Abstract
The photosynthetic bacterium Rhodobacter capsulatus contains two [NiFe]hydrogenases: an energy-generating hydrogenase, HupSL, and a regulatory hydrogenase, HupUV. The synthesis of HupSL is specifically activated by H2 through a signal transduction cascade comprising three proteins: the H2-sensing HupUV protein, the histidine kinase HupT, and the transcriptional regulator HupR. Whereas a phosphotransfer between HupT and HupR was previously demonstrated, interaction between HupUV and HupT was only hypothesized based on in vivo analyses of mutant phenotypes. To visualize the in vitro interaction between HupUV and HupT proteins, a six-His (His6)-HupU fusion protein and the HupV protein were coproduced by using a homologous expression system. The two proteins copurified as a His6-HupUHupV complex present in dimeric and tetrameric forms, both of which had H2 uptake activity. We demonstrated that HupT and HupUV interact and form stable complexes that could be separated on a native gel. Interaction was also monitored with surface plasmon resonance technology and was shown to be insensitive to salt concentration and pH changes, suggesting that the interactions involve hydrophobic residues. As expected, H2 affects the interaction between HupUV and HupT, leading to a weakening of the interaction, which is independent of the phosphate status of HupT. Several forms of HupT were tested for their ability to interact with HupUV and to complement hupT mutants. Strong interaction with HupUV was obtained with the isolated PAS domain of HupT and with inactive HupT mutated in the phosphorylable histidine residue, but only the wild-type HupT protein was able to restore normal H2 regulation.
The photosynthetic α-proteobacterium Rhodobacter capsulatus is endowed with an H2 uptake [NiFe]hydrogenase encoded by the hupSLC operon (7, 25). The hupS and hupL genes encode the small and large subunits of the enzyme, respectively, whereas hupC codes for an electron acceptor that is a membrane-integral cytochrome b. The activity of this hydrogenase allows the bacterium to grow under chemoautotrophic or photoautotrophic conditions. Its synthesis is activated under both aerobiosis and anaerobiosis by the presence of its substrate H2, either produced endogenously by nitrogenase (as a by-product) or added exogenously (12).
The regulatory cascade responding to H2 has been thoroughly studied in R. capsulatus (13, 16, 18, 47) and Ralstonia eutropha (3, 23, 26, 27), and comprises three proteins: HupUV, HupT, and HupR (HoxBC, HoxJ, and HoxA, respectively, in R. eutropha). In R. capsulatus, these proteins are encoded by four genes located on the chromosome in the locus hup. Whereas the hupT, hupU, and hupV genes form an operon (hupTUV), the hupR gene is located within the hyp genes, between hypB and hypC. We have shown, by Western blot analysis, that the intracellular level of HupR is stable under all the growth conditions tested (13). On the other hand, hupTUV transcription is regulated and is higher during heterotrophic growth than under autotrophic conditions, as assayed by phupT::lacZ fusions (16).
The first element identified in our laboratory was the HupR protein, a response regulator belonging to the family of the two-component regulatory systems, which presents the three domains typical of the NtrC subfamily (34). The hupR mutants were completely devoid of hydrogenase activity, even in the presence of H2, and could not grow under autotrophic conditions (34). Hence, HupR is an activator indispensable for hydrogenase gene expression. HupR directly controls hupSL transcription by binding to the promoter at a TTG-N5-CAA palindromic sequence centered at −152 nucleotides upstream from the transcriptional start site and is highly specific, as demonstrated by mutational analysis (13, 40). HupR belongs to a recently discovered class of enhancer-binding proteins, exemplified by the NtrC protein from R. capsulatus, that activate σ70 RNA polymerase instead of σ54 RNA polymerase (4).
Later, it was found that the product of the hupT gene is a histidine kinase able to autophosphorylate in presence of [γ-32P]ATP, and mutagenesis experiments showed the involvment of the hupT gene product in H2 regulation of HupSL synthesis (17, 18). Unexpectedly, the phenotypes of mutants defective either in the kinase gene or the gene of its cognate response regulator were opposite, and hupT mutants had high hydrogenase activities under all growth conditions tested, even in the absence of H2, due to a high level of hupSL expression, as assayed with phupS::lacZ fusions. This discrepancy was explained by the discovery that HupR is active under the unphosphorylated form, in contrast to what is generally found for the activators of two-component systems (13). Indeed, hupR mutants with the conserved phosphorylable aspartate residue either replaced by various amino acids or deleted exhibited high level of hydrogenase synthesis and activity. Thus, the histidine kinase HupT and the response regulator HupR form a two-component regulatory system, and phosphate transfer between these two partners was demonstrated in vitro (13). The same conclusion was also reached in the case of the HoxJ/HoxA system regulating the synthesis of the hydrogenases in R. eutropha (27).
The hupT gene is the first gene of a three-gene operon. The downstream hupU and hupV genes encode proteins homologous to the [NiFe]hydrogenases: HupU and HupV present 25 and 28% identity with the small and large subunits of HupSL hydrogenase of R. capsulatus, respectively (16). In particular, all of the residues able to bind the Fe-S clusters in HupS and the bimetallic center in the HupL active site of Desulfovibrio gigas [NiFe]hydrogenase (48) are conserved in HupUV. Recent data showed that this protein was able to catalyze the three typical reactions of the hydrogenases: namely H2 uptake and H2 evolution in presence of electron acceptor or donor, respectively, and hydrogen-deuterium exchange in their absence (46, 47). HupUV is a soluble protein and does not contain the N-terminal signal peptide, which addresses the hydrogenases to or across the membrane. It belongs to group 2 of the H2 uptake [NiFe]hydrogenases, which also includes the HupSL hydrogenases of cyanobacteria (44). These hydrogenases are soluble enzymes, and the sequences of their large subunit show several deletions compared to the sequences of the group 1 enzymes (44). However, unlike the energy-producing HupSL hydrogenase, the HupUV protein is not able to sustain cell growth. Analysis of hupUV mutants showed that the protein has a regulatory role and participates in the H2 regulatory pathway involving HupT. Indeed, hupUV mutants exhibit the same phenotype, although less strongly, as the hupT mutant (16). Because HupUV is able to bind H2, we have hypothesized that HupUV might be the direct H2 sensor of the H2 regulatory pathway, transferring the signal to the two-component HupT/HupR system, probably by interacting with the histidine kinase HupT. Formation of such complexes between the homologous proteins of R. eutropha (HoxBC/HoxJ) has recently been demonstrated (3).
