Abstract
The mechanism by which enzymes recognize the “uniform” collagen triple helix is not well understood. Matrix metalloproteinases (MMPs) cleave collagen after the Gly residue of the triplet sequence Gly∼[Ile/Leu]-[Ala/Leu] at a single, unique, position along the peptide chain. Sequence analysis of types I-III collagen has revealed a 5-triplet sequence pattern in which the natural cleavage triplets are always flanked by a specific distribution of imino acids. NMR and MMP kinetic studies of a series of homotrimer peptides that model type III collagen have been performed to correlate conformation and dynamics at, and near, the cleavage site to collagenolytic activity. A peptide that models the natural cleavage site is significantly more active than a peptide that models a potential but non-cleavable site just 2-triplets away and NMR studies show clearly that the Ile in the leading chain of the cleavage peptide is more exposed to solvent and less locally stable than the Ile in the middle and lagging chains. We propose that the unique local instability of Ile at the cleavage site in part arises from the placement of the conserved Pro at the P3 subsite. NMR studies of peptides with Pro substitutions indicate that the local dynamics of the three chains are directly modulated by their proximity to Pro. Correlation of peptide activity to NMR data shows that a single locally unstable chain at the cleavage site, rather than two or three labile chains, is more favorable for cleavage by MMP-1 and may be the determining factor for collagen recognition.
Keywords: Collagen, Kinetics, Matrix Metalloproteinase, NMR, Peptides
Introduction
The degradation of collagen, the major structural component of connective tissues in skin, bone, tendon, and ligament, is an integral part in many biological processes such as wound healing, cell migration, tissue remodeling, and organ morphogenesis (1–4). Accelerated breakdown of collagen may result in many diseases such as arthritis, tumor cell invasion, glomerulonephritis, and metastasis (5–7). Types I, II, and III collagens, also called interstitial collagens, are the most abundant (8–10), and contain a characteristic triple-helical conformation, which consists of three polyproline II-like helices supercoiled around a common axis (11, 12). The close packing of the three chains can only accommodate Gly as every third residue, generating the repetitive (Gly-Xaa-Yaa)n sequence pattern. The Gly residues are all buried in the center, and the structure is stabilized by interchain N-H (Gly) … C=O (Xaa) hydrogen bonds. The residues at the X and Y positions can be almost any amino acid, but they are frequently Pro and 4-hydroxyproline (Hyp2 or O), respectively.
The triple helical structure allows collagen to be degraded by only a few proteinases including a group of matrix metalloproteinases (MMPs) (5, 13). These MMPs (MMP-1, MMP-2, MMP-8, MMP-13, MMP-14, MMP-18) can bind and cleave interstitial collagens at a unique locus approximately three-fourths away from the N terminus of the collagens (14). The cleavage site is after the Gly residue in the sequence of Gly∼[Ile/Leu]-[Ala/Leu]. Interestingly, there are dozens of other sites in the collagens that contain Gly∼Ile/Leu bonds but they are not hydrolyzed (14). The mechanism by which MMPs recognize the single scissile bond in the repeating Gly-Xaa-Yaa triplets and uniform triple helical conformation is still poorly understood.
It has been shown that the amino acid sequence alone is not sufficient for the high specificity of collagen recognition by MMPs (15–17). The unique triple helical conformation of collagen must also play an important role, as indicated by significant differences in processing of native collagen and denatured collagen. Native collagen can mainly be degraded by some MMPs, while denatured collagen is susceptible to a variety of proteinases (15–19). Collagen is cleaved at a single site, but denatured collagen is cleaved at multiple sites containing the sequence Gly∼[Ile/Leu]-Yaa (17). Also, native collagen is a much better substrate than denatured collagen for some MMPs (17). It is thus believed that the triple helical conformation of collagen is crucial for MMP recognition. It has been suggested that the variation in sequence around the cleavage site would lead to a change in the triple helical conformation and/or dynamics of collagen at the collagenase cleavage site, and this change would be identified by MMPs (14, 17, 20).
Sequence analysis, biological assays, structural, and computational approaches have been used to investigate the basis of the specificity of MMP cleavage of collagen (21–27). Early investigation of collagen sequences suggested that the distribution of weak helix triplets, charged residues as well as the side-chain volume concentration could provide recognition information for MMPs (23). The weak helix hypothesis, which got the most attention, proposed that the cleavage site region is locally unstable because of deficiency in imino acids, and therefore the scissile bond could be exposed to the enzyme attack (21–23). Types I and III collagens can be cleaved by trypsin and α-chymotrypsin around the collagenase cleavage site, suggesting relaxation of the helix in this region (24, 25). The correct distribution of imino acids has been shown to be important for cleavage, as mutation of Ile776 or Gln774 and Ala777 to Pro residues in type I collagen eliminated MMP-1 and MMP-8 activity, suggesting that extra imino acids at the cleavage sites may make the sites more rigid and inaccessible for MMPs (26, 27). NMR and x-ray structures of MMP-1 and MMP-8 revealed that the active site cleft in the catalytic domain of the enzyme was too narrow to accommodate a triple helix (28–31). It was proposed that the triple helix needs to unwind locally so that a single chain can enter into the active site of MMPs or the active site of MMPs undergoes large conformational changes (32, 33). Computational simulations of type I collagen also indicated that the α2 chain is locally unfolded with disrupted hydrogen bonds N-terminal to the cleavage sites and simulations of peptide models of type III collagen show that the real cleavage site is more exposed to solvent and more flexible than the other four potential cleavage sites (33, 34). Taken together, these studies suggest that the conformation and dynamics at the collagenase cleavage site is critical for collagen recognition by MMPs.
NMR studies on peptide models of collagen are powerful approaches for probing the conformation and dynamics of individual chains of the triple helix directly at the specific residue level (35–39). Here we use NMR to characterize the differences in conformation and dynamics at the natural Gly∼Ile-Ala cleavage site versus potential cleavage sites. It has been shown that peptide models of the collagenase cleavage sites can be hydrolyzed by MMPs in a similar fashion to native collagens, indicating that they can serve as good biological models for MMP recognition (40–44). A series of homotrimer peptides modeling the natural cleavage site and potential but noncleavable sites in type III collagen are designed and investigated by NMR. Correlation of NMR conformation and dynamics of the peptides to collagenolytic activity provides evidence that increased dynamics and decreased local stability of Ile in just a single chain is important for increased MMP-1 kinetics. We present evidence that the neighboring imino acids, particularly the conserved Pro at the P3 subsite, modulate the local conformation and dynamics of the unique Ile at the natural cleavage site and thus the specificity of the recognition by MMPs.
EXPERIMENTAL PROCEDURES
Sample Preparation
The peptide sets were synthesized by the Tufts University Core Facility (Boston, MA) with 15N and/or 13C-labeled amino acids at selective positions for NMR characterization. Peptides T3–778, T3–778[IT-PO], and T3–778[P-A] were selectively 15N labeled at positions of Gly16, Ile17, Gly22, Ala23, and Gly31. Peptide T3–778[I-L] was selectively 15N labeled at positions Gly16, Leu17, Gly22, Ala23, and Gly31, while peptide T3–785 was selectively 15N/13C doubly labeled at positions Gly15, Leu16, Ala17, Gly18, and Gly24 (Table 1). The NMR samples for all peptides were prepared in 10% D2O/90% H2O at pH 3.1 with concentrations of 6 mm.
