Abstract
Activation of the ionotropic P2RX7 nucleotide receptor by extracellular ATP has been implicated in modulating inflammatory disease progression. Continuous exposure of P2RX7 to ligand can result in apoptosis in many cell types, including monocytic cells, whereas transient activation of P2RX7 is linked to inflammatory mediator production and the promotion of cell growth. Given the rapid hydrolysis of ATP in the circulation and interstitial space, transient activation of P2RX7 appears critically important for its action, yet its effects on gene expression are unclear. The present study demonstrates that short-term stimulation of human and mouse monocytic cells as well as mouse osteoblasts with P2RX7 agonists substantially induces the expression of several activating protein-1 (AP-1) members, particularly FosB. The potent activation of FosB after P2RX7 stimulation is especially noteworthy considering that little is known concerning the role of FosB in immunological regulation. Interestingly, the magnitude of FosB activation induced by P2RX7 stimulation appears greater than that observed with other known inducers of FosB expression. In addition, we have identified a previously unrecognized role for FosB in osteoblasts with respect to nucleotide-induced expression of cyclooxygenase-2 (COX-2), which is the rate-limiting enzyme in prostaglandin biosynthesis from arachidonic acid and is critical for osteoblastic differentiation and immune behavior. The present studies are the first to link P2RX7 action to FosB/AP-1 regulation in multiple cell types, including a role in nucleotide-induced COX-2 expression, and support a role for FosB in the control of immune and osteogenic function by P2RX7.
Keywords: AP-1 Transcription Factor, ATP, Cyclooxygenase (COX) Pathway, Fos, Innate Immunity, Purinergic Agonists, Purinergic Receptor, Signal Transduction, Monocytic Cells, Osteoblasts
Introduction
The progression of many immune-based diseases involve the transcriptional up-regulation of proinflammatory genes (1–7). Consequently, certain therapies directed toward these pathologies have been designed to target transcription factor activation. One key regulator of proinflammatory gene expression is the dimeric transcription factor complex AP-15 (1), and recent studies suggest that AP-1 activation is critical for propagating inflammation during short-term lung injury (3), rheumatoid arthritis (4, 5), and asthma (6) and in osteoimmunologic processes (7). Depending on the composition of the AP-1 dimer, its activation can induce the expression of numerous genes, including various proinflammatory enzymes and mediators, such as tumor-necrosis factor-α (TNF-α), interleukin-1 (IL-1), interferons, and metalloproteinases (4). Currently, decoy oligonucleotides targeting AP-1 are being tested as potential anti-inflammatory agents (6), and it is, thus, important to determine which AP-1 subunits are activated in various immunopathologies.
One class of mediators known to be present at high concentrations in many inflammatory microenvironments is comprised of extracellular nucleotides (8–11), which function as ligands for nucleotide receptors on the surface of a wide array of cell types. Interestingly, recent studies have revealed a role for nucleotide receptors as immune sensors of the disruption of homeostasis (12, 13). In this context, depending on the concentration and duration of extracellular ATP (eATP) exposure, nucleotide receptors influence the magnitude of a mounted innate immune response. In particular, activation of the ionotropic P2RX7 nucleotide receptor by eATP has been implicated in modulating inflammatory disease progression (12, 14, 15).
Numerous reports have revealed a central role for P2RX7 in modulating lipopolysaccharide (LPS)-induced activation of monocytic cells (e.g. monocytes and macrophages), such as the processing and/or release of IL-1β, IL-6, IL-8, and IL-18, the expression of inducible nitric-oxide synthase, the production of nitric oxide, and the generation of reactive oxygen species (14, 16–19). Thus, P2RX7 activity affects a wide range of inflammatory events, especially with regard to monocytic cell function.
Evidence for an in vivo role of P2RX7 in the progression of inflammatory disease has been obtained using animal models of arthritis wherein P2RX7 knock-out mice exhibit a reduced incidence and severity of anti-collagen-induced arthritic symptoms when compared with their littermate controls (20). Also, analyses of the skeletal system of P2RX7-null mice has revealed that stimulation of periosteal bone growth by mechanical loading is markedly attenuated in these animals when compared with wild-type controls (21). Notably, mechanical stimulation has been linked to ATP release from osteoblasts, the bone-forming cells of the skeletal system (22, 23), and P2RX7 is expressed by both osteoblasts and osteoclasts (cells responsible for the resorption of mineralized bone) (24, 25), both of which are important in bone homeostasis and have an intimate relationship with the immune system (7). For instance, osteoblast stimulation has been linked to prostaglandin E2 (PGE2) release, which is important for osteoclast differentiation and bone resorption (7) as well as in the pathogenesis of rheumatoid arthritis (26). In this context, eATP-induced PGE2 release from osteoblasts appears P2RX7-dependent (21). Thus, there is strong evidence supporting a role for P2RX7 in osteoblast function, but the molecular mechanisms behind its activity remain largely unknown.
Upon prolonged ligand exposure, P2RX7 activation has been associated with the passage of small molecules (<900 Da) through the plasma membrane via the formation of nonspecific pores (27). As a consequence, continuous exposure (e.g. >30 min in human embryonic kidney (HEK)-293 cells heterologously expressing P2RX7 (28) or >20 min in lymphocyte populations expressing P2RX7 (29)) to a high concentration of ligand results in eventual apoptosis (30). In contrast, transient P2RX7 activation (e.g. <30 min in HEK-293 cells heterologously expressing P2RX7 (28) or <20 min in macrophages (31)) results in calcium influx, phosphatidylserine exposure, and changes in cell morphology, including membrane blebbing and microvesicle shedding, without inducing cell death (28, 32). Interestingly, a subset of cells expressing endogenous P2RX7, including microglia and osteoblasts, do not undergo apoptosis after prolonged agonist stimulation (33, 34). Because cell death has been a major focus in the study of P2RX7 action, the capacity of P2RX7 to mediate other responses, such as changes in gene transcription, are less well defined and yet are likely important for cells that do not undergo apoptosis after P2RX7 activation. Such issues may be key in disease progression, and it is noteworthy that studies of global gene expression in P2RX7-stimulated peripheral blood mononuclear cells reveal a difference in gene expression between pulmonary tuberculosis patients and control patients (35). In sum, P2RX7 stimulation likely mediates important alterations in gene expression, yet the molecular mechanisms by which these events occur are not well defined.
We have previously shown that P2RX7-dependent activation of the cAMP response element-binding protein (CREB) leads to the expression of the AP-1 family member, c-Fos, in cells heterologously expressing P2RX7 (36). This observation coupled with a study revealing that nucleotide treatment of cells heterologously expressing P2RX7 induces early growth factor-1 biosynthesis (37) supports a role for P2RX7 activation in the induction of protein expression that is not dependent on co-stimulation with other immune modulators such as LPS. However, the induction of protein expression after either short-term or continuous stimulation of endogenous P2RX7 is poorly understood. In this regard the present study demonstrates that P2RX7 activation leads to the expression of the AP-1 family members FosB, c-Fos, and JunB in both a murine macrophage cell line and in primary human peripheral blood monocytes. This report is the first study to implicate P2RX7 in the activation of FosB and JunB in any system. These observations are also novel in that induction of FosB and JunB expression only requires transient stimulation of monocytic cells with P2RX7 agonists, which would circumvent P2RX7-mediated apoptosis of these cells. The induction of FosB after P2RX7 stimulation is noteworthy given its large magnitude (10–20-fold over basal versus 2–4-fold induction of FosB by PMA) and high sensitivity (as low as 10 μm BzATP compared with ≥100 μm BzATP for most other reported P2RX7-mediated events) when compared with other known inducers of this transcription factor. Short-term stimulation of P2RX7 was observed to induce FosB and JunB binding to AP-1 consensus oligonucleotides after BzATP treatment, indicating that these proteins are transcriptionally active in monocytic cells. The activation of FosB was not restricted to monocytic cells, as treatment of an osteoblastic cell line (MC3T3-E1) with P2RX7 agonists was also able to elicit FosB expression. A role for P2RX7-induced FosB in the induction of gene expression was also revealed, as treatment of MC3T3-E1 cells with a P2RX7 agonist could induce the FosB-dependent expression of COX-2, the rate-limiting enzyme in the biosynthesis of prostaglandins such as PGE2, from arachidonic acid.
