Abstract
The farAB operon of Neisseria gonorrhoeae encodes an efflux pump which mediates gonococcal resistance to antibacterial fatty acids. It was previously observed that expression of the farAB operon was positively regulated by MtrR, which is a repressor of the mtrCDE-encoded efflux pump system (E.-H. Lee and W. M. Shafer, Mol. Microbiol. 33:839-845, 1999). This regulation was believed to be indirect since MtrR did not bind to the farAB promoter. In this study, computer analysis of the gonococcal genome sequence database, lacZ reporter fusions, and gel mobility shift assays were used to elucidate the regulatory mechanism by which expression of the farAB operon is modulated by MtrR in gonococci. We identified a regulatory protein belonging to the MarR family of transcriptional repressors and found that it negatively controls expression of farAB by directly binding to the farAB promoter. We designated this regulator FarR to signify its role in regulating the farAB operon. We found that MtrR binds to the farR promoter, thereby repressing farR expression. Hence, MtrR regulates farAB in a positive fashion by modulating farR expression. This MtrR regulatory cascade seems to play an important role in adjusting levels of the FarAB and MtrCDE efflux pumps to prevent their excess expression in gonococci.
Neisseria gonorrhoeae is a strictly human pathogen that causes the sexually transmitted disease gonorrhea. Gonococci often infect mucosal sites bathed in fluids containing host-derived antimicrobial hydrophobic agents (HAs), such as free fatty acids (FAs) and bile salts. Gonococcal resistance to certain antimicrobial HAs has been attributed to the mtrCDE (for “multiple transferable resistance”)-encoded efflux pump, which exports a number of host-derived antimicrobial HAs (e.g., bile salts, antibacterial peptides, and FAs) (11, 28, 35). The mtr locus consists of three tandemly linked genes (mtrCDE) encoding cell envelope proteins that are divergently transcribed from the mtrR gene, positioned 250 bp upstream (11). The MtrR protein is a transcriptional repressor belonging to the TetR family of proteins and plays a critical role in modulating transcription of the mtrCDE operon. Thus, mutations within the mtrR coding region or the intergenic region between mtrR and mtrCDE enhanced mtrCDE expression, leading to elevated resistance to HAs (11, 12).
A recent study by Lee and Shafer(15) revealed a second efflux pump system that can modulate levels of gonococcal resistance to a subset of HAs. This system was termed far (for “FA resistance”) because it confers resistance to long-chain FAs. The far system was found to be responsible for the mtr-independent mechanism of FA resistance, which was previously observed in a number of clinical isolates obtained from homosexual men with rectal infections (22). The far system is composed of the FarA membrane fusion protein and the FarB cytoplasmic membrane transporter protein. This efflux pump requires the MtrE protein (5) as the outer membrane channel to export antibacterial FAs from inside the cell (15). Although the mtr- and the far-encoded systems independently mediate gonococcal resistance to host-derived antimicrobial HAs, their expression is controlled by MtrR. In contrast to that of the mtr system, expression of farAB was positively associated with the presence of a functional MtrR protein. However, the results indicated that this regulation was indirect because MtrR did not bind to the farAB operon (15).
At the amino acid sequence level, the farAB system is similar to the emrAB efflux pump system of Escherichia coli, which provides resistance to uncoupling agents and certain antibiotics (17). The emrAB operon is negatively controlled by the product of the emrR gene, which is located upstream of emrA (18). EmrR belongs to the MarR family of transcriptional regulatory proteins, which control a variety of biological functions, including resistance to antimicrobial agents (e.g., antibiotics, organic solvents, and oxidative stress agents) (1, 24). In addition to EmrR, at least two MarR family proteins are involved in the resistance of E. coli (MarR) and Pseudomonas aeruginosa (MexR) to antimicrobial agents by modulating the expression of efflux pump systems. MarR is a negative regulator of the marRAB operon in E. coli (21, 32). Mutations in the marR gene or certain inducing conditions cause the overexpression of the MarA activator, resulting in activation of a number of genetic loci, including the acrAB efflux pump system, and enhance bacterial resistance to antimicrobial agents (10, 27). In P. aeruginosa, inactivation of the MexR repressor results in the overexpression of the mexA-mexB-oprM efflux pump system, which is a major determinant for resistance to a broad range of antimicrobials (29, 31, 37, 40). The analysis of the crystal structures for MarR and MexR suggested that MarR family proteins bind to DNA as dimers through a conserved helix-turn-helix motif (2, 3). This binding often occurs through recognition of inverted or direct repeat sequences (6, 21, 39).