TABLE 1.
Triple helical peptides modeling the collagenase cleavage sites and their activities
Residues that are 15N labeled are underlined; each peptide has five labeled residues including a Gly at the central position in its (GPO)5/(GPO)3 C-terminal end; mutated residues relative to peptide T3–778 are bolded in peptides T3–778[I-L], T3–778[IT-TO], and T3–778[P-A].
| Peptide | Sequence | kcat/Km |
|---|---|---|
| m−1s−1 | ||
| T3–778 | (GPO)4-GPL-GIA-GIT-GAR-(GPO)5GY | 1253 ± 249 |
| T3–778[I-L] | (GPO)4-GPL-GLA-GIT-GAR-(GPO)5GY | 1562 ± 130 |
| T3–778[IT-PO] | (GPO)4-GPL-GIA-GPO-GAR-(GPO)5GY | 883 ± 134 |
| T3–778[P-A] | (GPO)4-GAL-GIA-GIT-GAR-(GPO)5GY | 125 ± 50 |
| T3–785 | PO(GPO)2-GIT-GAR-GLA-GPO-(GPO)3GY | 173 ± 81 |
MMP Kinetics
Peptides and enzymes were dissolved in enzyme assay buffer (50 mm Tris·HCl, pH 7.5, 100 mm NaCl, 10 mm CaCl2, 0.02% NaN3, 0.05% brij-35). MMP-1 was mixed with varying concentrations of each substrate and incubated at 20 °C. At varying time points the reaction was stopped by the addition of 25 mm EDTA, and the reaction product was determined by RP-HPLC analysis using an Agilent HP1100 HPLC system equipped with a C8 column. Melting temperatures were determined by circular dichroism spectroscopy. The temperature used for hydrolysis and NMR studies is 5–30 °C below the melting temperature for each peptide (data not shown).
NMR Spectroscopy
NMR experiments were performed on a Varian 500 MHz spectrometer or a Varian 600 MHz spectrometer with a cryogenic probe. 1H-15N heteronuclear single quantum coherence (HSQC) (45) and three-dimensional 15N-edited NOESY-HSQC experiments (46–48) with a mixing time of 50 ms were carried out for assignments of NMR resonances at 20 °C. Three-dimensional HNHA experiments were performed with an H-H coupling period of 25 ms at 20 °C to measure 3JHNHα coupling constants, which provided information about phi angles (49). Relaxation R1, R2 and heteronuclear NOE measurements (50–52) were done at 20 °C. All data were processed using the FELIX 2004 software package (MSI, San Diego, CA) or NMRPipe (53), and analyzed with FELIX 2004 or NMRView (54).
Hydrogen Exchange Experiments
Hydrogen exchange experiments were performed at 10 °C and pD 2.3–2.8 for the peptide sets (2.74 for T3–778, 2.34 for T3–778[I-L], 2.53 for T3–778[IT-PO], 2.56 for T3–778[P-A] and 2.41 for T3–785), where the pD is corrected for the glass electrode solvent isotope artifact (55). All samples were equilibrated in H2O at 10 °C for a minimum of 48 h to ensure that the monomer:trimer equilibrium is reached. The samples were then lyophilized, re-dissolved in 100% D2O and quickly transferred to the spectrometer which was equilibrated at 10 °C. A series of HSQC spectra were acquired every 5 min (64 t1 increments and 2 scans per increment) (except every 10 min for T3–778) immediately after the sample was placed in the probe to allow the measurements of fast-exchanging amide protons, which are replaced by deuterons within several hours. Then the HSQC spectra were acquired every 40 min to monitor the slowly exchanging amide protons. The hydrogen exchange rates kex were determined by a non-linear least squares fit using the equation I(t) = (I0 − I∞)exp(−kext), where It, I0, and I∞ are the intensities at time t, at time 0 and at a final time point when the exchange is completed. The protection factor P is defined as p = kint/kex, where kint is the intrinsic exchange rate of amide protons in the monomer state at a specific pH and temperature (56). The error of kint was estimated by assuming an experimental uncertainty of ±0.1 °C and ±0.02 pH units. The error of the protection factor P is calculated as follows in Equation 1.
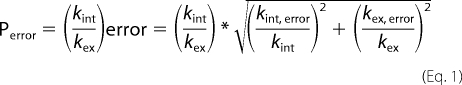 |
Local Stabilities
The hydrogen exchange process under native conditions is proposed to follow a two step model (57–59) in Equation 2,
where kcl and kop are the rate constants for structural closing and opening, respectively, and kint is the intrinsic rate constant at which the amide protons in the open form, or random coil state, exchange with deuterons. There is equilibrium between the open state and closed state for amide protons, while the exchange from the open form N-H to N-D is irreversible. Therefore, the measurable rate constant kex can be derived as follows in Equation 3.
When kcl ≫ kop and kcl ≫ kint (the EX2 exchange conditions), kex can be defined by a simpler Equation 4,
where Kop is the equilibrium constant for structural opening and can be related to the free energy for NH unfolding in Equation 5,
where R is the gas constant and T is the temperature. Therefore, assuming the EX2 exchange, ΔGHX can represent the local conformational stabilities for each residue and can be calculated from the values of the protection factor P derived from hydrogen exchange measurements.
RESULTS
Sequence Analysis of Collagenase Cleavage Sites in Collagen 5-Triplet Model of the Collagenase Cleavage Sites
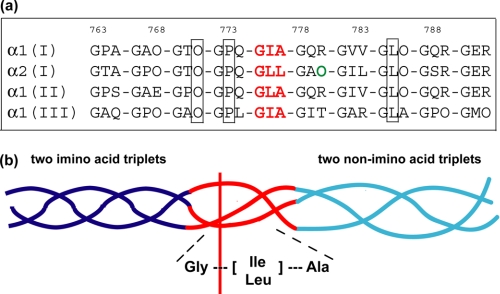
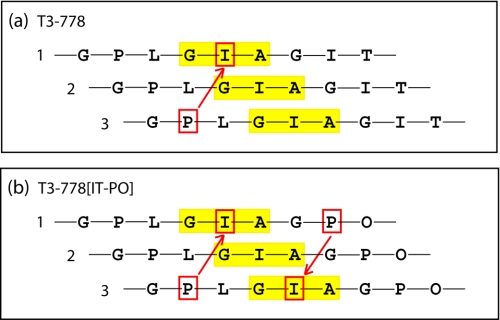
The amino acid sequences around the collagenase cleavage sites in human types I-III collagen were examined to evaluate if they share any unique features (Fig. 1a). Sequences of types I α(1) and α(2), type II and type III collagens were compared and show three conserved residues in addition to the Gly at the cleavage sites: one 4-hydroxyproline (Hyp) and one Pro in the two triplets immediately preceding or N-terminal to the cleavage site and one Leu in the third triplet following or C-terminal to the cleavage sites (Fig. 1a). The α1 chain of type I and the α1 chain of type II collagens share the most sequence similarity for the 6 residues surrounding the collagenase cleavage sites (GPQGIAGQR for type I α(1) and GPQGLAGQR for type II). The α1 chains of type I and type III collagens contain the same cleavage triplet GIA and the two triplets immediately following the cleavage sites do not have any imino acids. However, the α2 chain of type I collagen contains the cleavage triplet Gly∼Leu-Leu, which has a large residue Leu (rather than a small Ala) following the scissile bond as well as a Hyp in the following triplet.