In this paper, we describe the homologous overexpression and purification of the HupUV metalloprotein. Using the property of HupUV to oxidize H2, we have visualized its interaction with HupT by staining for hydrogenase activity on native gels and have shown that the interaction is affected by the presence of H2. We have also assessed the affinity of the two interacting proteins by surface plasmon resonance. Moreover, we have shown that HupUV interacts with the PAS domain of the histidine kinase HupT and that HupUV-HupT interaction in vivo is required for hupSL downregulation in the absence of H2. These results complement previous data showing that HupUV and HupT are involved together in the repression of hydrogenase expression in the absence of H2.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. R. capsulatus strains were grown heterotrophically at 30°C under anaerobiosis in the light or under aerobiosis in the dark. RCV medium (49) was supplemented with the following N and C sources: MG medium (7 mM glutamate, 30 mM dl-malate) and MN medium (7 mM ammonium sulfate, 30 mM dl-malate). Escherichia coli strains were grown at 37°C in Luria-Bertani medium. Antibiotics were added at concentrations of 100 (ampicillin), 25 (kanamycin), and 10 (tetracycline) mg/liter for E. coli and 10 (kanamycin) and 1 (tetracycline) mg/liter for R. capsulatus.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Gibco BRL |
| BL21(DE3)pLysS | F−ompT hsdSB (rB− mB−) dcm gal λ(DE3) pLysS(Cmr) | Novagen |
| R. capsulatus | ||
| B10 | Wild type | 28 |
| BSE8 | Hupc ΔhupT Kanr | 18 |
| JP91 | Hup−hupSL | 11 |
| RCM1 | Nif− ΔnifHDK Kanr | 50 |
| Plasmids | ||
| pPCR-Script SK(+) | Apr | Stratagene |
| pFRK-I | Apr Bler Gmr Kmr; fruP fusion vector | 15 |
| pET-15b | Apr; T7 promoter, expression vector | Novagen |
| pRK290 | Tcr; broad-host-range vector | 14 |
| pRK2013 | Kmr; mobilizes pRK290 | 14 |
| pPHU231 | Tcr; pRK290 with a 388-bp HaeII insert containing pUC18 polylinker | P. Hübner, unpublished |
| pPHU231ΔNcoI | Tcr; pPHU231 with NcoI site deleted | This work |
| pPHU236 | Tcr; broad-host-range lacZ fusion vector | 20 |
| pNF3 | Kmr Smr Spr; nif expression vector | 33 |
| pSE35 | Apr; pET-15b with 1.5-kb insert containing wild-type hupT | 17 |
| pSE70 | Apr; pET-15b with 1.5-kb insert containing mutated H217N hupT | 17 |
| pSE50 | Apr; pET-15b with 3.2 kb NdeI-SalI insert from pKES3 | 16 |
| pSE101 | Apr; pPCR-Script SK(+) with 0.33-kb PCR fragment (primers SE1-SE3, template pNF3) | This work |
| pSE102 | Tcr; pnif expression vector; pPHU231ΔNcoI with 0.35-kb HindIII-PstI insert from pSE101 | This work |
| pSE109 | Tcr; pSE102 with 3.3-kb NcoI-EcoRV insert from pSE50 | This work |
| pSE117 | Tcr; pSE102 with 4.6-kb BstEII-BamHI insert with hupUV-hypF genes | This work |
| pAC390 | Apr; pSE35 with hupT having a 0.3-kb BssHII-StuI deletion | This work |
| pAC394 | Tcr; pPHU231 with a 5.4-kb HindIII insert with pfru::hupT | This work |
| pAC395 | Tcr; pPHU231 with a 5.4-kb HindIII insert with pfru::hupTH217N | This work |
| pAC396 | Tcr; pPHU231 with a 5.1-kb HindIII insert with pfru::ΔPAShupT | This work |
| pAC397 | Apr; pSE35 with hupT having a 0.8-kb StuI deletion | This work |
DNA manipulation and bacterial mating.
DNA preparation and cloning were performed generally as described by Sambrook et al. (36). Restriction enzymes were used as indicated by the manufacturers. Triparental matings were made with the plasmid helper pRK2013 as described previously (9).
Construction of plasmids for homologous expression of hupUV in R. capsulatus.
A pRK290 derivative plasmid was constructed for expression in R. capsulatus genes under the control of the nitrogenase structural gene promoter, pnifHDK. A 330-bp fragment corresponding to the promoter was amplified by PCR with the pNF3 plasmid (33) as a template; the two oligonucleotides used as primers were SE1 (5′-AAGCTTCATCCCGCGCGATGAAG), which created the HindIII site (in boldface) and SE3 (5′-CCATGGGGTGGCTCCTTGGGGTT), which created the NcoI site (in boldface). The PCR product was cloned in the plasmid pPCR-Script SK(+). The resulting plasmid, pSE101, was cut with HindIII and PstI to obtain a 0.35-kb fragment, which was subsequently cloned into pPHU231ΔNcoI digested with the same enzyme, leading to pSE102, the pnifH-driven expression vector. (Plasmid pPHU231ΔNcoI is a derivative of pPHU231 in which the NcoI site has been eliminated after hydrolysis with the NcoI enzyme, treatment with Klenow enzyme, and ligation.)
To coproduce the six-His (His6)-HupU and HupV proteins, the his6-hupU-hupV (3.3-kb) fragment was excised from plasmid pSE50 cut with NcoI and EcoRV; it was then cloned into pSE102 cut with NcoI and ScaI, leading to plasmid pSE109. To coproduce HupUV with HypF, a 4.6-kb BstEII-BamHI fragment containing part of hupV and hypF was cloned in pSE103 in place of a 2.6-kb BstEII-BamHI fragment containing only part of hupV, giving the plasmid pSE117.
Purification of the His6-HupUV protein.
Because gene expression is driven by the promoter of the nifHDK genes on pSE109, the R. capsulatus strains were grown under conditions leading to high activity of the promoter—i.e., in the absence of both oxygen and NH4+. Therefore, cells were grown under anaerobic conditions in the light and in MG (malate-glutamate) medium in 1-liter flat flasks to ensure maximal illumination. When the optical density at 660 nm (OD660) reached 0.9 to 1, cells from 6 × 1 liter of cultures were harvested by centrifugation at 4°C, washed once in ice-cold 20 mM Tris-HCl (pH 8)-200 mM NaCl, and centrifuged again. The cells were resuspended in 75 ml of ice-cold IMAC-5 (20 mM Tris-HCl [pH 8], 0.5 mM NaCl, 10% glycerol, 5 mM imidazole) containing 1 mM phenylmethylsulfonyl fluoride, and disrupted by passage twice through a French press cell (Thermo Spectronic) at 15,000 lb/in2 (followed by 30 s of sonication). The cytosolic fraction was then isolated by centrifugation at 200,000 × g twice during 1 h at 4°C in a 60 Ti rotor (Beckman Corp.). After filtration through a 0.45-μm-pore-size filter, the supernatant fluid was loaded at a rate of 0.8 ml/min on a 5-ml HiTrap chelating HP column (Amersham Pharmacia Biotech) whose matrix (iminodiacetic acid) was previously charged with Ni2+. The column was washed with 30 ml of the equilibration buffer IMAC-5, 35 ml of IMAC-50, 20 ml of IMAC-100, and 20 ml of IMAC-200 (the numbers correspond to the millimolar imidazole concentration). The His6-HupUV complex was eluted with both IMAC-100 (15 ml, pool 1) and IMAC-200 (7.5 ml, pool 2). The two pools were dialyzed separately against IMAC-5 buffer overnight at 4°C and then loaded onto a 1-ml HiTrap chelating HP column charged in Ni2+. The column was washed with 4 ml of IMAC-5 and 4 ml of IMAC-50, and the proteins were eluted in 1 ml of IMAC-250. The proteins were dialyzed against a mixture of 20 mM Tris-HCl (pH 8), 150 mM NaCl, and 15% glycerol and then frozen as beads in liquid nitrogen and stored at −80°C. The purity of the HupUV preparation was assayed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and estimated at 90% for the pool 1 with a yield of about 0.3 mg of complex per liter of culture.