EXPERIMENTAL PROCEDURES
Materials
Unless otherwise specified, reagents for cell culture were purchased from Mediatech (Herndon, VA). All nucleotides were obtained from Sigma (St. Louis, MO), LPS (Escherichia coli, serotype 0111:B4), phorbol 12-myristate 13-acetate (PMA), and anisomycin were also purchased from Sigma. Ionomycin was obtained from Calbiochem (San Diego, CA). Antibodies used for immunoblotting β-tubulin, c-Fos, and Fos-B were purchased from Cell Signaling Technology (Danvers, MA); pan-reactive extracellular signal-regulated kinases 1/2 (ERK-1/2) was purchased from Upstate Biotechnology (Waltham, MA); anti-growth factor receptor-bound protein 2 (Grb2), anti-COX-2, anti-JunB, and anti-FosB (for electrophoretic mobility shift assay (EMSA)) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The active ERK1/2 kinase (MEK-1/2) inhibitor U0126 and an AP-1 consensus oligonucleotide probe were purchased from Promega (Madison, WI). The P2RX7 inhibitor A438079 was purchased from Tocris (Ellisville, MO). Dominant-negative CREB vectors were purchased from Clontech (Mountain View, CA), and the empty vector control plasmid was constructed by excising the CREB sequence from the S133A dominant-negative CREB plasmid.
Cell Culture
Murine RAW 264.7 macrophages were obtained from ATCC (Manassas, VA), whereas P2RX7-defective RAW cells were generated as previously described (36). Both RAW 264.7 cell lines were maintained in RPMI supplemented with 5% cosmic calf serum (Hyclone Logan, UT), 2 mm sodium pyruvate, 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin. HEK-293 cells were obtained from ATCC, whereas HEK-293 cells transfected with the pIREShyg (empty vector control) and pIRES/hP2RX7 expression vectors were generated as previously described (38). All HEK-293 cell lines were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 2 mm sodium pyruvate, and 100 units/ml penicillin/streptomycin. Murine MC3T3-E1 osteoblastic cells were cultured in α-minimal essential medium supplemented with 10% fetal bovine serum. All cells were cultured in 10-cm tissue culture dishes at 37 °C in a humidified atmosphere with 5% CO2.
Isolation of Human Blood-derived Monocytes
Heparinized blood was drawn from healthy adult volunteers at the University of Wisconsin Hospitals and Clinics (Madison, WI) in compliance with the requirements of the University of Wisconsin Health Sciences, Human Subjects Committee protocol. Blood-derived monocytes were purified from the heparinized blood as previously described (39) and were maintained in RPMI medium supplemented with 10% fetal bovine serum, 2 mm sodium pyruvate, 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin.
Cell Viability
RAW 264.7 macrophages were plated at 3 × 104 cells/well in 96-well plates and incubated at 37 °C for 16–24 h. After stimulation, cell viability was determined using the Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer's specifications. This proliferation assay assesses metabolic activity by measuring the conversion of a tetrazolium compound to a soluble formazan as catalyzed by dehydrogenase enzymes found in metabolically active cells. Soluble formazan can be quantified spectrophotometrically at 490 nm.
Quantitative PCR
To determine FosB mRNA expression, cells were treated as indicated in the figure legend and lysed in TRIzol (Invitrogen). RNA was extracted according to the manufacturer's protocols, and the levels of FosB mRNA were measured using quantitative RT-PCR. Primers directed toward murine FosB or 18 S (loading control) were designed using Beacon Design software. FosB primer, forward (AAGTGTGCTGTGGAGTTC) and reverse (ATGTTGGAAGTGGTCGA); 18 S primer, forward (GGACACGGACAGGATTGACAG) and reverse (ATCGCTCCACCAACTAAGAACG). Polymerase chain reactions were detected by SYBR green iQ supermix dye (Bio-Rad) and were performed using a My-iQ (Promega) real time thermal cycler (57–58 °C annealing temp, 40 cycles).
Immunoblotting
After isolation, human blood-derived monocytes were plated at 2 × 106 cells/well in 12-well CoStar (Corning Inc.) plates, washed after 2 h with HBSS, and incubated at 37 °C for 16–24 h. RAW 264.7 macrophages and HEK-293 cells were plated at a density of 3 × 105 - 5 × 105 cells/well in 24-well tissue culture plates and incubated at 37 °C for 16–24 h. For treatments that had short-term stimulation, cells were treated for 5 min, then washed twice with HBSS and incubated at 37 °C with fresh complete RAW media for the indicated times. After cell treatment and subsequent cell lysis with SDS sample buffer (20 mm Tris, pH 6.8, 2 mm EDTA, 1 mm Na3VO4, 2 mm DTT, 2% SDS, and 20% glycerol), the protein content of each sample was determined using the Micro-BCA protein assay (Pierce) according to the manufacturer's specifications. Immunoblotting was performed as previously described (36), and each primary antibody was used according to the manufacturer's protocols. The membranes were visualized using the Epichemi II darkroom (UVP, Upland, CA) equipped with a 12-bit cooled CCD camera. Image processing and analyses were performed using ImageJ 1.33u (NIH). To control for variations in protein loading, the membranes were re-probed with antibodies that react with Grb2, β-tubulin, or pan-ERK1/2. For immunoblot normalization, band density from the primary protein of interest was divided by the band density from its respective loading control.
EMSA
RAW 264.7 cells were plated at 1 × 106 cells/well in 6-well tissue culture plates and incubated for 18–24 h before the start of the experiment. After the cells were treated, nuclear extracts were prepared as described previously (40). Supernatants were collected, and protein concentrations were determined using the Bio-Rad protein assay according to manufacturer's specifications. Seven micrograms of nuclear protein extract per treatment were incubated with incubation buffer (40) at 4 °C for 15 min followed by an incubation of the extracts with and without 2 μg of antibody directed toward FosB, or JunB (for supershift) for 20 min at room temperature. Extracts were then incubated for 20 min with the AP-1 oligonucleotide probe labeled with 1 × 105 cpm of [γ-32P]ATP. The complexes were then separated on 4% non-denaturing polyacrylamide gels, and the migration of the labeled oligonucleotide was visualized by autoradiography.
Transfection of Dominant-negative CREB Constructs
HEK-293 cells heterologously expressing human P2RX7 (HEK/P2RX7) were plated at 1 × 105 cells/well on collagen-I (Sigma)-coated 24-well plates one day before transfection. The cells were then transiently transfected with either pCMV empty vector (designated as MT), pCMV-CREB133 (designated as S133A), or pCMV-KCREB (designated as KCREB) dominant-negative plasmids using FuGENE HD (Roche Applied Science, Indianapolis, IN) at an 8:2 transfection reagent:DNA ratio according to the manufacturer's specifications. Twenty-four hours post-transfection, the cells were treated, lysed in SDS sample buffer, and examined for FosB expression by immunoblotting.
FosB Knockdown
One day before transfection MC3T3-E1 cells were plated at 1.5 × 105 cells/well in 6-well plates. The cells were subsequently transiently transfected with a pRS vector containing HuSH-29 shRNAs (Origene, Rockville, MD) that was either scrambled (Scr (TR30012), 5′-GCACTACCAGAGCTAACTCAGATAGTAC T-3′) or targeted toward FosB (FosB_1 (TI351773), 5′-CTGTCTTCGGTGGCTCCTTCGGCAGTCC-3′, and FosB_2 (TI351774), 5′-ACACAGTGAAGTTCAAGTCCTCGGCGACC-3′) using TransIT-3T3 (Mirus, Madison, WI) according to the manufacturer's specifications. Seventy-two hours post-transfection the cells were treated, lysed in SDS sample buffer, and assayed for FosB and COX-2 expression via immunoblotting.
Statistical Analysis
Student's two-tailed paired t tests (Figs. 1, 2, 4 (A and B), 5, 6, and 9) or two-way analysis of variance using the Holm-Sidak method (Figs. 3, 4C, and 7) were used to calculate the statistical differences between samples. The threshold of significance was set at p values < 0.05.
FIGURE 1.