Since we previously observed that the farAB operon was regulated indirectly by the MtrR repressor, we sought to elucidate the molecular mechanism by which farAB is regulated via a MtrR-dependent mechanism. Accordingly, we asked if there is a regulatory protein in gonococci that directly controls expression of the farAB operon. We now report on the identification of a regulatory protein (FarR) in gonococci that belongs to the MarR family and show that it directly controls expression of farAB. Furthermore, we found that the MtrR repressor modulates expression of farR and is consequently implicated in positive regulation of the farAB operon. Thus, MtrR appears to be of importance in differentially controlling two efflux pumps, those encoded by mtrCDE and farAB, in gonococci.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strain TOP10 (Invitrogen, Carlsbad, Calif.) was used in all cloning experiments. N. gonorrhoeae strain FA19 was used as the primary gonococcal strain. E. coli strains were grown in Luria-Bertani broth at 37°C. Gonococcal strains were grown on gonococcal medium base (GCB) agar(Difco Laboratories, Detroit, Mich.) containing glucose and iron supplements at 37°C under 3.8% (vol/vol) CO2 (33).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference(s) |
|---|---|---|
| N. gonorrhoeae | ||
| FA19 | Wild type | 8, 9, 11, 33 |
| EL12 | FA19 (pLFAB1) | This study |
| EL24 | FA19 farR::Kmr | This study |
| EL26 | FA19 farR::Kmr (pLFAB1) | This study |
| EL27 | FA19 marR2::Kmr | This study |
| EL29 | FA19 marR2::Kmr (pLFAB1) | This study |
| EL33 | FA19 (pLFAR1) | This study |
| EL35 | FA19 farR::Kmr (pLFAR1) | This study |
| EL37 | FA19 ΔmtrR (pLFAR1) | This study |
| KH11 | FA19 ΔmtrR | 11 |
| E. coli TOP10 | (F−mcrA Δ(mrr-hsdRMS-mcrBC)φ80 lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (StrrendA1 nupG) | Invitrogen |
| Plasmids | ||
| pLES94 | pUC18 derivative allowing chromosomal lacZ fusion at proAB site in N. gonorrhoeae; Apr Cmr | 36 |
| pLFAB1 | pLES94 derivative carrying 300-bp upstream sequence of farA at BamHI site | This study |
| pLFAR1 | pLES94 derivative carrying 305-bp upstream sequence of farA at BamHI site | This study |
| pBAD-TOPO | pUC-derived protein expression vector carrying C-terminal six-His tag; Apr | Invitrogen |
| pBFAR1 | pBAD-TOPO derivative carrying 441-bp coding sequence of farR | This study |
Efficiency-of-plating (EOP) analysis.
After overnight growth on GCB agar plates with or without supplementation of palmitic acid (150 μg/ml; Sigma, St. Louis, Mo.), the CFU of gonococci were determined from GCB-palmitic acid plates and compared with the CFU on GCB plates as previously described (30).
Construction of an insertional mutation in marR1 (farR) and marR2.
A 956-bp region encompassing the marR1 (farR) gene was prepared by PCR using oligonucleotide primers marR2 and marR4 (Table 2). This PCR product was cloned into the pBAD-TOPO vector (Invitrogen) according to the manufacturer's instructions. The resulting construct was digested with StuI, which cleaved the plasmid at a unique restriction site located at the 5′-end sequence of the insert. The nonpolar aphA-3 cassette (23) was digested with SmaI from pUC18K and cloned into the StuI site of the construct so that it would be in the same transcriptional orientation as farR. This recombinant plasmid was introduced into E. coli TOP10 by transformation. Transformants were selected with kanamycin (50 μg/ml; Sigma) after gene expression was induced with 0.002% (vol/vol) arabinose. The plasmid was then purified from the host E. coli and used to transform N. gonorrhoeae strain FA19 as described previously (11). The transformants were selected on GCB agar plates containing kanamycin (50 μg/ml). An insertional mutation in the marR2 gene was also created with the kanamycin cassette as described above, with modifications. Briefly, two steps of PCR were carried out to create a SmaI restriction site in the middle of marR2. In the first step, two PCRs amplified each half of marR2. One reaction encompassed the upstream sequence and the 5′-end region of marR2, and the other one included the 3′ end of marR2 and its downstream sequence. SmaI restriction sites were introduced at the 3′ end of the 5′ region with primer R2#5 and at the 5′ end of the 3′ section of the marR2 gene with primer R2#6 (Table 2). An 1,800-bp fragment encompassing the entire marR2 gene in which the SmaI site was created was then amplified with primers R2#1 and R2#2 (Table 2). The resulting DNA was cloned into the pUC18 vector, and the aphA-3 cassette was introduced into the created SmaI site of the marR2 gene.