Construction of plasmids for production of mutant HupT proteins.
The plasmids pSE35 and pSE70, which overproduce HupT and H217N HupT, respectively, have already been described (17). The plasmid pAC390 was obtained as follows. The hupT gene was cut from pSE35 by XbaI-BamHI restriction and cloned in pUC18 restricted by the same enzymes. The sequence corresponding to the PAS domain was deleted by digestion with BssHII-StuI. The deleted plasmid was recovered from an agarose gel, treated with Klenow fragment, and ligated, thus conserving the reading frame. For the plasmid pAC397, the plasmid pSE35 was digested with StuI to delete a 0.8-kb fragment and religated. The plasmids pAC397 and pAC390 contained the 5′ and 3′ ends of hupT, respectively, which were cloned into the expression plasmid pET-15b in order to overproduce the PAS domain of the HupT protein and the PAS-deleted HupT protein.
Purification of HupT proteins.
Wild-type and mutated HupT proteins were produced with an N-terminal His6 tag and were thus purified on a HiTrap chelating column (Amersham Pharmacia Biotech). Cultures of BL21(DE3)pLysS harboring pSE35 (wild-type HupT), pSE70 (H217N HupT), pAC390 (PAS-deleted HupT), and pAC397 (PAS domain), induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and harvested at an OD600 of ca 0.9 to 1.0, were treated as described previously (17), except that cells were broken twice in a French pressure cell instead of being sonicated.
Complementation of the hupT mutant with mutated hupT genes.
The genes coding for HupT, H217N HupT, and PAS-deleted HupT were excised from the plasmids pSE35, pSE70, and pAC390 by NdeI-EcoRI digestion. The plasmid pFRK-I, containing a fructose-activated promoter, pfru, from R. capsulatus, had a 1.7-kb NdeI-EcoRI fragment deleted (deletion of the Bler Kmr cartridge) that was replaced by the three fragments described above. Then, HindIII fragments containing each of the three genes cloned downstream from the fru promoter were cut and cloned onto the broad-host-range plasmid pPHU231, opened at the HindIII site. The resulting plasmids (pAC394, pAC395, and pAC396) were introduced into the wild-type R. capsulatus B10 strain or the hupT mutant BSE8 by conjugation, and the transformed strains were grown under various conditions with and without 3 mM fructose. Hydrogenase activities were assayed in cells harvested at an OD660 of ca 1.8 under anaerobiosis and ca 1.0 under aerobiosis.
Western immunoblotting.
Proteins were separated on SDS-acrylamide gels (24), and the gels were transfered on nitrocellulose membranes (Protran BA83; Schleicher & Schuell) in Laemmli buffer containing 20% ethanol. The membranes were blocked for at least 1 h in the presence of 5% nonfat milk in phosphate-buffered saline (PBS) buffer containing 0.1% Tween 20 (T-PBS) and then washed three times with T-PBS and incubated for 1 h with monoclonal antipolyhistidine-horseradish peroxidase conjugate (Sigma) (1/3,000 dilution) in T-PBS. After three washes, the proteins were then revealed with the ECL enhanced chemiluminescence reagent (Amersham Biosciences).
Surface plasmon resonance experiments.
Interactions between the complex His6-HupUV and the HupT protein were analyzed in real time at 25°C with a BIAcore instrument. Purified HupT was diluted to 60 μg/ml in 10 mM sodium acetate (pH 5) and coupled to the carboxymethylated dextran surface of a sensor chip, CM5 (Biacore AB, Uppsala, Sweden), by using an N-hydroxysuccinimide-1-ethyl-3-(3-dimethylaminopropyl) carbodiimide coupling mixture. The binding was directly visualized by the resonance unit (RU) increase on the sensorgram; the amount of immobilized HupT protein was about 4,000 RU. The chip was then deactivated by ethanolamine to avoid further binding. The running buffer for protein interaction contained 20 mM Tris-HCl (pH 8), 150 mM NaCl, and 5% glycerol and was filtered through a 0.2-μm-pore-size filter. It circulated at a flow rate of 5 μl/min in the BIAcore processing unit. Sensorgrams were analyzed by nonlinear least-square curve fitting, allowing calculation of the apparent dissociation constant, KD, from the ratio of the two kinetic constants (koff/kon), as described previously (2).
Hydrogenase activity.
Hydrogenase activity was assayed in 20 mM Tris-HCl buffer (pH 8), with either 0.15 mM methylene blue (MB) or 2 mM benzyl viologen (BV) as an artificial electron acceptor for whole cells or cell extracts, respectively (10). In native gels, hydrogenase activity was revealed by incubating the gels under H2 for 10 to 20 min in 20 mM Tris-HCl buffer (pH 8), containing 2 mM BV. The reaction was stabilized by adding 1 mM triphenyltetrazolium chloride (TTC). Specific activities were expressed in micromoles of MB or BV reduced per hour per milligram of protein.
Miscellaneous.
Protein concentrations were determined by using bicinchoninic acid reagent (Bio-Rad) with bovine serum albumin as a standard. For whole cells of R. capsulatus, proteins were estimated from the empirical relationship OD660/5 = 1 mg of protein/ml (31). Native acrylamide gels were made like SDS-polyacrylamide gels, except that SDS and a concentration gel were omitted, and they were run in 0.5× Laemmli buffer. Before loading, samples were supplemented with 10% glycerol-0.005% bromophenol blue in 50 mM Tris-HCl buffer (pH 8). Apparent molecular masses of native proteins were determined by running in parallel several gels with different acrylamide concentrations as described by Bryan (5).
RESULTS
Homologous overexpression of the hupUV genes in R. capsulatus.
To study in vitro protein interactions, purified proteins were required. The histidine kinase HupT has previously been obtained by overexpression in E. coli of the hupT gene cloned in the expression vector pET-15b (17). However, all attempts to overexpress hupUV in the same way failed; the proteins obtained in E. coli were mainly in an insoluble form and inactive, as is usually observed with heterogeneously expressed hydrogenases. For this reason, we constructed a new plasmid, pSE109, able to express the hupUV genes in R. capsulatus, in order to provide the proteins required for the maturation of the regulatory hydrogenase. In this plasmid, the hupUV genes were expressed from the strong nifH promoter of R. capsulatus. The plasmid was introduced either into the wild-type B10, into the hupSL mutant JP91 devoid of HupSL synthesis, or into the ΔnifHDK mutant RCM1, since nitrogenase synthesis consumes a lot of energy. Similar yields of HupUV were obtained in the B10 and JP91 backgrounds, and the use of the ΔnifHDK mutant only slightly increased the production of HupUV proteins (data not shown). A plasmid was also constructed to coexpress the hypF gene with hupUV, since the HypF protein has been shown to be indispensable for the maturation of HupUV in R. capsulatus (8) or HoxBC in R. eutropha (6). Surprisingly, this strain did not produce more HupUV than SE109. Routinely, we chose to express hupUV genes in JP91 in order to avoid interference with the hydrogenase activity from HupSL.