FIGURE 1.
a, natural collagenase cleavage sites in human types I-III collagens. The triplet containing the cleavage scissile bond is colored in red. Conserved residues, including two imino acids in the two triplets immediately preceding the cleavage sites, are highlighted in the box. The two triplets immediately following the cleavage sites do not contain any imino acids, except one Hyp (colored in green) in the α2 chain of type I collagen. b, 5-triplet model of the collagenase cleavage sites in the α1 chain of types I-III collagen. The cleavage triplet GIA/GLA requires the presence of two imino acid triplets preceding the cleavage site as well as two non-imino acid triplets following the cleavage site. The cleavage triplet GLL in the α2 chain of type I collagen shows a different pattern.
The cleavage triplet Gly∼Ile-Ala in the α1 chains of type I and type III collagens is 100% conserved across different species spanning from rat to canis (supplemental Fig. S1). The cleavage triplet Gly∼Leu-Ala in type II collagen also has Leu 100% conserved. However, the cleavage triplet Gly∼Leu-Leu in the α2 chain of type I collagen has three occurrences of the scissile bond Gly∼Leu converted to Gly∼Ile or Gly∼Val and it has the conserved Hyp in the preceding two triplets substituted by Ala 4 out of 8 times (supplemental Fig. S1). The triplet Gly∼Leu-Leu occurs only once in human types I-III collagens, while Gly∼Ile-Ala and Gly∼Leu-Ala each occurs 9 times. Compared with the α1 chains, the α2 chain of type I collagen also has the least imino acid triplets (supplemental Fig. S2). The rarity and less conserved Gly∼Leu-Leu, the presence of Hyp following the cleavage triplet despite the poorer content of imino acids in the α2 chain, suggest that the cleavage triplet Gly∼Leu-Leu in the α2 chain of type I collagen may display a dissimilar pattern for collagenase recognition, compared with other cleavage triplets Gly∼Ile-Ala/Gly∼Leu-Ala in the α1 chains of type I and the chains of types II and III. A 5-triplet model of the collagenase cleavage sites could be built for the α1 chains of types I-III collagen (Fig. 1b). The cleavage triplet Gly∼Ile-Ala/Gly∼Leu-Ala requires the presence of two imino acid containing triplets N-terminal to the cleavage site as well as two non-imino acid containing triplets C-terminal to the cleavage site. Previous systematic sequence analysis led to a proposal that all the information necessary for efficient collagenolysis is contained in a 25-residue stretch, which is distinguished by a tight triple helix with high imino acid content prior to the cleavage site and a loose triple helix with low imino acid content following the cleavage site (14, 20). Here we propose that 15 key residues may be enough to define collagen recognition by MMPs.
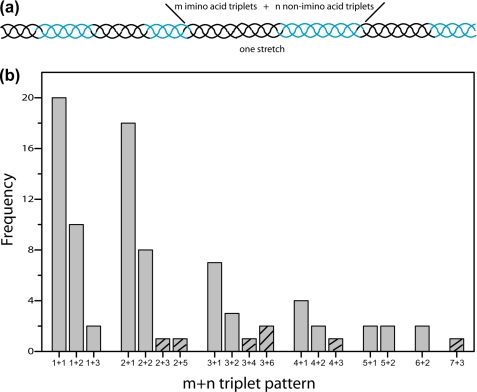
Distribution Analysis of Imino Acid Triplets Followed by Non-imino Acid Triplets
To test the hypothesis that the 5-triplet model is sufficient for hydrolysis, the amino acid sequences across all of human types I-III collagens are analyzed. The 5-triplet model highlights the importance of the juxtaposition of imino acids to non-imino acids in recognition by MMPs. The model can be described as a region containing two imino acid containing triplets followed by three non-imino acid containing triplets. The distribution of imino acid triplets and non-imino acid triplets in interstitial collagens was analyzed, to see if any other regions besides the natural cleavage region display the unique features of the 5-triplet model. The cumulative frequency plot of the imino acid containing triplets was first done, and the almost linear shape indicated that the imino acid triplets are pretty evenly distributed across collagen and there is no obvious region extremely rich or poor in imino acids (supplemental Fig. S2).
Further analysis of collagen sequences was done to identify whether there is any pattern in the distribution of imino acid triplets and non-imino acid triplets (Fig. 2). Collagen sequences were categorized into stretches of m imino acid triplets followed by n non-imino acid triplets (m+n triplet pattern). Type III collagen contains 87 such stretches (excluding 8 terminal triplets): 30 stretches contain only one imino acid triplet followed by one or two non-imino acid triplets while only 7 stretches contain at least 2 imino acid triplets followed by at least 3 non-imino acid triplets (Fig. 2). Similar results were obtained for other collagens: the α1 chains of types I and type II collagen only have 9 and 5 stretches containing at least 2 imino acid triplets followed by at least 3 non-imino acid triplets, including one stretch containing the natural cleavage site. The frequency analysis of the m+n triplet pattern indicated that imino acid distribution could limit the potential cleavage sites to only a few candidates. If charge is also considered, as no charged residue is required in the first non-imino acid triplet, only two stretches can be found in the α1 chains of types I-III collagens, while one of them is the natural cleavage site.
FIGURE 2.
Distribution of imino acid triplets followed by non-imino acid triplets in type III collagen. Collagen sequences are categorized into stretches of m imino acid triplets followed by n non-imino acid triplets (m+n triplet pattern) (a); Frequency of m+n triplet pattern in human type III collagen (b). Type III collagen contains 87 such stretches (excluding 8 terminal triplets), while only 7 stretches (striped bar) contain at least 2 imino acid triplets followed by at least 3 non-imino acid triplets and could be potential collagenase cleavage sites.
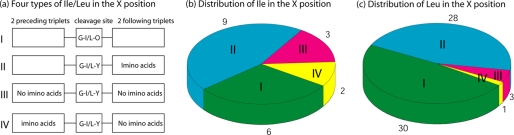
Environment around Ile/Leu in the X Position
There are 30 Ile and 66 Leu in the α1 chains of types I-III collagens. 67% of the Ile and 94% of the Leu are located in the X position, forming Gly∼Ile or Gly∼Leu bonds, which could potentially be cleaved by MMPs. The Ile/Leu in the X position are categorized according to the presence or absence of surrounding imino acids, to see if the Ile/Leu at the cleavage site is different from those at non-cleavage sites (Fig. 3). Four types of Ile/Leu in the X position could be identified: Ile/Leu immediately followed by a Hyp in the Y position (green); Ile/Leu followed by two triplets at least one of which contains imino acids (cyan); Ile/Leu preceded by two triplets at least one of which contains no imino acids (red); Ile/Leu with two preceding imino acid triplets and no imino acids in the Y position or in the two following triplets (yellow) (Fig. 3). 15 out of 20 Ile and 58 out of 62 Leu have a Hyp in the Y position or some imino acids in the two following triplets. Only one Ile in the α1 chains of type I and type III collagens and one Leu in type II collagen follow the last pattern (yellow) and they are the real collagenase cleavage sites. In other words, the 5-triplet model of GIA/GLA with two preceding imino acid triplets and two following non-imino acid triplets could 100% correctly predict the unique collagenase cleavage sites in all the α1 chains of types I-III collagens.