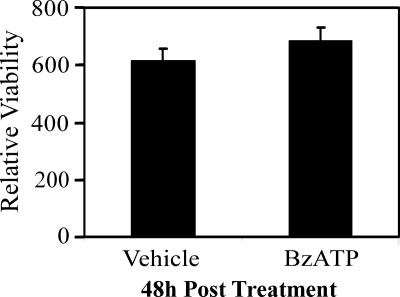
Effect of short-term P2RX7 stimulation on cell viability. RAW 264.7 macrophages were stimulated with either vehicle (2.5 mm HEPES) or 250 μm BzATP for 5 min. The cells were subsequently washed with HBSS and incubated at 37 °C with fresh complete media for 48 h. Cell viability was assessed spectrophotometrically by measuring the absorbance of formazan at 490 nm. The data are presented as the means ± S.D. of triplicate measurements from an experiment representative of three independent experiments. The difference in viability between vehicle- and BzATP-treated cells was not significant (p = 0.17).
FIGURE 2.
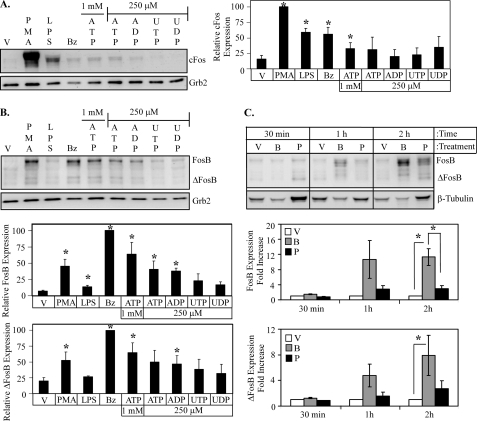
Extracellular nucleotides induce Fos expression in primary human monocytic cells. Isolated peripheral human blood monocytes were stimulated with either 2.5 mm HEPES (vehicle (V)), 1 μg/ml LPS, 1 μg/ml PMA, 250 μm BzATP (Bz), 1 mm ATP, or 250 μm ATP, ADP, UTP, or UDP for 5 min. The cells were subsequently washed with HBSS and incubated at 37 °C with fresh complete monocyte media for 2 h and immunoblotted for c-Fos (A) or FosB (B) expression. The results of at least three independent experiments were collated and represent relative c-Fos, FosB, and ΔFosB protein expression (mean ± S.E.). C, human peripheral blood monocytes were stimulated with either vehicle (V), 250 μm BzATP (B), or 1 μg/ml PMA (P) for 5 min., washed, then lysed after the indicated times and immunoblotted for FosB/ΔFosB expression. The results from four independent experiments were summarized and represent a -fold increase in FosB/ΔFosB protein expression (mean ± S.E.). *, p < 0.05.
FIGURE 4.
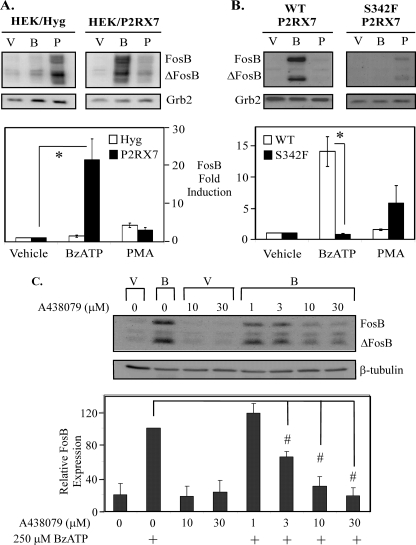
Nucleotide-induced FosB expression is dependent on P2RX7 function/expression. HEK-293 cells transfected with the pIREShyg or pIRES/hP2RX7 expression vectors (A) or RAW 264.7 macrophages endogenously expressing either wild-type or non-functional (S342F) P2RX7 (B) were stimulated with vehicle (V), 250 μm BzATP (B), or 1 μg/ml PMA (P) for 5 min, washed, then lysed after 2 h and immunoblotted for FosB expression. The results of three independent experiments were summarized and represent -fold increase of FosB protein expression (mean ± S.E.); *, p < 0.03. C, RAW 264.7 macrophages were pretreated for 30 min with either vehicle (water) or the P2RX7 inhibitor A438079 at concentrations from 1 to 30 μm. The cells were subsequently treated with either vehicle (2.5 mm HEPES) or 250 μm BzATP for 2 h. The results of three independent experiments were combined and are represented as a percent of BzATP-induced FosB protein expression (mean ± S.E.); #, p < 0.05 compared with vehicle-pretreated BzATP-induced FosB expression. For all panels in Fig. 4, stimulus-induced ΔFosB protein expression for all blots resulted in comparable -fold induction as FosB expression.
FIGURE 5.
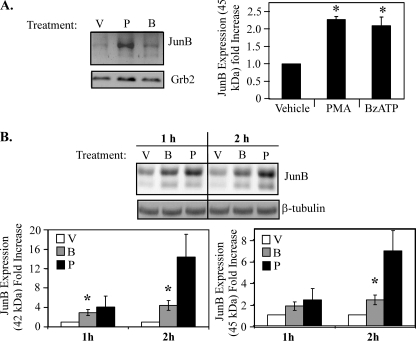
P2RX7 agonists induce JunB expression. Isolated peripheral human blood monocytes (A) or RAW 264.7 macrophages (B) were stimulated with either vehicle (V), 1 μg/ml PMA (P), or 250 μm BzATP (B) for 5 min, washed, lysed after the indicated times, and immunoblotted for JunB expression. Results of the three independent experiments were collated and -fold induction of JunB expression (45 kDa) was graphed ± S.E. *, p < 0.05. Similar results were seen for the 42-kDa isoform of JunB. Results of the three independent experiments were summarized as the mean ± S.E. and the -fold increase in JunB expression and was plotted. *, p < 0.03 compared with vehicle-treated cells.
FIGURE 6.
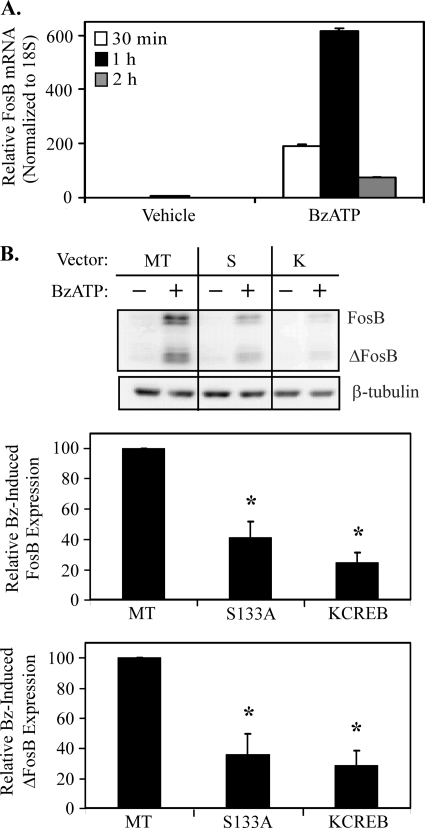
Attenuation of CREB activation leads to down-regulation of P2RX7 agonist-induced FosB protein expression. A, RAW 264.7 macrophages were treated with vehicle or 250 μm BzATP for the times indicated. Cells were lysed in TRIzol, and FosB and 18 S mRNA abundance was assessed via quantitative PCR. A representative graph from one of two identical experiments displaying the induction of FosB mRNA expression (normalized to 18 S) is shown. B, HEK-293 cells stably expressing P2RX7 were transiently transfected with pCMV empty vector (MT), pCMV-CREB133 (S), or pCMV-KCREB (K) CREB dominant-negative vectors. Twenty-four hours post-transfection, cells were treated with vehicle (2.5 mm HEPES) or 250 μm BzATP for 2 h and immunoblotted for total FosB protein expression. The results of five independent experiments were collated and are represented as a percent of BzATP-induced FosB/ΔFosB expression over basal FosB/ΔFosB expression (mean ± S.E.); *, p < 0.005 as compared with MT-transfected BzATP-treated FosB/ΔFosB expression.
FIGURE 9.