The HupU protein was synthesized from plasmid pSE109 as a fusion protein with an N-terminal His6 tag, in order to purify the protein by affinity chromatography on an Ni2+-charged column. On the SDS-acrylamide gel presented Fig. 1, we observed that the His6-HupU protein copurified with HupV after one step of purification and that both proteins were present in an equimolar ratio, proving that HupU and HupV form a soluble complex in vivo. A typical purification is shown in Fig. 1. Most of the proteins were not retained on the HiTrap column or were eluted with 50 mM imidazole buffer. The HupUV protein eluted at 100 to 200 mM imidazole. From 6 liters of photosynthetic culture and after two successive affinity columns, we obtained about 1.5 mg of pure HupUV protein. This complex was active and exhibited a specific H2 uptake activity in the presence of BV of about 300 μmol of reduced BV/h/mg of protein. The purification factor was 72, and the yield was 24%. Attempts to remove the His6 tag by thrombin treatment were unsuccessful (data not shown), probably due to the inaccessibility of the cleavage site at the N terminus of HupU.
FIG. 1.
Purification of HupUV protein on HiTrap chelating column. Twenty microliters of each fraction (except for fraction 6, which was 10 μl) was loaded onto a 12% SDS-acrylamide gel. Lanes 1, soluble fraction; 2, nonretained fraction; 3, wash with 50 mM imidazole buffer; 4, pool of the fractions eluted at 100 mM imidazole; 5, pool of the fractions eluted at 200 mM imidazole; 6, pool of fraction 4 purified on a second HiTrap column and elution with 250 mM imidazole. Molecular masses of protein markers are indicated on the left.
On a native gel, the HupUV protein always appeared as two enzymatically active bands, with molecular masses of about 80 and 170 kDa, indicating that HupUV exists in a dimeric form (HupUV) and a tetrameric form (HupUV2) (Fig. 2). Both forms were in equilibrium, and the relative proportion varied depending on the protein concentration in the preparation. The tetramer was predominant when the protein was concentrated. When the two forms were separated by gel filtration on Sephadex G100, the protein was diluted, and the major peak represented the dimeric form (data not shown).
FIG. 2.
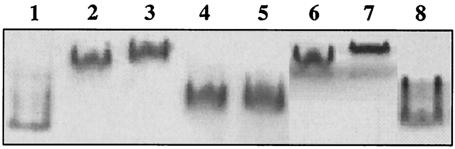
In vitro interaction between the HupUV and HupT proteins. The proteins (40 pmol of HupUV and 120 pmol of HupT) were incubated for 15 min at 30°C in a buffer containing 20 mM Tris-HCl (pH 8), 0.15 M NaCl, and 10% glycerol. The proteins were then run on a native 7.5% acrylamide gel in 0.5× Laemmli buffer, and the gel was revealed either by hydrogenase activity staining (A) or with anti-His6 tag antibodies (B). (C) Effect of H2 on the HupUV-HupT interaction. HupUV was first incubated with H2 during 15 min, and then HupT was added, and the incubation lasted for 15 additional min before the complexes were loaded on the gel.
Formation of HupUV/HupT complexes.
The interaction between HupUV and HupT was studied in vitro by first incubating the proteins together at 30°C and then loading them onto a native gel to separate and analyze the complexes. A typical interaction experiment is shown in Fig. 2. Two gels were run in parallel and revealed either with BV in the presence of H2 for hydrogenase activity (Fig. 2A) or with anti-His6 tag antibodies for the estimation of the amount of each form (Fig. 2B). Figure 2A showed that both forms of HupUV were enzymatically active, and the comparison of the two gels suggested that each form exhibited nearly the same hydrogenase specific activity. When HupT was incubated with HupUV, a new active band appeared with a higher molecular mass, representing a HupUV/HupT complex, and the amount of free HupUV decreased. The interaction between HupT and HupUV was fast and was usually complete within 5 min (data not shown). In the complex, HupUV remained active, as evidenced by the reduction of BV in the gel in presence of H2. Both the dimer and tetramer participated in the formation of the complex (the active band of the dimer disappeared totally); however, it is not yet known whether different complexes were formed with dimeric and tetrameric HupUV. The molecular mass of the complex was determined by comparison with marker proteins of high molecular mass and was about 300 kDa, suggesting the formation of a HupT2/HupUV2 complex. The HupUV-HupT interaction was not affected by the phosphate status of HupT, because the complex was the same either in the presence or absence of Mg-ATP (data not shown). Addition of H2 to HupUV before or during the incubation with HupT decreased the amount of formed complex or rendered the complex unstable (Fig. 2C). This effect of H2 was specific, and reduction of HupUV by dithionite did not change the HupUV-HupT interaction.
BIAcore experiment.
We further used BIA (Biomolecular Interaction Analysis) technology to monitor the HupT-HupUV interactions in real time. HupT was immobilized, via its native NH2 groups, on a carboxymethylated sensor chip. The binding of the HupUV protein to the surface-attached HupT during sample injection resulted in an increase in the signal corresponding to the association curve, shown on the sensorgram in Fig. 3. At the end of the injection, the sample was replaced by a continuous flow of buffer, and the decrease in signal reflected dissociation of HupUV from the surface-bound complex, giving the rate of dissociation of the complex on the surface (Fig. 3). We recorded sensorgrams at different HupUV concentrations, ranging from 1 to 10 μM. Kinetic rate constants for the binding and dissociation can be obtained by fitting the results to mathematical descriptions of interaction models. The apparent dissociation constant, KD, for HupT/HupUV was about 430 nM.
FIG. 3.
Analysis by surface plasmon resonance spectroscopy of HupT-HupUV interactions. HupT was immobilized on the sensor chip as described in Materials and Methods. Seventy-five microliters of HupUV (1 μM) was injected, and association was directly visualized on the sensorgram by the RU increase as a function of time. The dissociation phase was recorded when the sensor chip was flushed with buffer without added protein. The equations used to calculate the apparent dissociation constant, KD, by nonlinear least-square curve fitting are indicated.
To regenerate the chip (that is to increase the rate of dissociation of the complexes), we tried to change the pH of the buffer (pH 10.2 or 5.8), to increase the salt concentration (up to 500 mM NaCl), and to eliminate the glycerol; none of these changes had any significant effect on the dissociation rate. Finally, the regeneration of the chip was carried out by injection of 5 μl of 0.05% SDS.
When HupT was injected into the flowing buffer instead of HupUV, no association was observed between the immobilized and soluble proteins. Furthermore, no binding was observed when bovine serum albumin (2 or 4 μM) or HupR (1 or 2 μM) was tested (data not shown). In the opposite experiment (fixation of the complex HupUV on the sensor chip followed by the injection of HupT), no interaction was observed (data not shown). This lack of interaction could be explained by the random orientation of the protein during its covalent immobilization on the sensor chip, which might render its interacting domain less accessible to the partner.
Overproduction of mutated HupT proteins.