FIGURE 3.
Analysis of Ile/Leu in the X-position of the α1 chain of types I-III collagen. Ile/Leu in the X-position can be classified into four categories by the distribution of imino acids (a): Ile/Leu immediately followed by a Hyp (O) in the Y position (Type I, green); Ile/Leu followed by two triplets at least one of which contains imino acids (Type II, cyan); Ile/Leu preceded by two triplets at least one of which contains no imino acids (Type III, red); other Ile/Leu with two preceding imino acid triplets and no imino acids in the Y position or in the following two triplets (Type IV, yellow) (b, c). There is only one Ile in the α1 chain of types I and III collagens and one Leu in type II collagen following the pattern IV (yellow) and they are the real collagenase cleavage sites.
Peptide Design of Collagenase Cleavage Sites in Type III Collagen
Five peptides modeling the collagenase cleavage site and the neighboring region in type III collagen were assayed for biological activity and studied by NMR to determine the conformation and dynamics. In the collagenase cleavage site region in type III collagen (772–801: GPOGAOGPLGIAGITGARGLAGPOGMOGPR), only two triplets away from the natural cleavage triplet GIA, there is another potential cleavage triplet GLA that is not cleaved. Therefore, two peptides (T3–778 and T3–785) were designed to model the natural cleavage site and a potential but non-cleavable site (Table 1). Peptide T3–778 incorporates 4 triplets (GPLGIAGITGAR) from the cleavage site, while peptide T3–785 includes 4 triplets (GITGARGLAGPO) just following the cleavable site and containing a potentially cleavable GLA site. Peptide T3–785 has been studied previously by NMR, x-ray and computation (33, 52, 60). The two peptides share the same two triplets GITGAR and they have similarities in the other triplets (GPL versus GPO). However, the two peptides show significant differences in the position of the imino acid triplet relative to the cleavage triplet GIA/GLA: the imino acid triplet GPL is located preceding GIA while GPO is following GLA. For both peptides, GPO triplets were added at the two ends to enhance the formation of a triple helix.
As stated in the 5-triplet model, for the site to be cleaved in the α1 chains of types I-III collagens, it requires a cleavage triplet GIA/GLA with two preceding imino acid triplets and two following non-imino acid triplets (Fig. 1b). To investigate the effect of the preceding imino acid triplets, a peptide T3–778[P-A] was designed to substitute the conserved Pro at the P3 subsite with Ala (Table 1). To investigate the effect of the following non-imino acid triplets, a peptide T3–778[IT-PO] was designed with the Ile-Thr at the P4 ' and P5 ' subsites mutated to Pro-Hyp. The new sequence GPLGIAGPOGAR modeled by peptide T3–778[IT-PO] is found in type I collagen at a site just 20 residues away from the natural cleavage site and represents a potential cleavage site. To investigate if there is any difference between GIA and GLA, a peptide T3–778[I-L] with the Ile mutated to Leu was designed. The series of T3–778 peptides contain five selectively 15N-labeled residues: Gly16, Ile17, Gly22, Ala23, and Gly31 (Leu17 instead of Ile17 for peptide T3–778I-L). The peptide T3–785 contains five selectively 15N/13C doubly labeled residues: Gly15, Leu16, Ala17, Gly18, and Gly24. All the peptides have a residue Tyr added at the C-terminal for concentration determination (Table 1).
According to our 5-triplet model, incorporating 5 triplets in our model peptides may be a better peptide design for MMP recognition studies. However, triple helical peptides behave like large molecules and a longer peptide would decrease the NMR resolution significantly. Therefore, 4 triplets were included in our model peptides; the two non imino acid triplets following the cleavage site, the cleavage site and one imino acid rich triplet preceding the site. Although the second preceding imino acid-rich triplet was not included (GAO for T3–778), the N-terminal region of the peptide is imino acid-rich due to the flanking GPO sequence and may be considered to model this feature and represent a strong model for the collagenase cleavage site in type III collagen. The original sequence, GPOGAOGPLGIAGITGAR, has two imino acid-rich triplets, GPOGAO, N-terminal to the GPL triplet. The final (GPO)4 clamp at the N terminus of the peptide may compensate for the GAO triplet that is found immediately N-terminal to the GPL triplet, and the additional GPO triplet is directly modeled via the (GPO)4 clamps.
Hydrolysis Studies of the Model Collagenase Cleavage Peptides
The kinetics for peptide hydrolysis by MMP-1 were measured for the triple helical peptides described above (Table 1). The kcat/Km values for peptides T3–778 and T3–778[I-L] are the largest, indicating that both peptides modeling the natural cleavage sites are most active and GLA is not significantly different from GIA. Compared with T3–778, the kcat/Km values for peptides T3–778[IT-PO] and T3–778[P-A] were decreased by 30% and 90%, indicating that both substitutions, particularly the Pro-Ala mutation at the P3 subsite, leads to very decreased activities. Peptide T3–785, which models the potential but non-cleavable site, was much less active than peptide T3–778, as indicated by its kcat/Km, which is only one-sixth of the value of peptide T3–778.
It is interesting to compare the relative activities in the trimer state versus the monomer state to determine whether the hydrolysis arises from the sequence alone or from the triple helical features. Two monomer peptides M3–778 and M3–778[IT-PO], which model the same sequences as T3–778 and T3–778[IT-PO] but do not include the GPO triplets, were designed to investigate the activities of those sequences in the monomer state (Table 2). Kinetic analysis of MMP-1 activity showed that M3–778[IT-PO] was hydrolyzed with a far greater kcat/Km value than M3–778 (Table 2). In contrast, triple helical peptide T3–778 was more rapidly hydrolyzed by MMP-1 than T3–778[IT-PO] (Table 1), indicating that MMP-1 activity toward the single chains GPLGIAGITGA and GPLGIAGPOGA was reversed when the same sequences were in a triple helical context. These kinetics suggest that sequence is not the only critical component in modulating activity, but rather that triple helical conformation and dynamics are important for recognition.
TABLE 2.