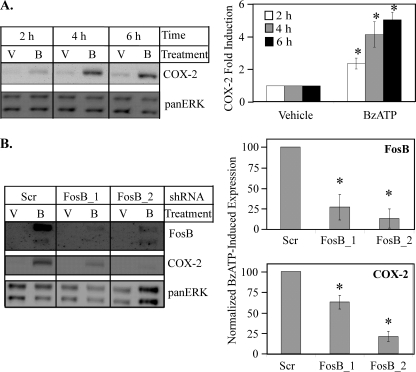
Attenuation of FosB expression leads to down-regulation of P2RX7 agonist-induced COX-2 protein expression. A, MC3T3-E1 cells were stimulated with either vehicle (V) or 250 μm BzATP (B) for the indicated times and immunoblotted for total COX-2 protein expression. Results of at least four independent experiments were collated, and -fold induction of COX-2 expression was graphed ± S.E. *, p < 0.03. B, MC3T3-E1 cells were transiently transfected with shRNA with a scrambled sequence (Scr) or two different sequences directed toward FosB (FosB_1 and FosB_2). Seventy-two hours post-transfection, cells were treated with vehicle (V) or 250 μm BzATP (B) for 4 h and immunoblotted for total FosB and COX-2 protein expression. The results of at least three independent experiments were collated and are represented as a percent of BzATP-induced FosB expression (top panel) or BzATP-induced COX-2 expression (bottom panel) (mean ± S.E.); *, p < 0.05 as compared with Scr-transfected BzATP-treated FosB or COX-2 expression.
FIGURE 3.
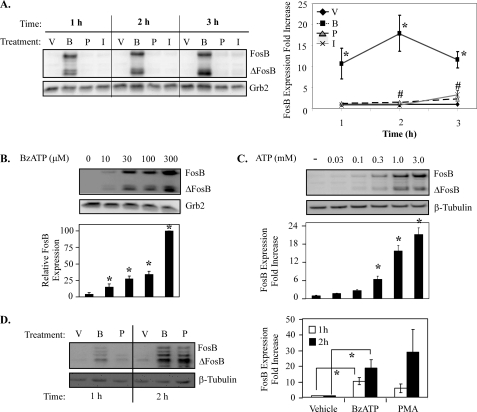
Extracellular nucleotides induce FosB induction in cell lines that express endogenous P2RX7. A, RAW 264. 7 macrophages were treated for 5 min with vehicle (V), 250 μm BzATP (B), 1 μg/ml PMA (P), or 1 μm ionomycin (I), washed, then lysed after the indicated times and immunoblotted for FosB/ΔFosB expression. The results of at least three independent experiments were combined and represent -fold increase in FosB protein expression (mean ± S.E.) *, p < 0.05 comparing BzATP- versus vehicle-induced FosB expression; #, p < 0.05 comparing PMA- versus vehicle-induced FosB expression. B and C, RAW 264.7 macrophages were treated with vehicle, BzATP at concentrations from 10–300 μm (B), or ATP at concentrations from 30–3000 μm for 5 min (C), washed, and incubated in complete media for 2 h, and lysates were blotted for FosB/ΔFosB expression. The results of at least three independent experiments were combined and represent relative FosB protein expression (mean ± S.E.). *, p < 0.05 compared with vehicle-induced FosB expression. D, MC3T3-E1 cells were treated with vehicle, BzATP, or PMA for the times indicated and immunoblotted for FosB expression. The results of at least three independent experiments were combined and represent relative FosB expression (mean ± S.E.). *, p < 0.03 compared with vehicle-induced FosB expression. For all panels in Fig. 3, stimulus-induced ΔFosB protein expression resulted in comparable -fold induction as FosB.
FIGURE 7.
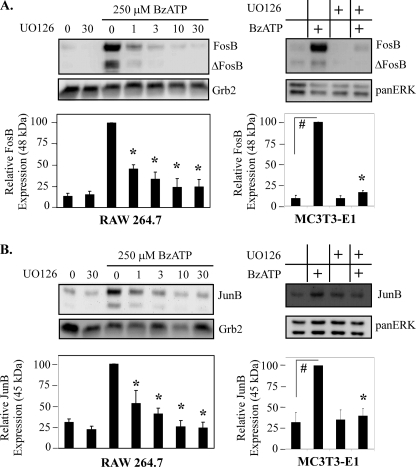
ERK1/2 kinase inhibitor attenuates P2RX7-induced AP-1 protein expression. RAW 264.7 macrophages or MC3T3-E1 osteoblastic cells were pretreated for 15 min with either vehicle (DMSO) or the MEK-1/2 inhibitor UO126 at the indicated μm concentrations (10 μm for MC3T3). The cells were subsequently treated with either vehicle (2.5 mm HEPES) or 250 μm BzATP for 2 h. A representative immunoblot displaying FosB (A) or JunB (B) protein expression is shown. The results of five independent experiments were combined and are represented as a percent of BzATP-induced AP-1 protein expression (mean ± S.E.). #, p < 0.003 Bz-induced FosB/JunB expression compared with vehicle control. *, p < 0.03 compared with vehicle pretreated BzATP-induced FosB/JunB expression.
RESULTS
Short-term Stimulation of P2RX7 Does Not Affect Macrophage Cell Viability
Although a subset of cells that express functional P2RX7 do not undergo apoptosis when continually stimulated with agonist (33, 34), it has been well recognized that prolonged stimulation of P2RX7 in most cell types, including monocytic cells, leads to cell death (30). However, more recent studies have provided evidence that cells can survive through a more physiologically relevant transient stimulation of this nucleotide receptor (28). In support of this model, we have observed that stimulation of RAW 264.7 macrophages with the P2RX7 agonist BzATP for 5 min followed by a media change permits the cells to remain viable for 48 h (Fig. 1) and 72 h (data not shown) after stimulation.
Extracellular Nucleotides Stimulate c-Fos and FosB Expression in Human Peripheral Blood Monocytes
We have recently observed that stimulation of P2RX7 leads to the transcriptional activation of CREB and the subsequent production of c-Fos in HEK-293 cells that exogenously express human P2RX7 (36). To evaluate if P2RX7 or other nucleotide receptors can induce the expression of c-Fos in primary cells, human peripheral blood monocytes were treated with various nucleotides and immunoblotted for c-Fos expression. As shown in Fig. 2A, treatment of peripheral blood monocytes with the P2RX7 agonists (i.e. millimolar levels of ATP and the pharmacological agonist BzATP) leads to the expression of c-Fos, although not to the extent of a high concentration of PMA, the prototypical AP-1 inducer. In contrast, stimulation of cells with other nucleotides that are more selective for the other nucleotide receptors did not lead to significant c-Fos protein expression. This observation supports our previous findings that nucleotide-induced c-Fos expression is mediated through P2RX7 (36).
CREB activation of AP-1 genes is not limited to c-Fos. Recent chromatin immunoprecipitation studies revealed that CREB is highly bound to CRE consensus site in the promoter of FosB, and RNAi-mediated knockdown of CREB down-regulates PMA-induced FosB gene expression (41). Furthermore, the expression of FosB mRNA has been observed to occur in a calcium-dependent manner (42). Considering that P2RX receptors induce a calcium influx upon activation, the induction of FosB expression by various nucleotides was also examined. Treatment of human peripheral blood monocytes with the P2RX7 agonists lead to the significant expression of FosB and its C-terminal truncated isoform, ΔFosB (Fig. 2B). Both FosB (45–48 kDa) and ΔFosB (33–37 kDa) produce multiple bands over time due to post-translational modifications (43), and these isoforms can create the banding pattern observed on the FosB immunoblot. Of the other nucleotides tested, only treatment of peripheral blood monocytes with ADP (the preferential agonist for certain P2RY nucleotide receptors) was also able to induce significant expression of FosB and ΔFosB. The P2RY agonists UDP and UTP did not significantly induce the expression of either FosB isoform. Interestingly, treatment of peripheral blood monocytes with LPS only produced a minimal amount of FosB but no detectable ΔFosB. Overall, the profile of nucleotide-induced FosB expression is consistent with P2RX7 activation.
To further investigate the induction of FosB expression in peripheral blood monocytes by P2RX7, a time course of BzATP- and PMA-induced FosB protein expression was performed. Short-term stimulation of P2RX7 leads to FosB expression as early as 1 h after stimulation and is significantly more robust than FosB/ΔFosB expression induced by a high concentration of PMA (1 μg/ml) 2 h post treatment (Fig. 2C).