TABLE 2.
Oligonucleotide sequences used in this study
| Primer | Sequence (5′-3′) | Locationa |
|---|---|---|
| farA26 | CCCAAAAGAGAGATGCCTTGC | −213; C strand of farA |
| farA26B | CCCAAAAGAGGGATTCCTTGC | −213; C strand of farA |
| farA52B | CCATAAGATTGGGATCCGAATTTCC | +34; NC strand of farA |
| farB1 | TTAGATACGACCATCGCCAACG | +79; C strand of farB |
| farB2 | ACAACGACGGTCATTGCC | +426; NC strand of farB |
| farRB1 | GCCGATGGGATCCCTTGTTG | +214; C strand of farR |
| farRB2 | GGTCATCAGGGGATCCCTTG | −69; NC strand of farR |
| KH9#2 | CGTTTCGGGTCGGTTTGACG | −12; NC strand of mtrR |
| KH9#3 | GACGACAGTGCCAATGCAACG | +71; NC strand of mtrC |
| marR2 | GTTTATGCACAGCATCACGG | +727; NC strand of farR |
| marR4 | CCGGAAGCCAAGTTTCGAGA | −924; C strand of farR |
| marR7 | ATGCCTACCCA ATCAAAACATGCG | +1; C strand of farR |
| marR8 | CGAGTTCAACGCATCCTCG | −438; NC strand of farR |
| R2#1 | CACACTGCTGATTCTGTTCGGC | −709; C strand of marR2 |
| R2#2 | CATCCACAGATAAGTGCCGAC | +1198; NC strand of marR2 |
| R2#5 | CAAAACCCTTGCCCGGGAAGGGTTGATTG | +191; C strand of marR2 |
| R2#6 | CAATCAACCCTTCCCGGGCAAGGGTTTTG | +220; NC strand of marR |
| RT1B | AAAATGCAGTTTGGATTCGAATGG | +11; C strand of degR |
Relative to translation start site of farA in the lacZ fusion. C, coding strand; NC, noncoding strand 45.
Construction and use of farAB-lacZ and farR-lacZ fusions in gonococci.
The farAB-lacZ and farR-lacZ fusions used in this study were prepared in pLES94 as previously described (36). Briefly, an approximately 300-bp sequence upstream of farA and farR was amplified by PCR using primers farA26B and farA52B and farRB1 and farRB2 (Table 2), respectively, and ligated into the BamHI site of pLES94. The ligation mixtures were introduced into E. coli TOP10 by transformation, and transformants were selected on LB agar plates supplemented with ampicillin (100 μg/ml; Sigma). After the orientation and sequence of the insert were checked and found correct, the resulting plasmids were used to transform strain FA19 to allow insertion of the farAB-lacZ or farR-lacZ fusion at the proAB site on the gonococcal chromosome (36). Transformants were selected on GCB plates containing chloramphenicol (1 μg/ml; Sigma). To create a farR mutation in the farR-lacZ fusion strain (EL33), a 2.5-kb DNA sequence encompassing the farR coding region, interrupted by a kanamycin resistance cassette, was amplified by PCR using oligonucleotide primers marR2 and marR4 from strain EL24 (farR::Kmr) and this product was introduced into strain EL33 by transformation. For the mtrR mutation in strain EL33, a 720-bp region containing the deleted mtrR gene (11) was amplified from DNA of strain KH11 (same as FA19 except ΔmtrR) with primers RT1B and KH9#3and transformed into EL33 as described above. Transformants were selected on GCB agar containing kanamycin (50 μg/ml) for the farR mutation and erythromycin (0.5 μg/ml) for the mtrR mutation.
