Abstract
AlgZ controls Pseudomonas aeruginosa alginate synthesis by activating algD, yet algZ expression is not detectable in nonmucoid strains. Mobility shift and Western blot assays revealed that algZ expression requires the sigma factor AlgT. The mapped algZ transcription start site revealed a consensus AlgT-dependent promoter that, when mutated, substantially reduced algZ transcription.
Individuals with cystic fibrosis (CF) are predisposed to pulmonary infections with a number of bacteria, including Pseudomonas aeruginosa (8). Whereas initial colonizing P. aeruginosa strains are nonmucoid, over the course of chronic infection, alginate-producing (mucoid) strains emerge. There is a distinct correlation between the appearance of mucoid P. aeruginosa and a worsening clinical prognosis for CF patients (8, 14). Control of alginate biosynthesis is complex and involves multiple genes. The algD gene, which is the first gene in the alginate biosynthetic operon and encodes GDP-mannose dehydrogenase, undergoes strong transcriptional activation in mucoid cells (6, 8). In a previous study, we discovered AlgZ (PA3385; reference 18), a protein that bound with high affinity and specificity to sequences upstream of the algD promoter and was essential for algD activation (3). Sequence analysis revealed that algZ encodes a protein of the ribbon-helix-helix family of DNA binding proteins (2). Members of this group include the Arc and Mnt repressors of bacteriophage P22, the methionine repressor protein MetJ, and NikR, a repressor of the high-affinity nickel uptake operon in Escherichia coli (2, 5, 15).
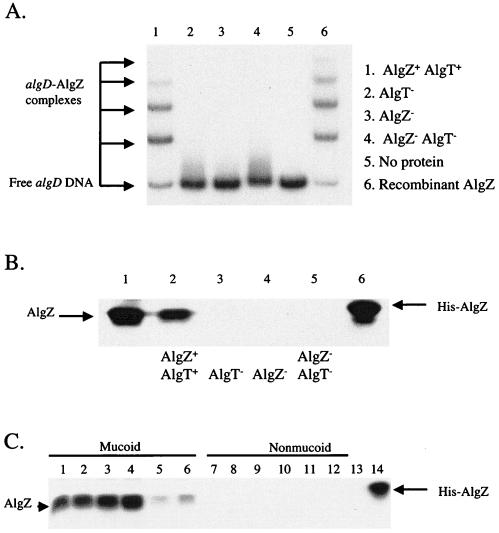
In earlier work, we observed that the expression or activity of algZ was correlated with the mucoid phenotype and dependent on the alternative sigma factor AlgT, which is also designated and annotated AlgU (8, 18). However, the mechanism by which AlgT controlled algZ was not investigated (3). An electrophoretic mobility shift assay that validates these data is depicted in Fig. 1A. In this experiment, extracts from mucoid strain FRD1 (13), as well as three isogenic mutants harboring either an algT::Tn501 allele (FRD440; reference 7), an algZ::xylE aacC1 allele (FRD1200), or the algT::Tn501 algZ::xylE aacC1 double mutation (FRD1202), were prepared and examined for binding to DNA upstream of the algD promoter. The algZ::xylE aacC1 mutation was constructed by previously described methods (16, 19), with pDJW588, a pEX18Ap-derived plasmid (10) with a 2.2-kb SmaI fragment containing xylE-aacC1 from pX1918G (10) inserted at the XhoI site within the algZ coding sequence (2). The algD DNA fragment was identical to that used in earlier AlgZ-algD DNA binding studies (2, 3). In addition, an extract from E. coli cells expressing recombinant AlgZ derived from BL21(DE3)/pPJ145 was examined for algD DNA binding activity. Plasmid pPJ145, which expresses wild-type AlgZ, was generated by PCR amplification of pDJW585 (2) with primers algZ9 (5′-CCCCCATATGCGCCCACTGAAACAGG-3′) and algZ23 (5′-GCGCTACGCGTGGGCGGCCGCGCTCAGGCCTGGG-3′) and subsequent cloning into pET29a (Novagen). All plasmids containing algZ originated from pDJW585, which is pUCP21T (17) harboring algZ derived from FRD1 on a 1.8-kb BamHI fragment. AlgZ present in the parental strain, FRD1, formed several protein-DNA complexes (Fig. 1A, lane 1). The migration of these complexes was identical to that observed with extracts of E. coli cells expressing recombinant AlgZ (Fig. 1A, lane 6). Previous competition studies indicated that this binding was highly specific (3). This is also evidenced by the fact that no binding was observed when an extract from an algZ mutant was tested (Fig. 1A, lane 3). Significantly, no