Monomeric peptides incorporating the sequences at the collagenase cleavage sites and their activities
| Peptide | Sequence | kcat/Km |
|---|---|---|
| m−1s−1 | ||
| M3–778 | Mca-GPL-GIA-GIT-GA-Lys(Dnp) | 5,637.2 ± 973.1 |
| M3–778[IT-PO] | Mca-GPL-GIA-GPO-GA-Lys(Dnp) | 61,717.5 ± 4,372.5 |
NMR Studies on Peptides Modeling the Collagenase Cleavage Site NMR Chain-specific Assignments
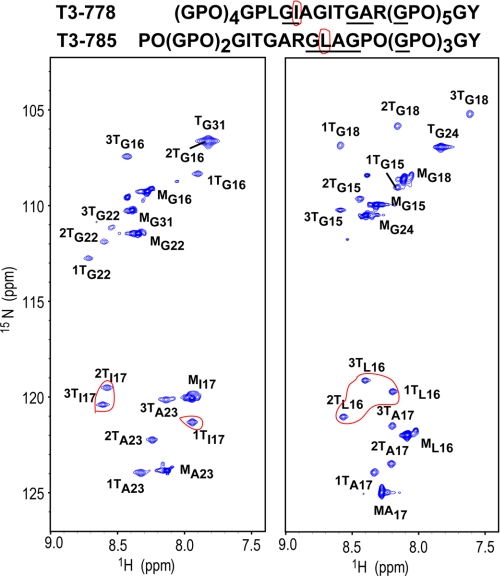
NMR 1H-15N HSQC spectra of the triple helical model peptides show that all labeled residues have trimer as well as monomer peaks, indicating that the peptides contain the typical triple helix conformation in equilibrium with the monomer conformation (Fig. 4, supplemental Fig. S3). Trimer resonances were assigned to specific chains from three-dimensional 15N-edited NOESY-HSQC experiments (46–48). For the active peptide T3–778, residues Gly16, Ile17, Gly22, and Ala23 have three well-dispersed trimer peaks as a result of the three non-equivalent chains, while residue Gly31 shows only a single resonance due to the repeating GPO environment. One unique feature of the peptide is that the Ile in the leading chain (1TI17) of the triple helix shows a distinct chemical shift from the Ile in the middle and lagging chains (Fig. 4). However, for the nearly inactive peptide T3–785, the chemical shift of the Leu in the leading chain (1TL16) is close to the Leu in the other two chains (Fig. 4).
FIGURE 4.
1H-15N HSQC spectra of active peptide T3–778 (left) and control peptide T3–785 (right). Peptide sequences are shown at the top with 15N/13C-labeled residues underlined. The red circle highlights the chemical shifts of residue Ile/Leu at the collagenase cleavage site. For active peptide T3–778, the chemical shift of Ile in chain 1 (1TI17) is different from Ile in chains 2 and 3. The peaks corresponding to the monomer and trimer state are denoted with a superscript M or T. The superscripted number 1, 2, and 3 corresponds to the leading, middle and lagging chains, respectively.
For the mutated T3–778 series (supplemental Fig. S3), peptide T3–778[I-L] shows almost the same spectrum as T3–778. Particularly, it shows a single Leu in the leading chain (1TL17) with a chemical shift distinct from the Leu in the other two chains. For peptide T3–778[IT-PO], the trimer resonances of Gly22 in all three chains and Ile17 in the middle and lagging chains (2TI17 and 3TI17) are significantly shifted upfield in the 15N dimension, whereas the chemical shifts of Gly16 in peptide T3–778[P-A] are mostly affected, when they are compared with original peptide T3–778 (supplemental Fig. S3). This indicates that the IT-PO and P-A mutations provide a different chemical environment for Gly16 or Ile17 at the collagenase cleavage site, which could play a significant role in the specificity of collagen recognition by an MMP.
Conformation of the Model Peptides
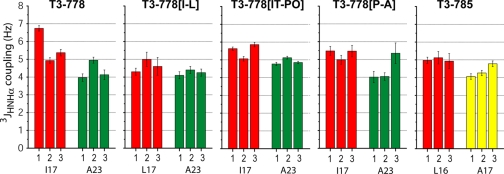
3JHNHα coupling constants that can be related to the dihedral angle ϕ were obtained for the trimers of the five peptides from HNHA experiments. Residues in the triple helical conformation typically contain phi angles from −55 to −75 degrees and have a corresponding J coupling value of 4–6 Hz (36, 39, 60). The J coupling values of all labeled Gly (Gly16, Gly22, and Gly31 for T3–778 series and Gly15, Gly18, and Gly24 for T3–785) were found to be between 4–6 Hz (data not shown). We have also measured the 3JHNHα coupling constants of residues Ile17/Leu17 in the X position at the cleavage site and of Ala23 also in the X position following the cleavage site in the T3–778 series of peptides as a control (Fig. 5). All of the Ile/Leu and Ala residues have J-coupling values that are relatively uniform between 4–6 Hz except for a single Ile in T3–778, 1TI17, which has a larger value of 6.74 (Fig. 5). As the residues in the triple helical conformation show small variations in J-coupling values, this higher value for the single Ile may be indicative of a distorted conformation. Ile17 in peptides T3–778[IT-PO] and T3–778[P-A] show a similar pattern to one another in J coupling values: 1TI17 and 3TI17 both have slightly larger values than 2TI17, suggesting that the IT-PO and P-A mutations have similar effects on the conformation of Ile at the cleavage site. In peptide T3–785, the Leu at the potential cleavage site have very uniform 3JHNHα coupling values of 5 Hz in three different chains as opposed to the active peptide T3–778 that has one unique Ile at the cleavage site.
FIGURE 5.
3JHNHα-coupling values of model peptides T3–778, T3–778[I-L], T3–778[IT-PO], T3–778[P-A], and T3–785. For the T3–778 series, Ile17/Leu17 and Ala23 are in the X position and colored in red and green, respectively. For T3–785, Leu16 in the X position is colored in red, and Ala17 in the Y position is colored in yellow.
Dynamics of the Model Peptides
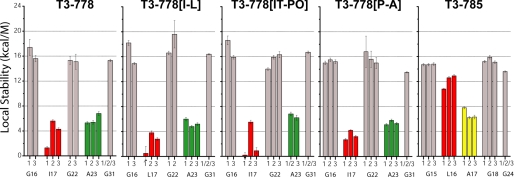
Hydrogen exchange experiments were performed on the five model peptides at all the labeled positions to explore the protection of amide protons from solvent (supplemental Table S1). Residue-specific local stabilities were calculated from the protection factors as described under “Experimental Procedures” (Fig. 6). It is manifest that the Gly residues have uniformly the highest local stabilities across different chains as well as different peptides. For the T3–778 series, all of the Gly residues including Gly16 at the cleavage site, Gly22 following the cleavage site and Gly31 in the GPO-repeating region have similar local stabilities on the order of 13.6 to 18.6 kcal/M, which are at least twice the values of the most stable Ile/Leu or Ala residues. The Gly residues in the control peptide T3–785, including Gly15, Gly18, and Gly24, behave the same as the Gly in the T3–778 series, indicating that local stabilities at the Gly residues between different peptides are comparable. These data suggest that all the Gly residues, located at, near or far from the cleavage site, are H-bonded and well protected from solvent.
FIGURE 6.
Local stabilities of model peptides T3–778, T3–778[I-L], T3–778[IT-PO], T3–778[P-A], and T3–785. Gly are colored in gray, Ile/Leu in the X position are colored in red, while Ala are colored green in the X position and yellow in the Y position.
