI] [PubMed] [Google Scholar]
- 11.Ng C. M., Cheng A., Myers L. A., Martinez-Murillo F., Jie C., Bedja D., Gabrielson K. L., Hausladen J. M., Mecham R. P., Judge D. P., Dietz H. C. (2004) J. Clin. Invest. 114, 1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habashi J. P., Judge D. P., Holm T. M., Cohn R. D., Loeys B. L., Cooper T. K., Myers L., Klein E. C., Liu G., Calvi C., Podowski M., Neptune E. R., Halushka M. K., Bedja D., Gabrielson K., Rifkin D. B., Carta L., Ramirez F., Huso D. L., Dietz H. C. (2006) Science 312, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohn R. D., van Erp C., Habashi J. P., Soleimani A. A., Klein E. C., Lisi M. T., Gamradt M., ap Rhys C. M., Holm T. M., Loeys B. L., Ramirez F., Judge D. P., Ward C. W., Dietz H. C. (2007) Nat. Med. 13, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arteaga-Solis E., Gayraud B., Lee S. Y., Shum L., Sakai L., Ramirez F. (2001) J. Cell Biol. 154, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez F., Arteaga-Solis E. (2008) in Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism (Rosen C. ed) 7th Ed., pp. 450–454, ASBMR Publications, Washington, D. C [Google Scholar]
- 16.Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., Ramirez F. (2006) J. Biol. Chem. 281, 8016–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitahama S., Gibson M. A., Hatzinikolas G., Hay S., Kuliwaba J. L., Evdokiou A., Atkins G. J., Findlay D. M. (2000) Bone 27, 61–67 [DOI] [PubMed] [Google Scholar]
- 18.Alliston T., Piek E., Derynck R.2008. in The TGFβ Family (Derynck R., Miyazono K. eds) pp. 667–723, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.Harada S., Rodan G. A. (2003) Nature 423, 349–355 [DOI] [PubMed] [Google Scholar]
- 20.Raggatt L. J., Partridge N. C. (2010) J. Biol. Chem. 285, 25103–25108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonadio J., Saunders T. L., Tsai E., Goldstein S. A., Morris-Wiman J., Brinkley L., Dolan D. F., Altschuler R. A., Hawkins J. E., Jr., Bateman J. F. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7145–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chipman S. D., Sweet H. O., McBride D. J., Jr., Davisson M. T., Marks S. C., Jr., Shuldiner A. R., Wenstrup R. J., Rowe D. W., Shapiro J. R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morello R., Bertin T. K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F. H., Vranka J., Bächinger H. P., Pace J. M., Schwarze U., Byers P. H., Weis M., Fernandes R. J., Eyre D. R., Yao Z., Boyce B. F., Lee B. (2006) Cell 127, 291–304 [DOI] [PubMed] [Google Scholar]
- 24.Murakami T., Saito A., Hino S., Kondo S., Kanemoto S., Chihara K., Sekiya H., Tsumagari K., Ochiai K., Yoshinaga K., Saitoh M., Nishimura R., Yoneda T., Kou I., Furuichi T., Ikegawa S., Ikawa M., Okabe M., Wanaka A., Imaizumi K. (2009) Nat. Cell Biol. 11, 1205–1211 [DOI] [PubMed] [Google Scholar]
- 25.Choi J. W., Sutor S. L., Lindquist L., Evans G. L., Madden B. J., Bergen H. R., 3rd, Hefferan T. E., Yaszemski M. J., Bram R. J. (2009) Plos Genet. 5, e1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sohaskey M. L., Jiang Y., Zhao J. J., Mohr A., Roemer F., Harland R. M. (2010) J. Cell Biol. 189, 511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delany A. M., Kalajzic I., Bradshaw A. D., Sage E. H., Canalis E. (2003) Endocrinology 144, 2588–2596 [DOI] [PubMed] [Google Scholar]