binding was detected when an algT mutant was examined (Fig. 1A, lane 2). This suggested that either the activity or the expression of algZ was AlgT dependent. To distinguish these, a His-tagged AlgZ protein was expressed from BL21(DE3)/pPJ138 cells, purified, and used to make AlgZ antibodies. Plasmid pPJ138 was constructed by cloning an algZ PCR fragment derived by amplification of pDJW585 with primers algZ9 (above) and algZ10 (5′-CCCCTCGAGGGCCTGGGCCAGCTCCGCATCG-3′) into pET29a. For antibody production, approximately 1 mg of His-tagged AlgZ derived from BL21(DE3)/pPJ138 was resolved by preparative sodium dodecyl sulfate (SDS)-15% polyacrylamide gel electrophoresis (PAGE), followed by brief staining with Coomassie blue, exhaustive destaining, and excision of the band representing AlgZ. This material was used as an immunogen by a commercial vendor (Covance, Denver, Pa.) to generate polyclonal antiserum in New Zealand White rabbits. Western blot analysis was performed essentially as previously described (11), with AlgZ antiserum at a 1:50,000 dilution and chemiluminescent reagents and in accordance with the procedures outlined by Amersham. This antiserum recognized the purified 14-kDa recombinant His-tagged AlgZ protein (Fig. 1B, lane 6), as well as a faster-migrating 12-kDa wild-type AlgZ protein in an extract of E. coli expressing native AlgZ (Fig. 1B, lane 1). AlgZ was detected in extracts derived from mucoid strain FRD1 but not in those from the algZ mutants (Fig. 1B, compare lanes 2 and 4). Additionally, there was no detectable AlgZ when an extract from the isogenic algT mutant was examined (Fig. 1B, lane 3). Taken together, the data in Fig. 1A and B strongly suggest that the absence of AlgZ DNA binding in the algT mutant is due to a lack of algZ expression and not modulation of the DNA binding activity of AlgZ.
FIG. 1.
(A) Analysis of AlgZ binding to algD by electrophoretic mobility shift assay. Approximately 2 pmol of a labeled algD fragment was left untreated (lane 5) or incubated with cell extracts derived from the following strains: lane 6, 100 ng of BL21(DE3)/pPJ145; lanes 1 to 4, 5 μg derived from FRD1 (Alg+), FRD440 (algT::Tn501), FRD1200 (algZ::xylE aacC1), and FRD1202 (algZ::xylE aacC1 algT::Tn501), respectively. (B) Western blot analysis of AlgZ. Extracts or purified AlgZ were resolved by SDS-15% PAGE and probed with antibodies to AlgZ. Lane 1, 1.0 μg of extract of E. coli BL21(DE3)/pPJ145; lanes 2 to 5, 35 μg of extract prepared from FRD1 (lane 2), FRD440 (lane 3), FRD1200 (lane 4), and FRD1202 (lane 5). Lane 6 contains 280 ng of purified His-tagged AlgZ from BL21(DE3)/pPJ138. (C) Western blot analysis of AlgZ in representative CF isolates. Extracts or purified AlgZ was resolved by SDS-15% PAGE and probed with antibodies to AlgZ. Lane 1 contains 25 μg of FRD1 extract. Lanes 2 to 6, contain 25 μg each of extracts prepared from mucoid CF isolates. Lane 7 contains 25 μg of FRD440 extract. Lanes 8 to 12 contain 30 μg of extracts prepared from nonmucoid CF isolates. Lane 13 contains 30 μg of FRD1200. Lane 14 contains 200 ng of purified, His-tagged AlgZ from BL21(DE3)/pPJ138.
Our earlier work showed that AlgZ binding activity was present in all of the mucoid strains examined yet absent in nonmucoid strains derived from CF patients (3). We used AlgZ antiserum to screen extracts from additional mucoid and nonmucoid P. aeruginosa strains derived from CF patients. These results confirm that AlgZ is highly expressed in mucoid strains derived from CF patients but absent in nonmucoid strains (Fig. 1C). This also indicates that the expression of AlgZ parallels the mucoid status of the cell and suggests that AlgZ-mediated activation of algD is conserved among mucoid CF isolates.