Ala23, located in the X position but not cleaved, can act as a good control for Ile17/Leu17 at the cleavage site. Ala23 is just 6-residues away from Ile17 and has a Gly-Pro-Hyp triplet adjacent to it. The Ala residues show uniform local stabilities across all the peptides as well as across all three chains within each peptide. Their local stabilities of 5.7 ± 0.6 kcal/m are significantly reduced relative to Gly, indicating poorer protection from solvent. It should be noted that the IT-PO mutation in peptide T3–778[IT-PO], which is just beside the Gly22-Ala23 site, does not affect the local stabilities of the three chains of Ala23, due perhaps to its imino acid rich C-terminal neighbor, which may impose a rigid environment on Ala23.
It is quite distinct that Leu16 at the potential cleavage site in the control peptide T3–785 behaves totally differently from the Ile17/Leu17 involved in the real scissile bond in the T3–778 series peptides. In T3–785, Leu16 in all three chains have equally high local stabilities in the range of 10.8 to 12.9 kcal/m, indicating the possible existence of indirect H-bonding and good protection from solvent. However, for the T3–778 series, Ile17/Leu17 shows variable and much lower local stabilities, indicating the loss of H-bonds and poor protection from solvent. Particularly, in the active peptide T3–778, Ile17 in the leading chain (1TI17) has an even lower local stability than the middle and lagging chains, indicating that a single chain Ile17 is extremely unprotected, almost as unprotected as the Ile in the monomer state. A single, unique Leu17 in the leading chain (1TL17) can also be observed for a similarly active peptide T3–778[I-L], suggesting that a single extremely unprotected Ile/Leu is the key for MMP hydrolysis.
It is interesting to note that the local stabilities of Ile/Leu at the cleavage site in the three different chains are affected differently by the IT-PO and P-A mutations. For peptide T3–778[IT-PO], Ile17 in the leading and lagging chains (1TI17 and 3TI17) have equally low local stabilities, while Ile17 in the middle chain (2TI17) has a higher local stability. However, for peptide T3–778[P-A], the local stabilities of Ile17 in all three chains are similar and higher. It suggests that the local stabilities of Ile/Leu at the natural cleavage site are chain-specific and are modulated by neighboring Pro.
DISCUSSION
The uniform rod-like shape of the triple helix and the repeating Gly-Xaa-Yaa sequence in fibrillar collagens create a challenge for understanding its remarkable specificity and multiplicity of interactions (1–5). MMPs cleave collagen after the Gly residue of the sequence Gly∼[Ile or Leu]-[Ala or Leu] at a single, unique, position along the peptide chain and interestingly, there are numerous additional sites in the triple helical regions of collagens that contain the same consensus tripeptide sequence but are not hydrolyzed (14, 61). This raises the question of how one Gly∼Ile-Ala triplet is distinguished from another in the 1000 residue chain. To understand the specific recognition of collagen by MMPs, we have designed peptide models of both cleavage and potential cleavage sites and correlated their MMP activities with conformation and dynamics determined from NMR experiments. We have chosen to study type III collagen as this is a homotrimer that can be modeled by self associating peptides amenable to NMR studies.
Sequence analysis of collagen has allowed the identification of a number of residues crucial for recognition. Earlier, Fields proposed a model in which 25 residues in the cleavage site region (P12-P13′) dictate collagenase behavior (14, 20). Our sequence analysis of types I, II, and III collagen over different collagen species identified a smaller unit of five triplets (residues P6-P9′) for the α1 chains that is unique in terms of having a cleavage triplet GIA/GLA with two imino acid containing triplets preceding the cleavage site and two non-imino acid triplets following the site. Among ∼80 potential cleavage Gly-Ile/Leu sites in all of the α1 chains of types I-III collagens, this sequence pattern was found 100% of the time in the real collagenase cleavage sites and can be used as a predictor of the cleavage site. Sequence analysis also showed that the α2 chain in type I collagen is different from the α1 chains in types I-III collagen in terms of imino acid distribution as well as the conservation of the cleavage triplet GLL, suggesting that the α2 chain may have a different pattern of recognition compared with the α1 chains.
It has been suggested that sequence variations around the cleavage site may cause small variations in structure and flexibility of collagen, which can be recognized by MMPs (14, 20, 52, 60). The differences in activities of native collagen and denatured collagen indicates that collagenolytic activities must be related to the unique triple helix of collagen (17). The reverse order of single chain activity versus triple helix activity of the model peptides studied here suggest that conformation and dynamics are the critical components in modulating activity for triple helical peptide models of collagen. However, experimental data about conformation and dynamics at the collagenase cleavage sites is still limited. The x-ray structure of peptide T3–785 suggests that the imino acid rich region has a different helical symmetry pattern from the imino and poor region (60). NMR studies of an unlabeled heteromeric triple helical peptide containing the cleavage site of type I collagen indicate that the cleavage site region has less ordered structure than the ends with GPO repeating triplets (62). Recent simulations of type III collagen have shown that the whole cleavage site is relatively exposed to solvent, and it samples structures that are more complementary to the MMP active site (34).
Here, NMR studies of selectively labeled peptide models of the collagenase cleavage sites are directly correlated to biological activity to define the conformational and dynamic features that are key to MMP-1 cleavage. Thus, we can evaluate the role of conformation and dynamics at the cleavage site itself, the role of the neighboring residues in modulating the cleavage site, and the role of the local stabilities of individual chains of the triple helix. Two triple helical peptides designed to model the natural cleavage site and a nearby non-cleavable site in type III collagen show significant differences in activities as well as conformation and dynamics. The peptide that models the natural cleavage site (T3–778) is significantly more active than the control peptide (T3–785) and shows clearly that the Ile in the leading chain (1TI17) is more dynamic and less locally stable than the Ile in the other two chains. The unique Ile in the active peptide may have a looser conformation and may be more exposed to solvent, and therefore more susceptible to enzyme attack. A peptide with an Ile-Leu mutation that is also similarly active to peptide T3–778 supports the existence of a more flexible residue after the Gly residue and more local instability. It should be noted that Ala23, which is also located in the X position and just 6-residues away from Ile17 at the cleavage site, shows uniformly higher local stabilities across all three chains, highlighting the uniqueness and the local nature of the dynamics of Ile17 in the leading chain (1TI17).
Two mechanisms have been proposed for collagen to be cleaved by MMPs. The first mechanism proposed that collagen is intact and the binding of MMPs unwinds collagen (32). The second mechanism suggests that collagen is passively activated and it has some unwound state which can be recognized by MMPs (63). Studies of type I collagen indicated that an inactive MMP-1(E200A) mutant binds and unwinds the triple-helical collagen first, which can then be cleaved by the catalytic domain of MMP-1 (32). However, computational studies indicated that the cleavage site of type I collagen could have one chain more flexible and looped out (63). Our result is the first experimental evidence that even in a homotrimer, the Gly-Ile site of one chain may be looser than the Gly-Ile sites in the other two chains. The site in this chain may be more susceptible to MMP cleavage. This could be consistent with the model suggesting that the site is already available for hydrolysis versus a need for unwinding after binding. However, our findings may be only characteristic of type III collagen, which is more susceptible to enzyme attack than types I and II collagen and which may have more local instabilities.