We next addressed the question of whether an enzymatically active and intact HupT protein is required for interaction with HupUV. For this purpose, we studied the interaction capabilities of three mutated HupT proteins in comparison with the wild-type protein. The histidine kinases, such as HupT, comprise several domains with specific roles. The N-terminal domain of HupT is characterized by the presence of a so-called PAS domain (39). These domains are generally involved in sensing various factors, such as light, O2, and redox changes, or in protein-protein interactions. To check whether the PAS domain of HupT is involved in the interaction with HupUV, a truncated protein (the PAS-deleted protein) and the isolated PAS domain were overproduced in E. coli from the strains AC390 and AC397, respectively. We also tested the H217N HupT protein, which is mutated in the conserved phosphorylable His217 residue, as previously described (17). Replacement of this residue leads to an inactive protein that is unable to autophosphorylate, and the mutated gene did not complement the hupT mutant (13).
The purified proteins were analyzed on native acrylamide gels. On these gels, mobility is affected by several factors, such as size and charge. Therefore, each protein was run on a series of gels of increasing acrylamide concentration, and the molecular masses were determined as described by Bryan (5). With wild-type HupT, two bands were observed, corresponding to the dimeric form (90 kDa) and the tetrameric form (180 kDa) (Fig. 4). The H217N HupT migrated exactly as the wild-type one, showing that dimerization of the mutant protein HupT was not affected. In native gels, PAS-deleted HupT appeared as two bands, larger than the wild-type HupT ones but still fitting well to the dimeric and tetrameric patterns. This is in agreement with the localization of dimerization determinants in the H region of the kinase transmitter domain (35). Furthermore, we found that the isolated PAS domain of HupT migrated at a molecular mass of about 70 kDa (Fig. 4) and thus was clearly in a dimeric form. Therefore, the PAS domain may contribute to the dimerization of the histidine kinase HupT, and the involvement of the N-terminal sensor domain in the dimerization of NtrB histidine kinase has recently been demonstrated (21, 29). The PAS domain might also increase the stability of the oligomeric form of the HupT protein, as already reported for dimeric proteins (39). In agreement with this is the observation that a lot of PAS-deleted protein remained in inclusion bodies, more than what was observed with the other three proteins.
FIG. 4.
Native acrylamide gel of the wild-type and deleted HupT proteins. Four micrograms of wild-type HupT, H217N HupT, PAS domain, and PAS-deleted HupT were run on a 7.5% native acrylamide gel and colored with Coomassie blue.
Requirement of the PAS domain for HupUV-HupT interaction.
To identify the domain of HupT required for complex formation with HupUV, the three mutated HupT proteins (H217N HupT, PAS-deleted HupT, and PAS domain), as well as the wild-type HupT, were incubated with HupUV. The complexes were then separated on native gels, and visualized by hydrogenase activity staining. As seen in Fig. 5, the H217N HupT (lanes 6 and 7) was able to interact with HupUV in the same way as wild-type HupT (lanes 2 and 3). The molecular masses of the complexes and the interactions with dimeric and tetrameric forms of HupUV were identical. Interestingly, the isolated PAS domain also interacted with the HupUV protein (Fig. 5, lanes 4 and 5). The complex was stable and migrated between the dimeric and tetrameric forms of HupUV (lane 1), suggesting an interaction of the PAS domain with a dimer of HupUV. On the other hand, no interaction was observed with the PAS-deleted HupT protein (Fig. 5, lane 8). This result demonstrated that interaction between HupUV and HupT is mediated by the PAS domain of HupT.
FIG. 5.
In vitro interaction between HupUV and various forms of the HupT protein. Conditions of incubation are the same as in Fig. 2. All samples contained 40 pmol of HupUV, and the additions were as follows. Lanes: 1, no further addition; 2, 60 pmol of wild-type HupT; 3, 180 pmol of wild-type HupT; 4, 80 pmol of PAS domain; 5, 160 pmol of PAS domain; 6, 60 pmol of H217N HupT; 7, 180 pmol of H217N HupT; and 8, 100 pmol of PAS-deleted HupT. After migration onto a 7.5% native acrylamide gel, the hydrogenase activity of HupUV was detected in the presence of H2 and 2 mM BV.
Complementation of the hupT mutant.
To determine whether the PAS-deleted HupT was able to restore normal regulation of hydrogenase synthesis in the hupT mutant BSE8, the mutated gene was expressed in R. capsulatus under control of the fructose-induced fru promoter. We used both wild-type hupT and hupTH217N genes as positive and negative controls, respectively. As expected (12), hydrogenase activities measured in absence of fructose in the wild-type strain B10 harboring the three plasmids were the highest when H2 was present, either in anaerobic photoheterotrophic cultures (MG medium, where nitrogenase evolves H2) or under aerobiosis after addition of exogenous H2 (Table 2); this results from the H2 stimulation of hydrogenase gene expression through the H2-responding cascade. Expression of the three hupT genes in the presence of 3 mM fructose did not significantly affect expression of hydrogenase in the wild-type strain B10, as shown in Table 2. Therefore, regulation of HupR activity in response to H2 availability was still effective, even when the mutated HupT proteins were expressed.
TABLE 2.
Hydrogenase activities of the wild-type B10 and hupT BSE8 strains from R. capsulatus, complemented with wild-type and mutant hupT genesa
| Growth condition | Fructose | Hydrogenase activity (μmol of reduced MB h−1 mg of protein−1) in strain:
|
|||||
|---|---|---|---|---|---|---|---|
| B10
|
BSE8
|
||||||
| pAC394 (wild-type HupT) | pAC395 (H217N HupT) | pAC396 (PASΔHupT) | pAC394 (wild-type HupT) | pAC395 (H217N HupT) | pAC396 (PASΔHupT) | ||
| Anaerobiosis | |||||||
| −H2 | − | 9 ± 1 | 15 ± 1 | 12 ± 1 | 26 ± 1 | 34 ± 2 | 54 ± 1 |
| + | 11 ± 2 | 11 ± 1 | 13 ± 2 | 7 ± 1 | 32 ± 1 | 48 ± 3 | |
| +H2 | − | 25 ± 1 | 28 ± 1 | 34 ± 4 | 29 ± 1 | 43 ± 1 | 40 ± 9 |
| + | 18 ± 1 | 18 ± 1 | 21 ± 2 | 25 ± 1 | 39 ± 4 | 27 ± 2 | |
| Aerobiosis | |||||||
| −H2 | − | 10 | 11 | NDb | 19 ± 1 | 66 ± 11 | 72 ± 4 |
| + | 5 | ND | ND | 7 ± 1 | 81 ± 5 | 61 ± 1 | |
| +H2 | − | 45 ± 5 | 36 ± 4 | 42 ± 5 | 58 ± 3 | 67 ± 6 | 65 ± 8 |
| + | 44 ± 2 | 36 ± 3 | 44 ± 9 | 50 ± 3 | 54 ± 9 | 60 ± 12 | |
Cells were grown overnight in 10-ml tubes under anaerobiosis under light (−H2, MN medium; +H2, MG medium) or in 20-ml flasks containing 3 ml of MN medium under aerobiosis (±10% H2) in the dark. Where indicated, fructose (3 mM) was added at the beginning of growth. Hydrogenase activity was assayed with methylene blue and was expressed as micromoles of reduced MB per hour per milligram of protein. The results represent the means of at least three independent measurements ± standard errors.