β-Gal assay.
Nonpiliated, transparent colony types were routinely grown on GCB agar or in GCB broth (Difco) as previously described (11). GCB-grown cells were harvested when the optical density of culture at 600 nm reached 0.3, and plate culture cells were taken after 20 h of growth. The cells were washed once in phosphate-buffered saline (pH 7.4) and frozen at −70°C overnight. The cell pellets were suspended in lysate buffer (1 mM dithiothreitol, 100 mM potassium phosphate, pH 7.8) and broken by repeated freeze-thaw cycles. The cell debris was removed by centrifugation at 15,000 × g for 10 min at 4°C. The amount of β-galactosidase (β-Gal) in the cell extracts was measured with a chemiluminescent β-Gal assay kit (Clontech, Palo Alto, Calif.) by following the manufacturer's instructions.
Purification of FarR and MtrR.
To overexpress and purify FarR, a 438-bp region containing the farR structural gene was amplified by PCR using oligonucleotides primers marR7 and marR8. This PCR product was cloned into pBAD (Invitrogen), producing a C-terminal fusion with a histidine tag. The recombinant plasmid was introduced by transformation into E. coli TOP10, and the resulting transformants were selected on LB plates supplemented with ampicillin (100 μg/ml). The E. coli strain containing pBFAR1 was grown in 200 ml of LB broth at 37°C with vigorous aeration. When the optical density of the culture at 600 nm reached 0.6, FarR expression was induced by the addition of 0.002% (wt/vol) arabinose for 3 h. Cells were harvested by centrifugation at 6,000 × g for 10 min and resuspended in 20 ml of binding buffer (1 mM phenylmethylsulfonyl fluoride, 5 mM imidazole, 20 mM phosphate, 500 mM NaCl, pH 7.8). After addition of lysozyme (100 μg/ml), cells were broken by repeated freeze-thaw and sonication cycles. Insoluble debris was removed by centrifugation at 3,000 × g for 15 min, and the supernatant was passed through a 0.8-μm-pore-size syringe filter (Millipore). All further purification was carried out by following the manufacturer's instruction (Invitrogen). The supernatant was loaded on a mini-Ni2+ affinity column that was equilibrated with native binding buffer (20 mM sodium phosphate, 500 mM sodium chloride, pH 7.8). After the column was washed with washing buffer (20 mM sodium phosphate, 500 mM sodium chloride, pH 6.0) several times, the FarR-His protein was eluted with 5 ml of each of five imidazole elution buffers that had increasing imidazole concentrations (50, 100, 200. 350, and 500 mM). All fractions were collected and subjected to electrophoresis on a sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) gel (14). A major peak of FarR was eluted with a minor peak of non-FarR material at about 350 to 450 mM imidazole. To remove this minor contamination, the FarR fraction was further purified by high-pressure liquid chromatography (HPLC; Jupiter 5μ C4, 300 A, 250 by 4.6 mm; Phenomenex). A major peak of FarR was collected in a solution of 70% acetonitrile-10% water-0.1% (vol/vol) trifluoroacetic acid and lyophilized. The lyophilized FarR protein was dissolved in water, dialyzed overnight against a buffer composed of 5 mM Tris (pH 8.0), 5 mM EDTA, 2 mM dithiothreitol, and 0.01% Triton X-100, and concentrated with a Centricon YM-3 centrifugal filter (Amicon; Millipore). The N-terminal amino acid sequence of FarR was analyzed by the automated Edman degradation method using cLC-Procise sequenator (Applied Biosystems, Foster City, Calif.). The MtrR-maltose binding protein (MBP) was purified as described previously (19).
Electrophoretic mobility shift assay (EMSA).