- 28.Xu T., Bianco P., Fisher L. W., Longenecker G., Smith E., Goldstein S., Bonadio J., Boskey A., Heegaard A. M., Sommer B., Satomura K., Dominguez P., Zhao C., Kulkarni A. B., Robey P. G., Young M. F. (1998) Nat. Genet. 20, 78–82 [DOI] [PubMed] [Google Scholar]
- 29.Bi Y., Nielsen K. L., Kilts T. M., Yoon A., A Karsdal M., Wimer H. F., Greenfield E. M., Heegaard A. M., Young M. F. (2006) Bone 38, 778–786 [DOI] [PubMed] [Google Scholar]
- 30.Chellaiah M. A., Kizer N., Biswas R., Alvarez U., Strauss-Schoenberger J., Rifas L., Rittling S. R., Denhardt D. T., Hruska K. A. (2003) Mol. Biol. Cell 14, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craft C. S., Zou W., Watkins M., Grimston S., Brodt M. D., Broekelmann T. J., Weinbaum J. S., Teitelbaum S. L., Pierce R. A., Civitelli R., Silva M. J., Mecham R. P. (2010) J. Biol. Chem. 285, 23858–23867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nistala H., Lee-Arteaga S., Smaldone S., Siciliano G., Carta L., Ono R., Sengle G., Arteaga-Solis E., Levasseur R., Ducy P., Sakai L. Y., Karsenty G., Ramirez F. (2010) J. Cell Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Kua H. Y., Hu Y., Guo K., Zeng Q., Wu Q., Ng H. H., Karsenty G., de Crombrugghe B., Yeh J., Li B. (2006) J. Cell Biol. 172, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carta L., Smaldone S., Zilberberg L., Loch D., Dietz H. C., Rifkin D. B., Ramirez F. (2009) J. Biol. Chem. 284, 5630–5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishina Y., Starbuck M. W., Gentile M. A., Fukuda T., Kasparcova V., Seedor J. G., Hanks M. C., Amling M., Pinero G. J., Harada S., Behringer R. R. (2004) J. Biol. Chem. 279, 27560–27566 [DOI] [PubMed] [Google Scholar]
- 36.Kamiya N., Ye L., Kobayashi T., Mochida Y., Yamauchi M., Kronenberg H. M., Feng J. Q., Mishina Y. (2008) Development 135, 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janssens K., ten Dijke P., Janssens S., Van Hul W. (2005) Endocr. Rev. 26, 743–774 [DOI] [PubMed] [Google Scholar]
- 38.Murakami T., Yamamoto M., Ono K., Nishikawa M., Nagata N., Motoyoshi K., Akatsu T. (1998) Biochem. Biophys. Res. Commun. 252, 747–752 [DOI] [PubMed] [Google Scholar]
- 39.Thirunavukkarasu K., Miles R. R., Halladay D. L., Yang X., Galvin R. J., Chandrasekhar S., Martin T. J., Onyia J. E. (2001) J. Biol. Chem. 276, 36241–36250 [DOI] [PubMed] [Google Scholar]
- 40.Karst M., Gorny G., Galvin R. J., Oursler M. J. (2004) J. Cell. Physiol. 200, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erlebacher A., Derynck R. (1996) J. Cell Biol. 132, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filvaroff E., Erlebacher A., Ye J., Gitelman S. E., Lotz J., Heillman M., Derynck R. (1999) Development 126, 4267–4279 [DOI] [PubMed] [Google Scholar]
- 43.Subramaniam M., Gorny G., Johnsen S. A., Monroe D. G., Evans G. L., Fraser D. G., Rickard D. J., Rasmussen K., van Deursen J. M., Turner R. T., Oursler M. J., Spelsberg T. C. (2005) Mol. Cell. Biol. 25, 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamoto M., Murai J., Yoshikawa H., Tsumaki N. (2006) J. Bone Miner. Res. 21, 1022–1033 [DOI] [PubMed] [Google Scholar]
- 45.Judge D. P., Biery N. J., Keene D. R., Geubtner J., Myers L., Huso D. L., Sakai L. Y., Dietz H. C. (2004) J. Clin. Invest. 114, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohammad K. S., Chen C. G., Balooch G., Stebbins E., McKenna C. R., Davis H., Niewolna M., Peng X. H., Nguyen D. H., Ionova-Martin S. S., Bracey J. W., Hogue W. R., Wong D. H., Ritchie R. O., Suva L. J., Derynck R., Guise T. A., Alliston T. (2009) PLoS One 4, e5275. [DOI] [PMC free article] [PubMed] [Google Scholar]