ND, not determined.
Inactivation of hupT leads to a derepression of hydrogenase synthesis in the absence of H2, and the levels of hydrogenase synthesis and activity are high under all growth conditions tested (17, 18). As shown in Table 2, in the hupT mutant BSE8, only the plasmid pAC394, which harbors the wild-type hupT gene, was able to restore normal regulation of hydrogenase synthesis. The complementation was observed when cells were grown in presence of fructose, which triggered the expression of the hupT gene. Partial restoration was also obtained in the absence of fructose, especially in aerobically grown cells, as previously noticed with hupUV mutants complemented with hupUV genes expressed from fru promoter (16). This was due to a significant expression of the protein even in absence of fructose, as observed by Western blotting (data not shown). Neither pAC395 (hupTH217N) nor pAC396 (PASΔhupT) could restore the repressing effect of HupT in the absence of H2 (Table 2). These in vivo experiments showed that (i) the PAS domain of HupT is necessary for transduction of the H2 signal, and (ii) in the absence of the PAS domain, the transmitter domain of HupT is unphosphorylated, since hydrogenase synthesis remained high in BSE8 harboring pAC396. Hence, this stressed the absolute requirement for a complete wild-type HupT protein for normal regulation.
DISCUSSION
H2 is the main activating signal for the expression of hydrogenases in several bacteria. The transduction pathway that responds specifically to H2 has been studied in Bradyrhizobium japonicum (41-43) and, in more detail, in R. eutropha (3, 23, 26, 27) and R. capsulatus (13, 16, 18, 40). This pathway comprises the three proteins HupUV, HupT, and HupR in R. capsulatus.
To study the in vitro interactions between these Hup proteins, we easily purified HupT and HupR after overproduction in E. coli. However, this method did not allow us to obtain active HupUV protein—probably because specific proteins are necessary to mature the metalloprotein. For this reason, we constructed an overexpression plasmid, based on the broad-host-range plasmid pPHU231, in which the hupUV genes were expressed from the nifH promoter of R. capsulatus. This promoter is very active, since in R. capsulatus grown under optimal conditions, the nitrogenase proteins could account for up to 20% of the total cell extract (22). To facilitate purification of HupUV, a His6 tag was inserted at the N terminus of HupU, and the presence of this tag did not impair hydrogenase activity of the protein, as previously reported (45). Thus, a one-step purification on an Ni2+-charged column was sufficient to obtain about 90% pure HupUV. Its H2 uptake activity was conveniently used to monitor the protein during the purification steps and to visualize it on native acrylamide gels. The HupUV specific activity for H2 uptake in presence of BV was 5 μmol/min/mg of protein compared to about 37 μmol/min/mg of protein for partially purified HupSL hydrogenase (38). On native acrylamide gel or during gel filtration, the HupUV protein appeared to occur in two forms corresponding to a dimer (HupUV) and a tetramer (HupUV2). Both forms seem to be equally active, since the amount of each form (as determined by immunoblots) corresponded to the level of activity (as determined in presence of BV). The homologous HoxBC protein from R. eutropha, purified after homologous expression from the strong promoter of the soluble hydrogenase operon, appeared also to occur as active dimeric and tetrameric forms (23), although the authors stated later that HoxBC is a tetramer (3).
As discussed above, we hypothesized that HupUV interacts with the histidine kinase HupT in order to regulate its phosphorylation state in response to H2 availability. And indeed, by incubating HupUV and HupT together, we did visualize the interaction directly on native acrylamide gels. The apparent molecular mass of the new band suggested that a HupT2/HupUV2 complex was formed, which was accompanied by a decrease of both forms of HupUV. However, our data did not allow us to determine which HupUV form interacted with HupT, since the binding of one form would displace the equilibrium between the two forms. Because HupT is largely dimeric, the protein can accommodate either two dimers or one tetramer of HupUV.
As observed with our H2 transduction cascade, in the NtrB/NtrC system, which regulates nitrogenase expression in response to intracellular nitrogen status, the signal is not sensed by the kinase NtrB but is sensed indirectly by the PII protein (reviewed in reference 1). It was recently shown, by cross-linking experiments and dissection of the NtrB kinase, that PII protein interacts with the kinase domain of the transmitter module of NtrB (21, 30, 32). The N-terminal sensor region contains a PAS domain that is thought to serve only to stabilize the conformation of the kinase (21). This is clearly different from what was observed for HupUV/HupT. Indeed, we found that the N-terminus PAS domain of HupT was absolutely required for the interaction with the regulatory hydrogenase. This was confirmed by the observation that the isolated PAS domain of HupT was able by itself to interact strongly with HupUV in vitro, leading to a complex with high H2 uptake activity in the presence of BV. This complex migrated on native gel at a position just below the tetrameric form of HupUV, so the dimeric PAS domain seems to accommodate only a dimer of HupUV. It is likely that the PAS domain of HupT interacts with HupU, the small subunit of the regulatory hydrogenase; however, attempts to purify the HupU subunit were unsuccessful due to the instability of the protein. The BIAcore experiment further indicated that the interaction might involve hydrophobic amino acid residues, because neither the presence of salts nor variation of the pH could affect the complex stability; only detergent could. Experiments are in progress to define the specific amino acid residues involved in the HupUV-HupT interface.
Based on hupT and hupUV mutant analysis, HupUV is thought to interact with HupT in the absence of H2 in order to favor its phosphorylation and thus inactivating the cognate response regulator HupR. Indeed, we showed earlier that in the absence of HupT, as well as in absence of HupUV, the level of hydrogenase synthesis was constitutively high, a consequence of HupR always being unphosphorylated (16, 18). Therefore, the autokinase activity of HupT depends on its interaction with HupUV, which only occurs in the absence of H2. On native gels, we observed that the H217N HupT interacted with HupUV exactly as the wild-type protein did, and there was no effect of the presence of Mg-ATP on HupUV/HupT complex formation and stability. Hence, the interaction of HupT with HupUV occurs independently of the phosphorylation status of the conserved histidine. In vivo, only the entire HupT protein was able to restore repression of hydrogenase synthesis in the absence of H2, presumably because the transduction pathway requires interaction between HupT and HupR as well as between HupUV and HupT.
As we expected, the presence of H2 affected the HupUV/HupT complexes, which were less stable and dissociated during electrophoretic migration (Fig. 2C). However we do not know how H2 is sensed, how it modulates this HupUV-HupT interaction, and consequently how the signal H2 is transduced from HupUV to HupT. Incubation of R. capsulatus HupUV with H2 had no effect on the stability or the dimer/tetramer equilibrium of HupUV that could affect the interaction. Therefore, since HupUV has the features of a [NiFe]hydrogenase, we could envisage that transduction involves electron transfer from HupUV to HupT. In the regulatory hydrogenase HoxBC of R. eutropha, no reduction of the three [4Fe-4S] clusters was observed in the presence of H2, and optical spectra revealed the presence of a redox-active chromophore that is reduced by H2 (3). Until now, we have found no indication for the presence of such a redox-active cofactor in R. capsulatus HupUV. It is possible that H2, by binding to the active Ni-Fe site of HupUV, produces a conformational change in the protein sufficient to decrease the interaction with the PAS domain of HupT; however, such conformational changes have not been observed between the oxidized and reduced forms of [NiFe]hydrogenases (19) that could be applied to HupUV. The question of the transduction mechanism is still open.