The farAB and farR promoter fragments were amplified by PCR from FA19 chromosomal DNA with oligonucleotides pairs farA26 and farA52B for the farAB promoter, farRB1and farRB2 for the farR promoter, KH9#2 and KH9#3 for the mtrR-CDE intervening region, and farB1 and farB2 for the farB coding region, (Table 2). The PCR products were end labeled with [γ-32P]dATP by using T4 polynucleotide kinase (New England Biolabs). Approximately 5 ng of the labeled DNA fragment was incubated with FarR or MtrR in 30 μl of reaction buffer (10 mM Tris-HCl [pH 7.5], 0.5 mM dithiothreitol, 0.5 mM EDTA, 4% [vol/vol] glycerol, 1 mM MgCl2, 50 mM NaCl, poly[dI-dC] [0.5 μg/ml], salmon sperm [200 μg/ml]) at room temperature for 25 min. For the competition assay, a nonlabeled target or irrelevant DNA was added in the binding reaction buffer. Samples were subjected to electrophoresis in a 4.5% native polyacrylamide gel at 4°C, followed by autoradiography (19).
RESULTS AND DISCUSSION
Identification of a regulatory protein (FarR) involved in negative regulation of the farAB operon.
Members of the MarR family of repressors (EmrR [E. coli], MarR [E. coli], and MexR [P. aeruginosa]) are involved in the regulation of efflux pump operons (3, 4, 6, 16, 18, 21, 29, 32, 39). In an attempt to identify a transcriptional regulatory protein that directly modulates farAB, we sought to identify a MarR-like protein(s) in gonococci. Since Neisseria meningitidis and N. gonorrhoeae are genetically closely related and since the complete annotation of the meningococcal genome sequencing database was available, we searched for a meningococcal marR-like gene(s) in the online database of N. meningitidis strain MC58 (www.tigr.org). This search identified two meningococcal open reading frames that encode transcriptional regulators of the MarR family (TIGR locus names: NMB1843 and NMB1585). Furthermore, sequences homologous to these genes, marR1 for NMB1843 and marR2 for NMB1585, were subsequently identified in the N. gonorrhoeae FA1090 genome sequence database (www.genome.ou.edu). When the putative products of marR1 and marR2 were compared with other MarR family regulatory proteins that modulate efflux pump operons, they were found to have 20 to 25% amino acid identity (data not presented).
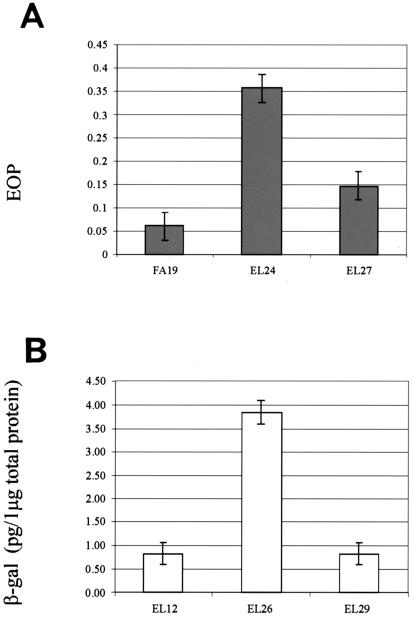
To determine whether the putative marR1- or marR2-encoded protein or both regulate farAB gene expression, we created an insertional mutation within the marR1 and marR2 genes in N. gonorrhoeae strain FA19 (see Materials and Methods for details). Transformants of strain FA19 containing the kanamycin resistance (Kmr) cassette in marR1 or marR2 were examined for their susceptibility to long-chain FAs. Since these types of FAs are very hydrophobic, with limited solubility, it was impossible to obtain a FA concentration higher than the MIC for the wild-type strain FA19. We therefore performed an EOP experiment using palmitic acid as described previously (30). CFU of transformants EL24 (marR1::Kmr) and EL27 (marR2::Kmr) were calculated from bacterial growth on GCB agar plates supplemented or not supplemented with palmitic acid (150 μg/ml). The results demonstrated that the marR1::Kmr mutation had a more significant (P = 0.01) impact on gonococcal susceptibility to palmitic acid than the marR2::Kmr mutation (P = 0.07). In this respect, the EOP of strain EL24 was sixfold higher than that of the parental strain, FA19 (Fig. 1A).
FIG. 1.
Effect of the marR1 mutation on FA resistance and farAB expression in N. gonorrhoeae FA19. (A) An EOP experiment was performed with strains FA19, EL24 (same as FA19 except marR1::Kmr) and EL27 (same as FA19 except marR2::Kmr) on GCB agar plates containing palmitic acid (150 μg/ml). EOPs are average values (± standard deviations [SD]) from at least three independent experiments. (B) Expression of farAB in EL12 (FA19[pLFAB1]) and its isogenic mutant strains EL26 (EL24[pLFAB1]) and EL29 (EL27[pLFAB1]). Shown are the amounts of β-Gal in cell extracts prepared as described in Materials and Methods from reporter strains EL12 and EL26, which contained the farAB::lacZ fusion. The results are averages of at least four independent experiments; error bars represent 1 SD.