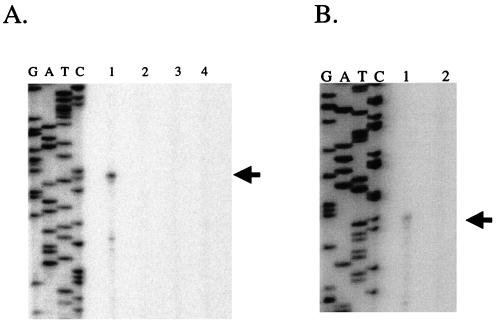
The above data suggest that AlgT controls algD expression and alginate synthesis in part through control of algZ. If this is the case, transcription of both algD and algZ should be reduced or eliminated in algT mutants. Data previously obtained from our laboratory and others have demonstrated that AlgT is essential for algD transcription (6, 9, 20). We performed primer extension experiments as outlined previously (20) to determine if the control exerted by AlgZ at algD was at the previously studied AlgT-dependent promoter (6, 20). RNA was harvested from parental mucoid strain FRD1, as well as isogenic algT, algZ, or algT algZ mutants, and analyzed for algD expression by primer extension. RNA from P. aeruginosa cells cultured in LBNS (10 g of tryptone and 5 g of yeast extract per liter) (A600 = 0.5) was purified by standard techniques (1). The oligonucleotide used for primer extension was algD1 (5′-AACAGGTTGAGTTTGTCCCT-3′, position +86 to position +66 relative to the start of algD transcription) (20) and was end labeled with polynucleotide kinase with [γ-32P]ATP as previously described (1). All detectable algD transcription originated from the previously mapped promoter (Fig. 2A, lane 1). Expression at this promoter was absolutely dependent on sigma factor AlgT (lane 2), as well as algZ (lanes 3 and 4). These data support the conclusions that AlgZ is an essential positive transcriptional activator of algD and that AlgZ controls algD through the previously characterized AlgT-dependent promoter.
FIG. 2.
(A) Primer extension analysis of algD. Oligonucleotide algD1 was end labeled and used in a primer extension experiment with 50 μg of total cellular RNA from the following strains: lane 1, FRD1 (Alg+); lane 2, FRD440 (algT::Tn501); lane 3, FRD1200 (algZ::xylE aacC1); lane 4, FRD1202 (algT::Tn501 algZ::xylE aacC1). The arrow represents the start site of algD transcription (20). The algD sequencing ladder (GATC) was produced from pDJW220 (20) with the same oligonucleotide (algD1) used for synthesis of the probe in the primer extension experiment. (B) Primer extension analysis of algZ. Oligonucleotide algZ15 (see Fig. 3A) was end labeled and used in a primer extension experiment with 50 μg of total cellular RNA from the following: lane 1, FRD1 (Alg+); lane 2, FRD440 (algT::Tn501). The arrow represents the start site of algZ transcription. The algZ sequencing ladder (GATC) was produced from pDJW585 with the same oligonucleotide (algZ15) used for synthesis of the probe in the primer extension experiment.
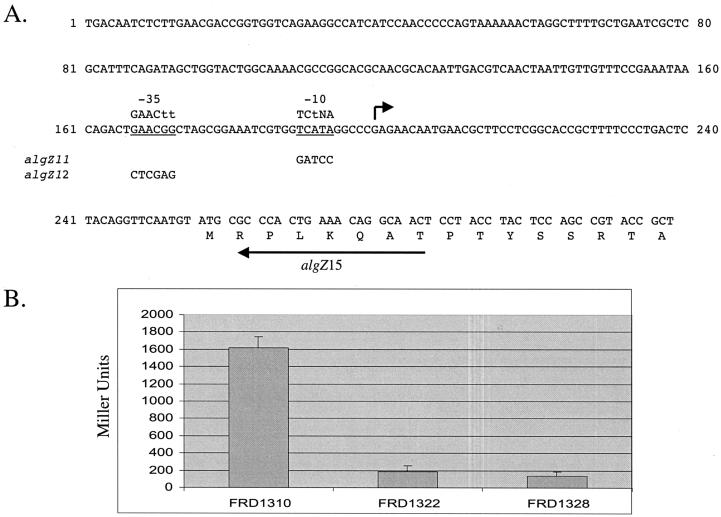
To test whether AlgT was responsible for algZ transcription and to map a promoter(s) required for algZ expression, primer extension analysis was performed on RNA derived from P. aeruginosa strains FRD1 and FRD440 (Fig. 2B). A primer positioned just downstream of the algZ translation initiation site (algZ15 [5′-GTTGCCTGTTTCAGTGGGC-3′]) (Fig. 3A) was end labeled, hybridized to RNA, and reverse transcribed. A primer extension product of 78 nucleotides was observed when RNA from parental (AlgT+) strain FRD1 was examined (Fig. 2B, lane 1). However, no primer extension signal was observed when RNA from the algT mutant FRD440 was examined (Fig. 2B, lane 2, arrow in Fig. 3A). This is consistent with data in Fig. 1B, which indicate no AlgZ protein present in algT mutants. Inspection of the sequences immediately upstream of the algZ transcription start site revealed a partial match with the consensus AlgT promoter recognition sequence (8), GAACTT 16/17 bp TCTNA (Fig. 3A). This suggested that the alternative sigma factor AlgT was required directly for expression from this promoter.
FIG. 3.