Nucleotide-induced FosB Expression Is Dependent on P2RX7
Because of the strong induction of FosB by P2RX7 agonists in primary human blood monocytes and given the limited supply of these cells and their limited ability to be molecularly manipulated, further characterization of this process was performed in cell lines known to express endogenous P2RX7. Considering P2RX7 activation leads to a rapid increase in intracellular free Ca2+, the effect of ionomycin, a calcium ionophore, on the expression of FosB in macrophages was also tested. As shown in Fig. 3A, treatment with BzATP for 5 min induces FosB expression in murine RAW 264.7 macrophages in as early as 1 h. The induction of FosB protein after short-term BzATP stimulation of RAW 264.7 macrophages was significantly greater than that induced by either PMA or ionomycin treatment, suggesting that increased intracellular Ca2+ is not the only factor leading to the robust BzATP-induced FosB expression that we observed. The weak induction by PMA or ionomycin did not appear to be a consequence of the washing conditions given that continuous treatment of RAW 264.7 macrophages with PMA or ionomycin without washing was still not as potent as BzATP at inducing FosB expression (data not shown). Similar to primary monocytes, 30 min of treatment with any of the stimuli did not induce FosB expression in RAW 264.7 macrophages (data not shown).
Because a maximal dose of P2RX7 agonist (250 μm BzATP) results in robust FosB expression in monocytic cells, we next determined the dose-dependence (ligand sensitivity) of this P2RX7-mediated induction of FosB. We observed that as little as 10 μm BzATP produces detectable FosB and ΔFosB expression (p < 0.02), illustrating that this response exhibits a high degree of sensitivity to P2RX7 ligands (Fig. 3B). To determine whether the same sensitivity of FosB expression to P2RX7 agonist could be recapitulated with its physiological ligand (ATP), a dose response was also performed using increasing concentrations of ATP. Similar to the effects observed with BzATP, ATP induces FosB and ΔFosB expression (p < 0.05) with a dose of ATP as low as 300 μm (Fig. 3C), which is quite low compared with many other P2RX7-initiated responses.
The endogenous expression of P2RX7 is not limited to monocytic cells, e.g. osteoblasts are also known to express P2RX7 (24, 25), and the activation of AP-1 in these cells is important in bone homeostasis (7). Thus, to determine whether P2RX7 agonists could induce FosB expression in osteoblasts, the osteoblastic cell line MC3T3-E1 was examined. Intriguingly, prolonged P2RX7 activation does not lead to apoptosis in osteoblasts (21); therefore, FosB induction after continuous stimulation of MC3T3-E1 cells with 250 μm BzATP was tested. As illustrated in Fig. 3D, treatment of MC3T3-E1 with BzATP cells leads to the induction of FosB as early as 1 h, indicating that P2RX7 agonist-induced FosB expression is not exclusive to monocytic cells. It should be noted that short term agonist exposure of P2RX7 in MC3T3 cells also induces FosB expression (supplemental Fig. 1).
Although BzATP is a potent agonist of P2RX7, BzATP can also activate several other P2 receptors at high concentrations (e.g. P2RX1 and P2RY11) (44). Therefore, it was critical to evaluate whether the nucleotide-induced FosB/ΔFosB expression observed in the present studies is the result of P2RX7 activation rather than the result of activation of other P2 receptors. Accordingly, the effect of BzATP on HEK-293 cells heterologously expressing P2RX7 was examined. HEK-293 cells are a suitable model system for studying P2RX7 given that these cells do not endogenously express P2RX receptors (45). After short-term BzATP stimulation, only HEK-293 cells expressing P2RX7 displayed FosB/ΔFosB protein induction (Fig. 4A). Similar to the results from the peripheral blood monocytes and RAW 264.7 macrophages, BzATP stimulation induces FosB expression more potently than high concentrations of PMA. This effect was not attributable to delayed kinetics, as longer stimulation of HEK-293 cells with PMA (up to 24 h) did not lead to a stronger FosB activation (data not shown).
To further ascertain if FosB expression induced by BzATP was mediated through endogenous P2RX7, a non-functional P2RX7 cell line was utilized. Specifically, RAW 264.7 macrophages possessing an endogenous mutant P2RX7 gene containing a serine-to-phenylalanine mutation in the second transmembrane domain (S342F) were used as a non-functional P2RX7 model because this mutation confers attenuated P2RX7 protein expression and function (18, 36, 38) while retaining the ability to respond to other stimuli (e.g. anisomycin) to the same extent as the P2RX7 wild-type-expressing cells (36). Strong FosB/ΔFosB protein expression was detected 2 h after a 5 min treatment of wild-type murine RAW 264.7 macrophages with BzATP (Fig. 4B), whereas BzATP treatment of macrophages containing the S342F P2RX7 mutant gene did not induce detectable FosB/ΔFosB expression. Additionally, pretreatment of RAW 264.7 macrophages with a P2RX7 selective inhibitor, A438079, dose-dependently attenuated BzATP-induced FosB/ΔFosB expression, further supporting that nucleotide-induced FosB is P2RX7-dependent (Fig. 4C).
P2RX7 Agonists Induce JunB Expression
For Fos proteins to become transcriptionally active AP-1 complexes, they must heterodimerize with Jun family proteins (c-Jun, JunB, or JunD), form active AP-1 complexes, and bind to AP-1 consensus sites 12-O-tetradecanoylphorbol-13-acetate response elements (TREs) present in the promoters of various genes (2). Similar to FosB, CREB is bound under basal conditions to the promoter of JunB, and RNAi-mediated knockdown of CREB down-regulates PMA-induced JunB gene expression (41). Therefore, the effect of the P2RX7 agonist BzATP on JunB expression was examined. Treatment of human peripheral blood monocytes with BzATP for 5 min induced an increase in protein expression of both the 42- and 45-kDa JunB isoforms at 1 h post-treatment (Fig. 5A). Stimulation of RAW 264.7 macrophages with BzATP also leads to an increase in protein expression of both the 42- and 45-kDa JunB isoforms (Fig. 5B). These data support a role for P2RX7 activation in the up-regulation of JunB expression.
Attenuation of CREB Activation Leads to Down-regulation of P2RX7 Agonist-induced FosB Protein Expression
The presence of dominant-negative CREB vectors attenuates P2RX7 agonist-induced c-Fos expression (36), and PMA-induced FosB is CREB-dependent in HEK-293 cells (41); thus, it is plausible that CREB participates in the regulation of P2RX7-induced FosB expression. To support a role for transcriptional regulation of FosB protein expression after P2RX7 stimulation, the effect of a P2RX7 agonist on FosB mRNA induction was evaluated. As shown in Fig. 6A, stimulation of RAW 264.7 macrophages with BzATP induces detectable induction of FosB mRNA transcript as early as 30 min post-treatment and peaks 1 h post-treatment. To test whether BzATP-induced FosB expression was downstream of P2RX7-dependent CREB activation, two distinct dominant-negative CREB constructs were utilized. The pCMV-CREB133 (S133A) vector has a serine-to-alanine mutation at the phosphorylation site necessary for CREB transactivation, whereas the pCMV-KCREB (KCREB) vector has a mutation in the DNA binding domain of CREB that when overexpressed appears to form an inactive dimer with endogenous CREB. The two dominant-negative CREB vectors and a pCMV empty vector (designated as MT) were transiently transfected into HEK-293 cells stably expressing P2RX7. Twenty-four hours after transfection, cells were treated with vehicle or 250 μm BzATP for 2 h and immunoblotted for FosB protein expression. In the presence of CREB dominant-interfering mutants, BzATP-induced FosB/ΔFosB protein expression was significantly attenuated (Fig. 6B) (p < 0.01). These data support an important role for CREB activation in P2RX7-dependent FosB protein expression.
ERK1/2 Inhibition Attenuates P2RX7 Agonist-induced FosB/JunB Expression
Because CREB activation after P2RX7 stimulation is ERK1/2-dependent (36) and both FosB and JunB are shown to be induced in a CREB-dependent manner in response to a proinflammatory stimulus (41), we next sought to establish if ERK1/2 activation is upstream of P2RX7 agonist-induced FosB and JunB expression. To evaluate if P2RX7 agonist-induced ERK1/2 activation leads to FosB and JunB expression, the pharmacological inhibitor UO126 was utilized to inhibit MEK1/2-ERK1/2 signaling in RAW 264.7 macrophages and MC3T3-E1 osteoblasts. These inhibitor-pretreated cells were subsequently treated with 250 μm BzATP, and cell lysates were immunoblotted for FosB and JunB expression. The results summarized in Fig. 7 show that UO126 significantly inhibits both BzATP-induced FosB and BzATP-induced JunB expression (Fig. 7, A and B; #, p < 0.01 compared with vehicle pre-treated BzATP-induced FosB expression; *, p < 0.03 compared with vehicle pre-treated BzATP-induced JunB expression). The inhibition of MEK 1/2 in peripheral blood mononuclear cells also significantly attenuated BzATP-induced FosB expression (supplemental Fig. 2), supporting a role for ERK1/2 activation in the expression of these proteins.