To ascertain if MarR1 or MarR2 regulates the farAB operon, a farAB-lacZ reporter fusion was constructed in strain FA19. For this purpose, 300 bp of the farAB upstream region, including the ATG start codon and the codons for the first seven amino acids, was amplified by PCR and cloned into pLES94, resulting in a translational lacZ fusion. The resulting recombinant plasmid was introduced into the proAB site on the gonococcal chromosome by homologous recombination (36). We performed a chemiluminescence β-Gal assay using stationary cells grown overnight on GCB agar to permit maximal expression of farAB. The results showed that the marR1 mutation (strain EL26) caused a fourfold increase (P = 0.02) in the amount of β-Gal production (Fig. 1B), while the marR2 mutation (strain EL29) did not (P = 0.5) impact farAB expression (Fig. 1B). On the basis of these results and the FA susceptibility data (Fig. 1A), we hypothesized that MarR1 negatively regulates farAB expression. Because MarR1 had the most significant impact on gonococcal susceptibility to FAs and farAB expression, we studied it in more detail. MarR1 was renamed FarR to signify its role in regulation of farAB expression.
DNA-binding properties of FarR.
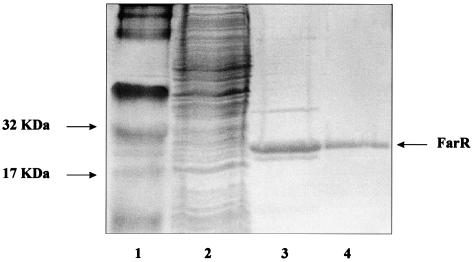
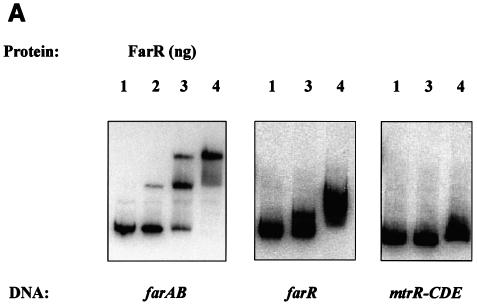
To determine whether FarR regulates the expression of farAB by directly binding to the farAB operon, FarR was purified. The farR coding sequence was cloned into the pBAD-TOPO vector to form a C-terminal fusion with a histidine tag with expression under the control of the arabinose-inducible promoter in E. coli TOP10. Crude cell extracts were prepared from a 200-ml culture and passed through a mini-Ni2+ affinity column. Analysis of fractions eluted from this matrix by SDS-PAGE revealed that the FarR-His fusion protein was slightly contaminated with a protein of about 40 kDa (Fig. 2, lane 3). This contaminating 40-kDa protein was removed from the FarR-His fusion protein by HPLC (Fig. 2, lane 4). The recovered FarR-His fusion protein was shown to have a molecular mass of 20 kDa when analyzed by HPLC, which is in good agreement with the predicted mass of 20.68 kDa (data not shown). N-terminal amino acid sequencing confirmed that the first nine amino acids of the recovered protein (MPTQSKHAS) were identical to the corresponding amino acid sequence predicted by DNA sequence analysis of farR (data not presented). The DNA-binding capacity of the FarR-His protein was studied by EMSA. The target DNA consisted of approximately 300 bp of the farA upstream region that included the farAB promoter (PfarAB). Using EMSA, we detected two potential FarR-DNA complexes. The first (form I) was observed at a level of 0.2 ng of FarR, and a slower-migrating complex (form II) was observed with increasing levels of FarR (1 to 5 ng) (Fig. 3A), suggesting that FarR binds to at least two sites in the upstream sequence of farA. To show the binding specificity of FarR, we performed competition assays. Addition of a 200-fold molar excess of a heterologous unlabeled DNA fragment containing 310 bp of the mtrR-CDE intervening region had no effect on binding (Fig. 3B). However, addition of unlabeled PfarAB inhibited the binding of FarR to the labeled PfarAB fragment. These results indicated that FarR binds to the farAB promoter in a specific manner.