(A) Upstream regulatory sequences at algZ. The sequences immediately upstream of the algZ coding region are depicted. The small arrow represents the position of the mapped algZ transcription start site, and the proposed AlgT promoter is underlined. Sequences on top of this represent the consensus AlgT promoter (8). Sequences under this represent mutant alleles algZ11 and algZ12. (B) algZ-lacZ transcription studies. Strains FRD1310 (FRD1 algZ-lacZ), FRD1322 (FRD1 algZ12-lacZ), and FRD1328 (FRD1 algZ11-lacZ) were cultured on LBNS plates and assayed for algZ-lacZ transcriptional activity.
To determine if the consensus AlgT promoter functioned in vivo, site-directed mutagenesis was used to disrupt the putative promoter. Two allelic variants were created that harbored mutations (Promega Altered Sites) in the putative −10 (algZ11) and −35 (algZ12) sequences (Fig. 3A). Promoter sequences from wild-type algZ, as well as algZ11 and algZ12, were amplified by PCR with primers algZ15 (5′-GTTGCCTGTTTCAGTGGGCG-3′) and algZ24 (5′-GGTGTAGACCAAGCTTGAAGGAGACTG-3′) and cloned into mini-CTX-lacZ (4) to form algZ-lacZ operon fusions. Plasmids harboring the algZ-lacZ fragments were integrated in single copy into the FRD1 chromosome at the neutral attB site as described elsewhere (4, 21), resulting in strains FRD1310 (FRD1 algZ-lacZ), FRD1322 (FRD1 algZ12-lacZ), and FRD1328 (FRD1 algZ11-lacZ). β-Galactosidase levels (Miller units [12]) were determined from FRD1310, FRD1322, and FRD1328 cells recovered from LBNS plates. Compared with the wild-type algZ-lacZ levels expressed in FRD1310, mutations in the −35 or −10 sequence resulted in a substantial reduction in algZ transcription (88.5 or 91.5%, respectively; Fig. 3B).
Taken together, these data provide evidence that expression of algZ requires the alternative sigma factor AlgT. Most likely, AlgT is directly involved in algZ promoter recognition and transcription initiation since a promoter that is similar to a consensus AlgT promoter was identified upstream of the mapped transcription start site and mutations in the −35 or −10 element significantly reduced but did not completely eliminate algZ transcription.
Acknowledgments
Public Health Service grants AI-35177 and HL-58334 (D.J.W.) supported this work.
We thank H. Schweizer for providing the tools for gene replacements and D. Ramsey for editing the manuscript.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, vol. 2, p. 4.1-4.2. Greene Publishing and John Wiley & Sons, New York, N.Y.
- 2.Baynham, P., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. [DOI] [PubMed] [Google Scholar]
- 3.Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. [DOI] [PubMed] [Google Scholar]
- 4.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-952. [DOI] [PubMed] [Google Scholar]
- 5.Chivers, P. T., and R. T. Sauer. 2000. Regulation of high affinity nickel uptake in bacteria: Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. [DOI] [PubMed] [Google Scholar]
- 6.Deretic, V., J. F. Gill, and A. M. Chakrabarty. 1987. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 11:4567-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn, J. L., and D. E. Ohman. 1988. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J. Bacteriol. 170:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershberger, C. D., R. W. Ye, M. R. Parsek, Z. D. Xie, and A. M. Chakrabarty. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative σ factor (σE). Proc. Natl. Acad. Sci. USA 92:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: applications for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 11.Ma, S., U. Selvaraj, D. E. Ohman, R. Quarless, D. J. Hassett, and D. J. Wozniak. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. β-Galactosidase assay in permeabilized cells, p. 493-495. In S. R. Maloy, V. J. Stewart, and R. K. Taylor (ed.), Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 13.Ohman, D. E., and A. M. Chakrabarty. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 33:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pier, G. B. 1998. Pseudomonas aeruginosa: a key problem in cystic fibrosis. ASM News 6:339-347. [Google Scholar]
- 15.Raumann, B. E., M. A. Rould, C. O. Pabo, and R. T. Sauer. 1994. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature 367:754-757. [DOI] [PubMed] [Google Scholar]
- 16.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 17.Schweizer, H. P., T. Klassen, and T. Hoang. 1996. Improved methods for gene analysis and expression in Pseudomonas spp., p. 229-237. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.), Molecular biology of Pseudomonas. ASM Press, Washington, D.C.
- 18.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsenk, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 19.Woolwine, S., and D. J. Wozniak. 1999. Identification of an Escherichia coli pepA homolog and its involvement in suppression of the algB phenotype in mucoid Pseudomonas aeruginosa. J. Bacteriol. 181:107-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wozniak, D. J., and D. E. Ohman. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyckoff, T. J. O., and D. J. Wozniak. 2001. Transcriptional analysis of genes involved in Pseudomonas aeruginosa biofilms. Methods Enzymol. 336:144-151. [DOI] [PubMed] [Google Scholar]