The transduction cascades responding to H2 have strong similarity in the photosynthetic bacterium R. capsulatus (13, 16, 18, 47) and the H2-oxidizing bacterium R. eutropha (3, 23, 26, 27). In particular, both bacteria possess an atypical two-component regulatory system (HupT/HupR or HoxJ/HoxA), in which the response regulator (HupR/HoxA) is active under the unphosphorylated form (13, 26). Also in both systems, the histidine kinase is able to interact directly with the H2 sensor, as demonstrated recently in R. eutropha (3). However, there are several interesting differences between the regulation patterns of R. capsulatus and R. eutropha. First, expression of hydrogenases in R. eutropha varies strongly according to the substrate (energy poor or energy rich): that is, hydrogenases are synthesized only when they are “needed” and in the presence of the substrate, H2 (37). In R. capsulatus, hydrogenase can be synthesized even when energy is abundantly produced (for example, during photoheterotrophic growth). In this case, the role of HupTUV proteins was to decrease hydrogenase synthesis, in the absence of its substrate, to a level acceptable for the cell. Concerning the transcription of hydrogenase structural genes, an RNA polymerase linked to a σ54 factor is involved in R. eutropha (37), whereas the housekeeping RNA polymerase is required for R. capsulatus hupSL transcription. This is in agreement with a central domain of HupR that is devoid of both ATPase activity and a σ54-interaction motif (13). Interestingly, the R. capsulatus NtrC protein regulating expression of nitrogenase also functions with a σ70 RNA polymerase (4). Several differences can be noted concerning the regulatory hydrogenases. Whereas both HupUV (47) and HoxBC (3) are able to produce H2 in the presence of D2 during H-D exchange reaction experiments, HD formation was detected only with HupUV, as previously demonstrated (45) and recently confirmed (A. L. De Lacey, personal communication). Therefore, there are differences at the molecular and mechanistic levels between these two proteins. It also seems that the oligomeric statuses of the two regulatory hydrogenases are quite different. As isolated, HupUV showed an equilibrium between the dimeric and tetrameric forms. To the contrary, HoxBC is mainly tetrameric. It should also be noted that isolated HoxBC is unstable under H2, whereas HupUV remains stable. A further difference concerns the histidine kinase. As discussed above, the “un-signal” status of the HupT protein appears to be the nonphosphorylated one, whereas in R. eutropha, HoxJ apparently remains phosphorylated in the absence of HoxBC, since synthesis of both hydrogenases is inhibited (26). Consequently different interactions between the H2-sensing proteins and the histidine kinases may occur in the two systems, and the respective mechanisms warrant further investigation.
Acknowledgments
We thank M. Satre and J. C. Willison for critical reading of the manuscript. We also thank A. L. De Lacey for the H-D exchange reaction experiment.
This work was supported by research grants from the Commissariat à l'Energie Atomique (CEA), the Centre National de la Recherche Scientifique (CNRS), and the Université Joseph Fourier (UJF) de Grenoble.
REFERENCES
- 1.Arcondéguy, T., R. Jack, and M. Merrick. 2001. PII signal transduction proteins: pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry, L., S. Mattei, B. Blot, R. Sadoul, M. Satre, and G. Klein. 2002. Biochemical characterization of two analogues of the apoptosis-linked gene 2 protein in Dictyostelium discoideum and interaction with a physiological partner in mammals, murine Alix. J. Biol. Chem. 277:21947-21954. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard, M., T. Buhrke, B. Bleijlevens, A. L. De Lacey, V. M. Fernandez, S. P. Albracht, and B. Friedrich. 2001. The H2 sensor of Ralstonia eutropha. Biochemical characteristics, spectroscopic properties, and its interaction with a histidine protein kinase. J. Biol. Chem. 276:15592-15597. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, W. C., and R. G. Kranz. 1998. A bacterial ATP-dependent, enhancer binding protein that activates the housekeeping RNA polymerase. Genes Dev. 12:1884-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan, J. K. 1977. Molecular weights of protein multimers from polyacrylamide gel electrophoresis. Anal. Biochem. 78:513-519. [DOI] [PubMed] [Google Scholar]
- 6.Buhrke, T., B. Bleijlevens, S. P. J. Albracht, and B. Friedrich. 2001. Involvement of hyp gene products in maturation of the H2-sensing [NiFe] hydrogenase of Ralstonia eutropha. J. Bacteriol. 183:7087-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cauvin, B., A. Colbeau, and P. M. Vignais. 1991. The hydrogenase structural operon in Rhodobacter capsulatus contains a third gene, hupM, necessary for the formation of a physiologically competent hydrogenase. Mol. Microbiol. 5:2519-2527. [DOI] [PubMed] [Google Scholar]
- 8.Colbeau, A., S. Elsen, M. Tomiyama, N. A. Zorin, B. Dimon, and P. M. Vignais. 1998. Rhodobacter capsulatus HypF is involved in regulation of hydrogenase synthesis through the HupUV proteins. Eur. J. Biochem. 251:65-71. [DOI] [PubMed] [Google Scholar]
- 9.Colbeau, A., A. Godfroy, and P. M. Vignais. 1986. Cloning of DNA fragments carrying hydrogenase genes of Rhodopseudomonas capsulata. Biochimie 68:147-155. [DOI] [PubMed] [Google Scholar]
- 10.Colbeau, A., B. C. Kelley, and P. M. Vignais. 1980. Hydrogenase activity in Rhodopseudomonas capsulata: relationship with nitrogenase activity. J. Bacteriol. 144:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colbeau, A., J. P. Magnin, B. Cauvin, T. Champion, and P. M. Vignais. 1990. Genetic and physical mapping of an hydrogenase gene cluster from Rhodobacter capsulatus. Mol. Gen. Genet. 220:393-399. [Google Scholar]
- 12.Colbeau, A., and P. M. Vignais. 1992. Use of hupS::lacZ gene fusion to study regulation of hydrogenase expression in Rhodobacter capsulatus: stimulation by H2. J. Bacteriol. 174:4258-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dischert, W., P. M. Vignais, and A. Colbeau. 1999. The synthesis of Rhodobacter capsulatus HupSL hydrogenase is regulated by the two-component HupT/HupR system. Mol. Microbiol. 34:995-1006. [DOI] [PubMed] [Google Scholar]
- 14.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duport, C., C. Meyer, I. Naud, and Y. Jouanneau. 1994. A new gene expression system based on a fructose-dependent promoter from Rhodobacter capsulatus. Gene 145:103-108. [DOI] [PubMed] [Google Scholar]