The activities of collagen have been shown to be regulated by neighboring residues around the cleavage sites, particularly imino acids (64). Our sequence analysis also shows that imino acids are generally favored preceding Gly-Ile/Leu sites, while they are not favored following the collagenase cleavage site. However, it is poorly understood how the imino acid distribution affects the conformation and dynamics at the collagenase cleavage sites and thus collagenolytic activities in type III collagen. Our studies on the active peptides T3–778 and T3–778[I-L] and its two imino acid mutations T3–778[P-A] and T3–778[IT-PO] indicate a correlation between collagenolytic activity and local stabilities arising from modulation of neighboring residues. The less active peptides T3–778[IT-PO] and T3–778[P-A] with Pro substitutions either preceding or following the cleavage site have either two or three Ile sites that are similar to one another with respect to local dynamics. T3–785, a peptide with a potential cleavage site, is almost inactive and has three equally protected residues in the X position. These data show that the MMP-1 activities of the peptides depend on the unique dynamics of Ile in the leading chain (1TI17) at the cleavage site, and the neighboring Pro residues play important roles in modulating the local dynamics at the Ile site within the 5-triplet region identified from sequence analysis.
To understand how the imino acids surrounding the natural cleavage site affect the uniqueness of Ile, the staggering between the three chains needs to be considered (Fig. 7). The staggering makes the three chains non-equivalent. For the most active peptide (T3–778), only the Ile in the first chain (1TI17) is near the Pro at the P3 subsite of chain 3, while the other two Ile, 2TI7 and 3TI17 in chains 2 and 3, do not have any close contact with Pro. For the IT-PO substituted peptide (T3–778[IT-PO]), the new Pro at the P4 ' subsite is close to one additional Ile in the third chain, making the 3TI17 and 1TI17 have similar local Pro environments. For the P-A-substituted peptide (T3–778[P-A]), removing the Pro at the P3 subsite removes its effect on the Ile in the first chain, making the Ile in all three chains share similar non-proline environments.
FIGURE 7.
Effect of neighboring Pro on Ile at the cleavage site. a, Pro at the P3 subsite in the lagging chain affects the Ile in the leading chain (1TI17); b, Pro at the P4 ' subsite in the leading chain affects the Ile in the lagging chain (3TI17). Leading, middle, and lagging chains are indicated as number 1, 2, and 3, respectively. The cleavage triplet GIA is indicated with yellow background. Relationship of Pro in one chain and Ile in the other chain is indicated in red arrows.
Our NMR studies suggest, for the first time, the key role of the Pro immediately preceding the Gly-Ile/Leu scissile bond in modulating the local dynamics of Ile/Leu at the cleavage site in the context of an imino acid poor C-terminal region in type III collagen. That Pro is conserved for types I-III collagens across eight different species and it has been shown to be important for collagenolytic activity. Our studies on peptide models of type III collagen indicate that the Pro at the P3 subsite defines a locally dynamic Ile/Leu in the leading chain (1TI17), while substituting it to Ala makes the Ile residues similarly stable in all three chains and makes the peptide almost inactive. Correlation of peptide activity to NMR data suggests that a single locally unstable chain at the cleavage site in type III collagen, rather than all three labile chains, is more favorable for collagen cleavage by MMPs. Similarly, type I collagen heterotrimer has a conserved Pro at the P3 subsite suggesting that one chain may have more flexibility at the Ile/Leu site than the other two chains. These data support the studies that have shown that only a single chain of collagen can be incorporated into the active site cleft of MMPs and that type I collagen is cleaved by MMPs one chain after the other (32, 34).
Supplementary Material
This work was supported, in whole or in part, by grants from the National Institutes of Health (GM45302, to J. B., CA98799, to G. B. F., and DE14318 COSTAR Program, to J. L. L.), the National Science Foundation (DBI-0403062 and DBI-0320746, to J. B.), and the Robert A. Welch Foundation (to G. B. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- Hyp
- hydroxyproline (three-letter code)
- MMP
- matrix metalloproteinases
- HSQC
- heteronuclear single quantum coherence
- O
- hydroxyproline (single-letter code).
REFERENCES
- 1.Visse R., Nagase H. (2003) Circulation Res. 92, 827–839 [DOI] [PubMed] [Google Scholar]
- 2.Brinckerhoff C. E., Matrisian L. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 207–214 [DOI] [PubMed] [Google Scholar]
- 3.Cawston T., Billington C., Cleaver C., Elliott S., Hui W., Koshy P., Shingleton B., Rowan A. (1999) Ann. N.Y. Acad. Sci. 878, 120–129 [DOI] [PubMed] [Google Scholar]
- 4.Baragi V. M., Qiu L., Gunja-Smith Z., Woessner J. F., Jr., Lesch C. A., Guglietta A. (1997) Scand. J. Gastroenterol. 32, 419–426 [DOI] [PubMed] [Google Scholar]
- 5.Sternlicht M. D., Werb Z. (2001) Annu. Rev. Cell Dev. Biol. 17, 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woessner J. F. (1998) in Matrix Metalloproteinases (Parks W. C., Mecham R. P. eds) Academic Press, San Diego [Google Scholar]
- 7.Riley G. P., Harrall R. L., Watson P. G., Cawston T. E., Hazleman B. L. (1995) Eye 9, 703–718 [DOI] [PubMed] [Google Scholar]
- 8.Shoulders M. D., Raines R. T. (2009) Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Rest M., Aubert-Foucher E., Dublet B., Eichenberger D., Font B., Goldschmidt D. (1991) Biochem. Soc. Trans. 19, 820–824 [DOI] [PubMed] [Google Scholar]
- 10.Kielty C. M., Grant M. E. (2002) in Connective Tissue and Its Heritable Disorders, Molecular, Genetic, and Medical Aspects (Royes P. M., Steinmann B. U. eds) Wiley Liss, New York [Google Scholar]
- 11.Rich A., Crick F. H. (1961) J. Mol. Biol. 3, 483–506 [DOI] [PubMed] [Google Scholar]
- 12.Ramachandran G. N. (1967) in Treatise on Collagen (Ramachandran G. N. ed) Academic Press, New York [Google Scholar]