AP-1 DNA Binding Activity Is Induced by a P2RX7 Agonist
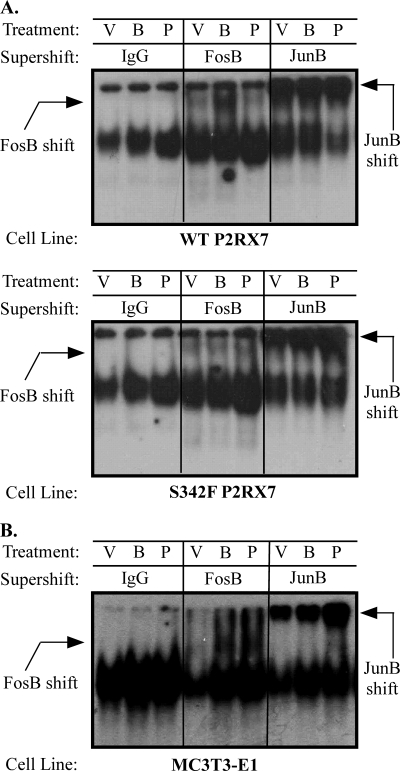
FosB and JunB are members of the AP-1 family of transcription factors that bind to TRE consensus sequences in the promoter regions of many genes that have been implicated in proinflammatory events. However, the role of ΔFosB in transcriptional activation is unclear. Several studies suggest that ΔFosB acts as a transcriptional repressor by binding to other AP-1 proteins and sequestering them from AP-1 elements (46), but other studies have shown that ΔFosB can induce gene expression (43, 47). Thus, to assess whether treatment of macrophages with a P2RX7 agonist results in enhanced AP-1 activity, EMSAs were performed using an oligonucleotide probe containing the consensus AP-1 binding site. As shown in Fig. 8, treatment of RAW 264.7 macrophages or MC3T3-E1 osteoblasts with BzATP or the AP-1 activator, PMA, enhanced protein binding to the AP-1 DNA consensus sequence. To evaluate whether FosB and/or JunB were components of the protein complex bound to the AP-1 DNA consensus sequence, supershift assays were performed. Both FosB and JunB antibodies induce a detectable supershift of the BzATP-treated nuclear lysates. In accordance with our FosB protein induction, PMA was only able to induce a noticeable FosB supershift in MC3T3-E1 nuclear lysates but can induce a detectable JunB shift in all cell types tested.
FIGURE 8.
P2RX7 stimulation induces AP-1 activation. RAW 264.7 macrophages endogenously expressing either wild-type or non-functional (S342F) P2RX7 (A) or MC3T3-E1 osteoblasts (B) were stimulated with either vehicle (V), 250 μm BzATP (B), or 1 μg/ml PMA (P) for the indicated times. Nuclear proteins were isolated and incubated in the presence of 2 μg of an antibody (Ab) against FosB or JunB proteins or an antibody isotype control. Nuclear extracts were then analyzed by EMSA with a radiolabeled AP-1 (TRE) DNA probe. Results are representative of at least three independent experiments.
FosB Knockdown Attenuates P2RX7 Agonist-induced COX-2 Expression
Monocytic cells are difficult to manipulate in terms of gene knockdown; thus, we focused on a cell system (MC3T3-E1 osteoblasts) that endogenously expresses P2RX7 but exhibits the capability of using molecular approaches such as shRNA to assess the role of FosB in mediating cellular responses to P2RX7 ligands. In this regard, P2RX7 is necessary for shear-stress-induced PGE2 production by MC3T3-E1 osteoblasts, and PGE2 synthesis is generally dependent on cyclooxygenase expression, the rate-limiting enzyme in prostaglandin biosynthesis from arachidonic acid. In the context of both macrophages and osteoblasts, COX-2 is the predominant enzyme responsible for prostaglandin production (48, 49), and stimulation of P2RX7 on primary human monocytes results in enhanced COX-2 expression (50). Because of the importance of COX-2 in the production of PGE2, we determined whether P2RX7 stimulation of osteoblasts induced COX-2 protein expression, which has not been previously determined. Murine MC3T3-E1 osteoblastic cells were treated with vehicle or 250 μm BzATP for 2–6 h and immunoblotted for COX-2 expression. As seen in Fig. 9A, BzATP significantly induces COX-2 expression in MC3T3-E1 cells as early as 2 h post-treatment.
To test whether BzATP-induced FosB expression is upstream of P2RX7 agonist-induced COX-2 expression, two distinct shRNA constructs toward FosB were utilized. MC3T3-E1 cells were transiently transfected with pRS vectors containing HuSH-29 shRNAs that were either scrambled (designated as Scr) or targeted toward FosB (designated as FosB_1 and FosB_2). Seventy-two hours after transfection, cells were treated with vehicle or 250 μm BzATP for 4 h and immunoblotted for FosB and COX-2 protein expression. As shown in Fig. 9B, the shRNA directed toward FosB was able to knock down BzATP-induced FosB and COX-2 protein expression (p < 0.05). These data support an important role for FosB activation in P2RX7-dependent COX-2 protein expression.
DISCUSSION
This study is the first to link the activation of a nucleotide receptor to FosB and JunB expression and reveals the induction of these proteins after a short-term stimulation of P2RX7. These findings have important physiological ramifications when considering that, given the rapid hydrolysis of ATP in vivo (51, 52) and, therefore, the transience with which stimulatory concentrations exist, short-term activation of P2RX7 can contribute to altered gene expression without inducing apoptosis.
Many of the studies examining the role of P2RX7 in modulating immune function have utilized a high micromolar concentration of the P2RX7 agonist, BzATP (100–250 μm) or millimolar concentration of ATP (1–5 mm). Here we show that as little as 10 μm BzATP or 300 μm ATP can induce a significant induction of FosB/ΔFosB expression in macrophages (Fig. 3, B and C), which highlights the sensitivity of the P2RX7 receptor regarding nucleotide-induced protein expression. This observation correlates well with previous reports indicating that fluid shear stress-induced activation of P2RX7 is important in osteoblast signaling (21), as fluid shear stress leads to a modest concentration of nucleotides in the microenvironment.
Earlier evidence for the general activation of AP-1 proteins by eATP has been reported in other cell types. Cotreatment of human fetal astrocytes with IL-1β and ATP was observed to induce greater AP-1 reporter gene activation than that from IL-1β alone, and treatment with P2 receptor antagonists inhibits cytokine-induced AP-1 activation (53). However, these effects are likely not a feature of P2RX7 activation because the potent P2RX7 agonist BzATP was not as effective as 100 μm ATP and 100 μm ADP in inducing AP-1 activation (53) and the P2 receptor antagonists used in these earlier studies do not act selectively on P2RX7. Administration of eATP (10–1000 μm) to human tonsilar B lymphocytes was found to increase c-Fos mRNA levels within 30 min (54). Furthermore, stimulation of Jurkat T cells with 3 mm ATP induces an increase in AP-1 DNA binding activity resulting from a modest increase in c-Jun and c-Fos expression (55). Nonetheless, these observations only indirectly suggest a role for P2RX7 in AP-1 activation through the use of a high concentration of eATP. Extracellular nucleotides have been linked to weak cFos mRNA expression in an osteoblastic cell line (56), but this induction was not significantly higher than serum treatment and was linked to P2RY activation. Our studies are the first to show direct evidence for P2RX7 stimulation in AP-1 activation by demonstrating a lack of FosB induction in macrophages expressing a non-functional P2RX7 or by macrophages treated with a P2RX7-selective inhibitor, as well as by demonstrating a gain of nucleotide-induced FosB expression in HEK-293 cells that heterologously express human P2RX7.