FIG. 2.
Expression and purification of FarR-His. Protein samples collected during the purification were analyzed on SDS-15% PAGE gels stained with Coomassie brilliant blue. Lane 1, molecular weight standard markers (arrows [left], 17- and 32-kDa markers); lane 2, cell lysate after induction; lane 3, pooled fraction after Ni2+ affinity chromatography; lane 4, purified FarR after HPLC purification. Arrow (right), location of the FarR-His monomer.
FIG. 3.
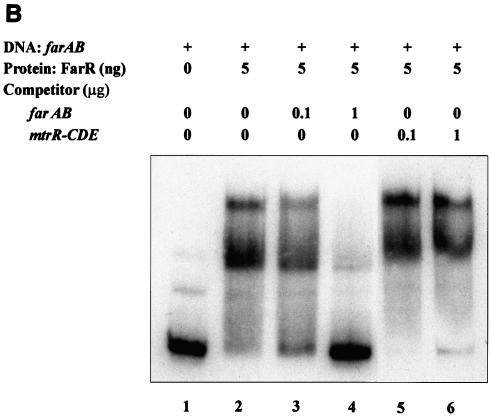
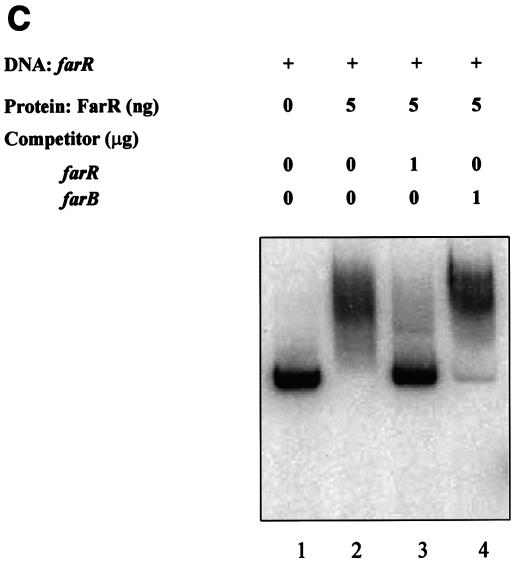
DNA-binding properties of FarR. Shown is the binding of the purified His-tagged FarR protein to target DNA sequences. (A) FarR binding to farAB, farR, and mtrR-CDE promoter regions. Lanes 1, free labeled probe; lanes 2, probe with 0.2 ng of FarR; lanes 3, probe with 1 ng of FarR; lanes 4, probe with 5 ng of FarR. The probe used is indicated at the bottom of each panel. (B and C) Competition assays. (B) 32P-labeled 300-bp DNA encompassing the farAB promoter region was incubated with 5 ng of FarR. This binding was competed with unlabeled farAB (300 bp) or the mtrR-CDE intergenic region (310 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 0.1 μg of farAB DNA; lane 4, FarR with 1 μg of farAB DNA; lane 5, FarR with 0.1 μg of mtrR-CDE DNA; lane 6, FarR with 1 μg of mtrR-CDE DNA. (C) The 32P-labeled 305-bp farR promoter region was incubated with 5 ng of FarR. This binding was competed with the unlabeled farR promoter (305 bp) or a DNA sequence containing the farB coding region (365 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 1 μg of farR DNA; lane 4, FarR with 1 μg of farB DNA.
Autoregulation of farR expression.
Regulatory proteins that control expression of efflux pump operons are often subject to autoregulation (20, 29). To explore this possibility, we conducted an EMSA using FarR and a DNA fragment containing the farR promoter. A retarded complex was observed at the concentration of 5 ng, indicating that FarR might regulate its own expression (Fig. 3A). The specificity of this binding was confirmed by performing a competition assay, the results of which are shown in Fig. 3C. This experiment revealed that an unlabeled farR promoter sequence but not a DNA sequence within the farB coding region could compete with farR binding to the labeled farR promoter sequence. To determine if FarR regulates its own expression, the farR gene was inactivated by a nonpolar kanamycin resistance cassette in the farR-lacZ fusion strain EL33, giving rise to strain EL35, and β-Gal activity was assessed. The results (Table 3) revealed that inactivation of farR in strain EL35 resulted in a greater-than-twofold increase in β-Gal activity, indicating that FarR represses its own expression.