- 13.Aimes R. T., Quigley J. P. (1995) J. Biol. Chem. 270, 5872–5876 [DOI] [PubMed] [Google Scholar]
- 14.Fields G. B. (1991) J Theor. Biol. 153, 585–602 [DOI] [PubMed] [Google Scholar]
- 15.Fields G. B., Van Wart H. E., Birkedal-Hansen H. (1987) J. Biol. Chem. 262, 6221–6226 [PubMed] [Google Scholar]
- 16.Welgus H. G., Jeffrey J. J., Eisen A. Z. (1981) J. Biol. Chem. 256, 9511–9515 [PubMed] [Google Scholar]
- 17.Welgus H. G., Jeffrey J. J., Stricklin G. P., Eisen A. Z. (1982) J. Biol. Chem. 257, 11534–11539 [PubMed] [Google Scholar]
- 18.Netzel-Arnett S., Fields G. B., Birkedal-Hansen H., Van Wart H. E. (1991) J. Biol. Chem. 266, 6747–6755 [PubMed] [Google Scholar]
- 19.Mallya S. K., Mookhtiar K. A., Gao Y., Brew K., Dioszegi M., Birkedal-Hansen H., Van Wart H. E. (1990) Biochemistry 29, 10628–10634 [DOI] [PubMed] [Google Scholar]
- 20.Lauer-Fields J. L., Juska D., Fields G. B. (2002) Biopolymers 66, 19–32 [DOI] [PubMed] [Google Scholar]
- 21.Miller E. J., Harris E. D., Jr., Chung E., Finch J. E., Jr., McCroskery P. A., Butler W. T. (1976) Biochemistry 15, 787–792 [DOI] [PubMed] [Google Scholar]
- 22.Highberger J. H., Corbett C., Gross J. (1979) Biochem. Biophys. Res. Commun. 89, 202–208 [DOI] [PubMed] [Google Scholar]
- 23.Gross J., Highberger J. H., Johnson-Wint B., Biswas C. (1980) in Collagenase in Normal and Pathological Connective Tissues (Woolley D. E., Evanson J. M. eds) John Wiley & Sons, New York [Google Scholar]
- 24.Ryhänen L., Zaragoza E. J., Uitto J. (1983) Arch Biochem. Biophys. 223, 562–571 [DOI] [PubMed] [Google Scholar]
- 25.Miller E. J., Finch J. E., Jr., Chung E., Butler W. T., Robertson P. B. (1976) Arch. Biochem. Biophys. 173, 631–637 [DOI] [PubMed] [Google Scholar]
- 26.Hasty K. A., Wu H., Byrne M., Goldring M. B., Seyer J. M., Jaenisch R., Krane S. M., Mainardi C. L. (1993) Matrix 13, 181–186 [DOI] [PubMed] [Google Scholar]
- 27.Wu H., Byrne M. H., Stacey A., Goldring M. B., Birkhead J. R., Jaenisch R., Krane S. M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5888–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Brick P., O'Hare M. C., Skarzynski T., Lloyd L. F., Curry V. A., Clark I. M., Bigg H. F., Hazleman B. L., Cawston T. E. (1995) Structure 3, 541–549 [DOI] [PubMed] [Google Scholar]
- 29.Moy F. J., Chanda P. K., Cosmi S., Pisano M. R., Urbano C., Wilhelm J., Powers R. (1998) Biochemistry 37, 1495–1504 [DOI] [PubMed] [Google Scholar]
- 30.Bode W., Fernandez-Catalan C., Tschesche H., Grams F., Nagase H., Maskos K. (1999) Cell Mol Life Sci 55, 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bode W., Reinemer P., Huber R., Kleine T., Schnierer S., Tschesche H. (1994) The EMBO J. 13, 1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung L., Dinakarpandian D., Yoshida N., Lauer-Fields J. L., Fields G. B., Visse R., Nagase H. (2004) EMBO J. 23, 3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stultz C. M. (2002) J. Mol. Biol. 319, 997–1003 [DOI] [PubMed] [Google Scholar]
- 34.Salsas-Escat R., Stultz C. M. (2010) Proteins 78, 325–335 [DOI] [PubMed] [Google Scholar]
- 35.Xiao J., Baum J. (2009) J. Am. Chem. Soc. 131, 18194–18195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Brodsky B., Baum J. (2009) J. Biol. Chem. 284, 20660–20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Brodsky B., Baum J. (2007) J. Biol. Chem. 282, 22699–22706 [DOI] [PubMed] [Google Scholar]
- 38.Madhan B., Xiao J., Thiagarajan G., Baum J., Brodsky B. (2008) J. Am. Chem. Soc. 130, 13520–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiagarajan G., Li Y., Mohs A., Strafaci C., Popiel M., Baum J., Brodsky B. (2008) J. Mol. Biol. 376, 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ottl J., Battistuta R., Pieper M., Tschesche H., Bode W., Kühn K., Moroder L. (1996) FEBS Lett. 398, 31–36 [DOI] [PubMed] [Google Scholar]
- 41.Lauer-Fields J. L., Tuzinski K. A., Shimokawa K., Nagase H., Fields G. B. (2000) J. Biol. Chem. 275, 13282–13290 [DOI] [PubMed] [Google Scholar]
- 42.Lauer-Fields J. L., Fields G. B. (2002) Biol. Chem. 383, 1095–1105 [DOI] [PubMed] [Google Scholar]
- 43.Lauer-Fields J. L., Sritharan T., Stack M. S., Nagase H., Fields G. B. (2003) J. Biol. Chem. 278, 18140–18145 [DOI] [PubMed] [Google Scholar]
- 44.Minond D., Lauer-Fields J. L., Nagase H., Fields G. B. (2004) Biochemistry 43, 11474–11481 [DOI] [PubMed] [Google Scholar]
- 45.Kay L. E., Keifer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 46.Messerle B. A., Wider G., Otting G., Weber C., Wüthrich K. (1989) J. Magn. Reson. 85, 608–613 [Google Scholar]
- 47.Marion D., Kay L. E., Sparks S. W., Torchia D. A., Bax A. (1989) J. Am. Chem. Soc. 111, 1515–1517 [Google Scholar]
- 48.Fesik S. W., Zuiderweg E. R. (1988) J. Magn. Reson. 78, 588–593 [Google Scholar]
- 49.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 50.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 51.Palmer A. G., 3rd. (1993) Curr. Opin. Biotechnol. 4, 385–391 [DOI] [PubMed] [Google Scholar]
- 52.Fan P., Li M. H., Brodsky B., Baum J. (1993) Biochemistry 32, 13299–13309 [DOI] [PubMed] [Google Scholar]
- 53.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 54.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 55.Glasoe P. K., Long F. A. (1960) J. Phys. Chem. 64, 188–189 [Google Scholar]
- 56.Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huyghues-Despointes B. M., Pace C. N., Englander S. W., Scholtz J. M. (2001) Methods Mol. Biol. 168, 69–92 [DOI] [PubMed] [Google Scholar]
- 58.Englander S. W., Krishna M. M. G. (2001) Nat. Struct. Biol.. 8, 741–742 [DOI] [PubMed] [Google Scholar]
- 59.Hvidt A., Nielsen S. O. (1966) Adv. Protein Chem. 21, 287–386 [DOI] [PubMed] [Google Scholar]
- 60.Kramer R. Z., Bella J., Mayville P., Brodsky B., Berman H. M. (1999) Nat. Struct Biol 6, 454–457 [DOI] [PubMed] [Google Scholar]
- 61.Fields G. B., Van Wart H. E. (1992) Matrix Suppl. 1, 68–70 [PubMed] [Google Scholar]
- 62.Fiori S., Saccà B., Moroder L. (2002) J. Mol. Biol. 319, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 63.Nerenberg P. S., Salsas-Escat R., Stultz C. M. (2008) Proteins 70, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 64.Nagase H., Fields G. B. (1996) Biopolymers 40, 399–416 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.