Although the most potent stimuli that we examined for FosB expression in human peripheral blood monocytes were P2RX7 agonists, ADP induces a significant level of FosB expression (Fig. 2B). These data support the previous finding that treatment of astrocytes with 100 μm ADP induces an increase in AP-1 activation (53). This activation may arise from the stimulation of P2Y receptors that utilize ADP as their preferential agonist, such as P2RY1, P2RY12, or P2RY13 (15). The possibility that other P2 receptor family members may also promote FosB expression is not surprising considering receptor-mediated Ca2+ influx has been shown to induce FosB expression (42). Further work is warranted to determine whether nucleotide-induced FosB expression can also be mediated by these P2Y receptors.
Our data also demonstrate that P2RX7 agonist-induced FosB and JunB expression is abrogated dose-dependently by the MEK1/2 antagonist UO126 (Fig. 7), suggesting that the MEK/ERK1/2 cascade is upstream of the expression of these AP-1 proteins in macrophages. In monocytic cells, a role for other mitogen-activated protein kinases in P2RX7-dependent AP-1 activation has been supported. Transactivation of c-Jun is augmented by the JNK1/2-dependent phosphorylation of serines 63 and 73, and studies from our laboratory and others have noted the activation of JNK1/2 after stimulation of monocytic cells with P2RX7 agonists (18, 40, 57). This finding correlates well with our unpublished observation that stimulation of macrophages with BzATP leads to an AP-1 supershift with an antibody directed toward c-Jun, supporting a role for c-Jun in P2RX7 agonist-induced AP-1 complex formation. Interestingly, P2RX7 agonist-induced JNK activation in RAW 264.7 is attenuated by N-acetylcysteine or ascorbic acid (18), implicating a potential role for reactive oxygen species in P2RX7-dependent AP-1 activation.
Stimulation of macrophages and osteoblasts with a P2RX7 agonist induces enhanced AP-1 protein binding and AP-1 supershifts (Fig. 8), supporting a role for short-term P2RX7 stimulation in the activation of AP-1 in macrophages. The activation of AP-1 may also be the mechanism by which P2RX7 augments the induction of protein expression in monocytic cells primed with proinflammatory stimuli such as LPS or TNF-α, e.g. this augmentation may occur via the activation of enhanceosomes that regulate the expression of these genes. Notably, the cytokines expressed in response to LPS that are potentiated by P2RX7 agonists, such as inducible nitric-oxide synthase and TNF-α, have AP-1 consensus sequences in their promoters (4). Considering that many of the LPS-induced mediators that P2RX7 has been shown to modulate are not induced by P2RX7 activation alone (16, 19), a regulatory role for P2RX7 in enhanceosome formation is highly plausible.
Although reporter assays and EMSAs have been primarily used to demonstrate activation of transcription factors by P2RX7 agonists, little is known about which genes these transcription factors up-regulate. To this end, our data support a role for P2RX7-mediated CREB activation in the expression of c-Fos (36) and FosB/ΔFosB (Fig. 7) and the subsequent induction of COX-2 in a FosB-dependent manner (Fig. 9B). The ability of P2RX7 activation to induce the expression of COX-2, which has been linked to numerous proinflammatory diseases, demonstrates the ability of P2RX7 stimulation to induce gene expression. Together, these observations reveal an important role for P2RX7 stimulation in transcriptional activation of both monocytic cells and osteoblasts in the absence of cell death.
Evidence that FosB is active in bone formation is provided by studies wherein transgenic mice, engineered to overexpress either FosB or ΔFosB, were found to exhibit markedly increased bone formation without affecting bone resorption (58, 59). Interestingly, the femurs from P2RX7 knockout mice show similar length but smaller diameter compared with respective age- and gender-matched wild-type controls (60). The P2RX7-null mice also displayed lower cortical bone mass as well as an attenuation of the periosteal bone formation rate, although osteoclast formation in the bone marrow of P2RX7-null mice appear normal (60). Furthermore, mechanical stress to bone induces the transcriptional induction of both FosB and ΔFosB in osteoblasts (61). Therefore, P2RX7-dependent FosB expression and subsequent COX-2 expression supports the notion that P2RX7 transcriptional activation has a role in bone formation, further implicating P2RX7 as a novel therapeutic target for the management of skeletal disorders.
In summary, these data reveal an important role for short-term stimulation of P2RX7 in the transcriptional regulation of AP-1 protein induction in monocytic cells and a role for FosB in P2RX7-dependent protein expression that is independent of the presence of a proinflammatory co-stimulus.
Supplementary Material
Acknowledgments
We thank the members of the Bertics laboratory, especially Dr. Mary Ellen Bates for her critical review of the manuscript and Arturo Guadarrama for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AI070503 and HL069116.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- AP-1
- activating protein-1
- eATP
- extracellular ATP
- P2RY
- P2Y receptor
- P2RX
- P2X receptor
- PGE2
- prostaglandin E2
- BzATP
- 2′(3′)-O-(4-benzoylbenzoyl)-ATP
- CREB
- cAMP response element-binding protein
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1.Foletta V. C., Segal D. H., Cohen D. R. (1998) J. Leukoc. Biol. 63, 139–152 [DOI] [PubMed] [Google Scholar]
- 2.Shaulian E., Karin M. (2002) Nat. Cell Biol. 4, E131–136 [DOI] [PubMed] [Google Scholar]
- 3.Guo R. F., Lentsch A. B., Sarma J. V., Sun L., Riedemann N. C., McClintock S. D., McGuire S. R., Van Rooijen N., Ward P. A. (2002) Am. J. Pathol. 161, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firestein G. S., Manning A. M. (1999) Arthritis Rheum. 42, 609–621 [DOI] [PubMed] [Google Scholar]
- 5.Aud D., Peng S. L. (2006) Nat. Clin. Pract. Rheumatol. 2, 434–442 [DOI] [PubMed] [Google Scholar]
- 6.Desmet C., Gosset P., Henry E., Garzé V., Faisca P., Vos N., Jaspar F., Mélotte D., Lambrecht B., Desmecht D., Pajak B., Moser M., Lekeux P., Bureau F. (2005) Am. J. Respir. Crit. Care Med. 172, 671–678 [DOI] [PubMed] [Google Scholar]
- 7.Takayanagi H. (2007) Nat. Rev. Immunol. 7, 292–304 [DOI] [PubMed] [Google Scholar]
- 8.Dubyak G. R. (1991) Am. J. Respir. Cell Mol. Biol. 4, 295–300 [DOI] [PubMed] [Google Scholar]
- 9.Gordon J. L. (1986) Biochem. J. 233, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnstock G. (1997) Neuropharmacology 36, 1127–1139 [DOI] [PubMed] [Google Scholar]
- 11.Pellegatti P., Falzoni S., Pinton P., Rizzuto R., Di Virgilio F. (2005) Mol. Biol. Cell 16, 3659–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Virgilio F. (2005) Purinergic Signal. 1, 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khakh B. S., North R. A. (2006) Nature 442, 527–532 [DOI] [PubMed] [Google Scholar]
- 14.Lister M. F., Sharkey J., Sawatzky D. A., Hodgkiss J. P., Davidson D. J., Rossi A. G., Finlayson K. (2007) J. Inflamm. (Lond) 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burnstock G. (2006) Pharmacol. Rev. 58, 58–86 [DOI] [PubMed] [Google Scholar]