- 16.Elsen, S., A. Colbeau, J. Chabert, and P. M. Vignais. 1996. The hupTUV operon is involved in negative control of hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 178:5174-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsen, S., A. Colbeau, and P. M. Vignais. 1997. Purification and in vitro phosphorylation of HupT, a regulatory protein controlling hydrogenase gene expression in Rhodobacter capsulatus. J. Bacteriol. 179:968-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsen, S., P. Richaud, A. Colbeau, and P. M. Vignais. 1993. Sequence analysis and interposon mutagenesis of the hupT gene, which encodes a sensor protein involved in repression of hydrogenase synthesis in Rhodobacter capsulatus. J. Bacteriol. 175:7404-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi, Y., H. Ogata, K. Miki, N. Yasuoka, and T. Yagi. 1999. Removal of the bridging ligand atom at the Ni-Fe active site of [NiFe]hydrogenase upon reduction with H2, as revealed by X-ray structure analysis at 1.4 A resolution. Struct. Fold Design 7:549-556. [DOI] [PubMed] [Google Scholar]
- 20.Hübner, P., J. C. Willison, P. M. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, P., M. R. Atkinson, C. Srisawat, Q. Sun, and A. J. Ninfa. 2000. Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein. Biochemistry 39:13433-13449. [DOI] [PubMed] [Google Scholar]
- 22.Jouanneau, Y., B. Wong, and P. M. Vignais. 1985. Stimulation by light of nitrogen synthesis in cells of Rhodopseudomonas capsulata growing in N-limited continuous cultures. Biochim. Biophys. Acta 808:149-155. [Google Scholar]
- 23.Kleihues, L., O. Lenz, M. Bernhard, T. Buhrke, and B. Friedrich. 2000. The H2 sensor of Ralstonia eutropha is a member of the subclass of regulatory [NiFe] hydrogenases. J. Bacteriol. 182:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Leclerc, M., A. Colbeau, B. Cauvin, and P. M. Vignais. 1988. Cloning and sequencing of the genes encoding the large and the small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol. Gen. Genet. 214:97-107. [DOI] [PubMed] [Google Scholar]
- 26.Lenz, O., and B. Friedrich. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95:12474-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenz, O., A. Strack, A. Tran-Betcke, and B. Friedrich. 1997. A hydrogen-sensing system in transcriptional regulation of hydrogenase gene expression in Alcaligenes species. J. Bacteriol. 179:1655-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marrs, B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 71:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Argudo, I., J. Martin-Nieto, P. Salinas, R. Maldonado, M. Drummond, and A. Contreras. 2001. Two-hybrid analysis of domain interactions involving NtrB and NtrC two-component regulators. Mol. Microbiol. 40:169-178. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Argudo, I., P. Salinas, R. Maldonado, and A. Contreras. 2002. Domain interactions on the ntr signal transduction pathway: two-hybrid analysis of mutant and truncated derivatives of histidine kinase NtrB. J. Bacteriol. 184:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer, J., B. C. Kelley, and P. M. Vignais. 1978. Nitrogen fixation and hydrogen metabolism in photosynthetic bacteria. Biochimie 60:245-260. [DOI] [PubMed] [Google Scholar]
- 32.Pioszak, A. A., P. Jiang, and A. J. Ninfa. 2000. The Escherichia coli PII signal transduction protein regulates the activities of the two-component system transmitter protein NRII by direct interaction with the kinase domain of the transmitter module. Biochemistry 39:13450-13461. [DOI] [PubMed] [Google Scholar]
- 33.Pollock, D., C. E. Bauer, and P. A. Scolnik. 1988. Transcription of the Rhodobacter capsulatus nifHDK operon is modulated by the nitrogen source. Construction of plasmid expression vectors based on the nifHDK promoter. Gene 65:269-275. [DOI] [PubMed] [Google Scholar]
- 34.Richaud, P., A. Colbeau, B. Toussaint, and P. M. Vignais. 1991. Identification and sequence analysis of the hupR1 gene, which encodes a response regulator of the NtrC family required for hydrogenase expression in Rhodobacter capsulatus. J. Bacteriol. 173:5928-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626-633. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schwartz, E., U. Gerischer, and B. Friedrich. 1998. Transcriptional regulation of Alcaligenes eutrophus hydrogenase genes. J. Bacteriol. 180:3197-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seefeldt, L. C., L. C. McCollum, C. M. Doyle, and D. J. Arp. 1987. Immunological and molecular evidence for a membrane-bound dimeric hydrogenase in Rhodopseudomonas capsulatus. Biochim. Biophys. Acta 914:299-303. [Google Scholar]
- 39.Taylor, B. L., and I. B. Zhulin. 1999.PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toussaint, B., R. de Sury d'Aspremont, I. Delic-Attree, V. Berchet, S. Elsen, A. Colbeau, W. Dischert, Y. Lazzaroni, and P. M. Vignais. 1997. The Rhodobacter capsulatus hupSLC promoter: identification of cis-regulatory elements and of trans-activating factors involved in H2 activation of hupSLC transcription. Mol. Microbiol. 26:927-937. [DOI] [PubMed] [Google Scholar]
- 41.Van Soom, C., J. Browaeys, C. Verreth, and J. Vanderleyden. 1993. Nucleotide sequence analysis of four genes, hupC, hupD, hupF and hupG, downstream of the hydrogenase structural genes in Bradyrhizobium japonicum. J. Mol. Biol. 234:508-512. [DOI] [PubMed] [Google Scholar]
- 42.Van Soom, C., P. de Wilde, and J. Vanderleyden. 1997. HoxA is a transcriptional regulator for expression of the hup structural genes in free-living Bradyrhizobium japonicum. Mol. Microbiol. 23:967-977. [DOI] [PubMed] [Google Scholar]
- 43.Van Soom, C., I. Lerouge, J. Vanderleyden, T. Ruiz-Arguëso, and J. M. Palacios. 1999. Identification and characterization of hupT, a gene involved in negative regulation of hydrogen oxidation in Bradyrhizobium japonicum. J. Bacteriol. 181:5085-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 45.Vignais, P. M., L. Cournac, E. C. Hatchikian, S. Elsen, L. Serebryakova, N. Zorin, and B. Dimon. 2002. Continuous monitoring of the activation and activity of [NiFe]hydrogenases by membrane-inlet mass spectrometry. Int. J. Hydrogen Energy 27:1441-1448. [Google Scholar]
- 46.Vignais, P. M., B. Dimon, N. A. Zorin, A. Colbeau, and S. Elsen. 1997. HupUV proteins of Rhodobacter capsulatus can bind H2: evidence from the H-D exchange reaction. J. Bacteriol. 179:290-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vignais, P. M., B. Dimon, N. A. Zorin, M. Tomiyama, and A. Colbeau. 2000. Characterization of the hydrogen-deuterium exchange activities of the energy-transducing HupSL hydrogenase and H2-signaling HupUV hydrogenase in Rhodobacter capsulatus. J. Bacteriol. 182:5997-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 49.Weaver, P. F., J. D. Wall, and H. Gest. 1975. Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105:207-216. [DOI] [PubMed] [Google Scholar]
- 50.Willison, J. C., J. Pierrard, and P. Hübner. 1993. Sequence and transcript analysis of the nitrogenase structural gene operon (nifHDK) of Rhodobacter capsulatus: evidence for intramolecular processing of nifHDK mRNA. Gene 133:39-46. [DOI] [PubMed] [Google Scholar]