- 16.Watters J., Sommer J., Fisette P., Pfeiffer Z., Aga M., Prabhu U., Guerra A., Denlinger L., Bertics P. (2001) Drug Dev. Res. 53, 91–104 [Google Scholar]
- 17.Guerra A. N., Gavala M. L., Chung H. S., Bertics P. J. (2007) Purinergic Signal. 3, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer Z. A., Guerra A. N., Hill L. M., Gavala M. L., Prabhu U., Aga M., Hall D. J., Bertics P. J. (2007) Free Radic. Biol. Med. 42, 1506–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solle M., Labasi J., Perregaux D. G., Stam E., Petrushova N., Koller B. H., Griffiths R. J., Gabel C. A. (2001) J. Biol. Chem. 276, 125–132 [DOI] [PubMed] [Google Scholar]
- 20.Labasi J. M., Petrushova N., Donovan C., McCurdy S., Lira P., Payette M. M., Brissette W., Wicks J. R., Audoly L., Gabel C. A. (2002) J. Immunol. 168, 6436–6445 [DOI] [PubMed] [Google Scholar]
- 21.Li J., Liu D., Ke H. Z., Duncan R. L., Turner C. H. (2005) J. Biol. Chem. 280, 42952–42959 [DOI] [PubMed] [Google Scholar]
- 22.Romanello M., Pani B., Bicego M., D'Andrea P. (2001) Biochem. Biophys. Res. Commun. 289, 1275–1281 [DOI] [PubMed] [Google Scholar]
- 23.Buckley K. A., Golding S. L., Rice J. M., Dillon J. P., Gallagher J. A. (2003) FASEB J. 17, 1401–1410 [DOI] [PubMed] [Google Scholar]
- 24.Ke H. Z. (2005) J. Bone Miner. Metab. 23, 84–89 [DOI] [PubMed] [Google Scholar]
- 25.Orriss I. R., Knight G. E., Ranasinghe S., Burnstock G., Arnett T. R. (2006) Bone 39, 300–309 [DOI] [PubMed] [Google Scholar]
- 26.McCoy J. M., Wicks J. R., Audoly L. P. (2002) J. Clin. Invest. 110, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.North R. A. (2002) Physiol. Rev. 82, 1013–1067 [DOI] [PubMed] [Google Scholar]
- 28.Mackenzie A. B., Young M. T., Adinolfi E., Surprenant A. (2005) J. Biol. Chem. 280, 33968–33976 [DOI] [PubMed] [Google Scholar]
- 29.Fernando K. C., Gargett C. E., Wiley J. S. (1999) Arch. Biochem. Biophys. 362, 197–202 [DOI] [PubMed] [Google Scholar]
- 30.Di Virgilio F., Chiozzi P., Falzoni S., Ferrari D., Sanz J. M., Venketaraman V., Baricordi O. R. (1998) Cell Death Differ. 5, 191–199 [DOI] [PubMed] [Google Scholar]
- 31.Di Virgilio F., Falzoni S., Chiozzi P., Sanz J. M., Ferrari D., Buell G. N. (1999) J. Leukoc. Biol. 66, 723–726 [DOI] [PubMed] [Google Scholar]
- 32.Pfeiffer Z. A., Aga M., Prabhu U., Watters J. J., Hall D. J., Bertics P. J. (2004) J. Leukoc. Biol. 75, 1173–1182 [DOI] [PubMed] [Google Scholar]
- 33.Li X., Qi X., Zhou L., Catera D., Rote N. S., Potashkin J., Abdul-Karim F. W., Gorodeski G. I. (2007) Gynecol. Oncol. 106, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brautigam V. M., Frasier C., Nikodemova M., Watters J. J. (2005) J. Neuroimmunol. 166, 113–125 [DOI] [PubMed] [Google Scholar]
- 35.Franco-Martínez S., Niño-Moreno P., Bernal-Silva S., Baranda L., Rocha-Meza M., Portales-Cervantes L., Layseca-Espinosa E., González-Amaro R., Portales-Pérez D. (2006) Clin. Exp. Immunol. 146, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavala M. L., Pfeiffer Z. A., Bertics P. J. (2008) J. Leukoc. Biol. 84, 1159–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefano L., Rössler O. G., Griesemer D., Hoth M., Thiel G. (2007) J. Cell. Physiol. 213, 36–44 [DOI] [PubMed] [Google Scholar]
- 38.Denlinger L. C., Sommer J. A., Parker K., Gudipaty L., Fisette P. L., Watters J. W., Proctor R. A., Dubyak G. R., Bertics P. J. (2003) J. Immunol. 171, 1304–1311 [DOI] [PubMed] [Google Scholar]
- 39.Korpi-Steiner N. L., Bates M. E., Lee W. M., Hall D. J., Bertics P. J. (2006) J. Leukoc. Biol. 80, 1364–1374 [DOI] [PubMed] [Google Scholar]
- 40.Aga M., Watters J. J., Pfeiffer Z. A., Wiepz G. J., Sommer J. A., Bertics P. J. (2004) Am. J. Physiol. Cell Physiol. 286, C923–C930 [DOI] [PubMed] [Google Scholar]
- 41.Ravnskjaer K., Kester H., Liu Y., Zhang X., Lee D., Yates J. R., 3rd, Montminy M. (2007) EMBO J. 26, 2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woszczek G., Chen L. Y., Nagineni S., Kern S., Barb J., Munson P. J., Logun C., Danner R. L., Shelhamer J. H. (2008) J. Allergy Clin. Immunol. 121, 215–221.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClung C. A., Ulery P. G., Perrotti L. I., Zachariou V., Berton O., Nestler E. J. (2004) Brain Res. Mol. Brain Res. 132, 146–154 [DOI] [PubMed] [Google Scholar]
- 44.Di Virgilio F., Baricordi O. R., Romagnoli R., Baraldi P. G. (2005) Curr. Drug Targets Cardiovasc. Haematol. Disord. 5, 85–99 [DOI] [PubMed] [Google Scholar]
- 45.Humphreys B. D., Dubyak G. R. (1998) J. Leukoc. Biol. 64, 265–273 [DOI] [PubMed] [Google Scholar]
- 46.Nakabeppu Y., Nathans D. (1991) Cell 64, 751–759 [DOI] [PubMed] [Google Scholar]
- 47.Kelz M. B., Chen J., Carlezon W. A., Jr., Whisler K., Gilden L., Beckmann A. M., Steffen C., Zhang Y. J., Marotti L., Self D. W., Tkatch T., Baranauskas G., Surmeier D. J., Neve R. L., Duman R. S., Picciotto M. R., Nestler E. J. (1999) Nature 401, 272–276 [DOI] [PubMed] [Google Scholar]
- 48.Mestre J. R., Mackrell P. J., Rivadeneira D. E., Stapleton P. P., Tanabe T., Daly J. M. (2001) J. Biol. Chem. 276, 3977–3982 [DOI] [PubMed] [Google Scholar]
- 49.Pavalko F. M., Norvell S. M., Burr D. B., Turner C. H., Duncan R. L., Bidwell J. P. (2003) J. Cell. Biochem. 88, 104–112 [DOI] [PubMed] [Google Scholar]
- 50.Aga M., Johnson C. J., Hart A. P., Guadarrama A. G., Suresh M., Svaren J., Bertics P. J., Darien B. J. (2002) J. Leukoc. Biol. 72, 222–232 [PubMed] [Google Scholar]
- 51.Lazarowski E. R., Boucher R. C., Harden T. K. (2003) Mol. Pharmacol. 64, 785–795 [DOI] [PubMed] [Google Scholar]
- 52.Picher M., Burch L. H., Boucher R. C. (2004) J. Biol. Chem. 279, 20234–20241 [DOI] [PubMed] [Google Scholar]
- 53.John G. R., Simpson J. E., Woodroofe M. N., Lee S. C., Brosnan C. F. (2001) J. Neurosci. 21, 4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Padeh S., Cohen A., Roifman C. M. (1991) J. Immunol. 146, 1626–1632 [PubMed] [Google Scholar]
- 55.Budagian V., Bulanova E., Brovko L., Orinska Z., Fayad R., Paus R., Bulfone-Paus S. (2003) J. Biol. Chem. 278, 1549–1560 [DOI] [PubMed] [Google Scholar]
- 56.Bowler W. B., Dixon C. J., Halleux C., Maier R., Bilbe G., Fraser W. D., Gallagher J. A., Hipskind R. A. (1999) J. Biol. Chem. 274, 14315–14324 [DOI] [PubMed] [Google Scholar]
- 57.Humphreys B. D., Rice J., Kertesy S. B., Dubyak G. R. (2000) J. Biol. Chem. 275, 26792–26798 [DOI] [PubMed] [Google Scholar]
- 58.Sims N. A., Sabatakos G., Chen J. S., Kelz M. B., Nestler E. J., Baron R. (2002) Bone 30, 32–39 [DOI] [PubMed] [Google Scholar]
- 59.Kveiborg M., Sabatakos G., Chiusaroli R., Wu M., Philbrick W. M., Horne W. C., Baron R. (2004) Mol. Cell. Biol. 24, 2820–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ke H. Z., Qi H., Weidema A. F., Zhang Q., Panupinthu N., Crawford D. T., Grasser W. A., Paralkar V. M., Li M., Audoly L. P., Gabel C. A., Jee W. S., Dixon S. J., Sims S. M., Thompson D. D. (2003) Mol. Endocrinol. 17, 1356–1367 [DOI] [PubMed] [Google Scholar]
- 61.Inoue D., Kido S., Matsumoto T. (2004) J. Biol. Chem. 279, 49795–49803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.