TABLE 3.
Effect of farR or mtrR mutation on farR expression in N. gonorrhoeaea
| Strain | Vector | β-Gal sp actb (ng/107 CFU) |
|---|---|---|
| EL33 (MtrR+ FarR+) | pLFAR1c | 248 ± 71.8 |
| EL35 (MtrR+ FarR−) | pLFAR1 | 718 ± 190.2 |
| EL37 (MtrR− FarR+) | pLFAR1 | 381 ± 125.6 |
N. gonorrhoeae (Opa− Pil−) fusion strains were grown to late log phase in GCB broth and assayed for β-Gal activities.
Data are averages of four experiments, each in duplicate, ± standard deviations. The differences between strains were significant (P values: EL33 versus EL35, 0.002; EL33 versus EL37, 0.009) as determined by Student's t test.
pLFAR1 containing a translational farAB-lacZ fusion was inserted into the chromosomal proAB site of N. gonorrhoeae.
MtrR binds to the FarR promoter, resulting in regulation of farAB.
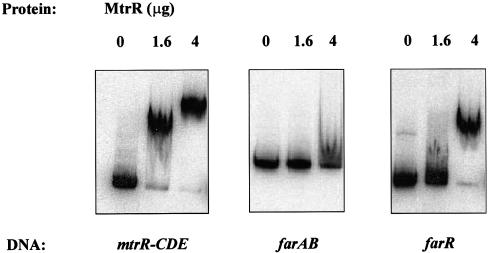
Previously, we observed that MtrR, a repressor of the mtr efflux system (19, 28), was involved in positive regulation of the farAB operon (15). The presence of both MtrR and its DNA-binding activity was shown to be required for the basal level of farAB expression and FA resistance in gonococci. However, because MtrR did not bind to the farAB promoter in a specific manner (Fig. 4), we hypothesized that MtrR may indirectly regulate the farAB operon through its capacity to regulate another gene, perhaps a transcriptional repressor of farAB (15). Accordingly, we asked whether MtrR modulates farR expression. We noted that the putative promoter region for the farR gene contained a sequence (5′-GATTAAAATATAACTATTAA-3′) resembling the mtrR-CDE intervening region. This sequence encompassed the −10 region of the mtrR promoter and the 13-bp inverted repeat sequence (homologous nucleotides are underlined), which was previously shown to be important for MtrR regulation of mtrCDE (12). However, this site only partially overlaps the MtrR-binding site previously identified by Lucas et al. (19), suggesting that the precise nucleotides for MtrR regulation remain unidentified. Therefore, to determine if MtrR binds to the farR promoter, we performed an EMSA using a purified MBP-MtrR fusion protein(19). MBP-MtrR-dependent gel shifts were observed for the mtrR-mtrCDE intergenic and the farR promoter regions (Fig. 4). The specificity of this binding was indicated by the finding that an unlabeled farR promoter DNA sequence could compete with the labeled complexes but that the farAB upstream sequence was unable to compete with this binding (data not shown).
FIG. 4.
MtrR binds to the DNA sequence upstream of farR. Shown is the binding of MBP-MtrR to target DNA sequences, the mtrR-CDE, farAB, and farR promoter regions. Lanes 1 (from left), free labeled probe; lanes 2, probe with 1.6 μg of MBP-MtrR; lanes 3, probe with 4 μg of MBP-MtrR. The probe used is indicated at the bottom.
To ascertain the in vivo effects of MtrR binding to the farR promoter, we constructed an mtrR deletion strain from a derivative of strain FA19 that contained a translational farR-lacZ fusion (strain EL37). The level of β-Gal activity, directly correlating with farR expression, in cells taken at late log phase of growth was assessed. The results demonstrated that deletion of mtrR resulted in a 54% increase in farR-lacZ expression compared with that for the parent strain (Table 3). These results indicate that MtrR negatively regulates farR expression. It is important to emphasize that the modest increase in farR expression due to the loss of MtrR is not an unusual phenomenon for efflux pump operons since inactivation of acrR and mexR was also shown to cause only a small increase in expression of the acrAB and mexAB-oprM efflux pump operons, respectively (20, 29). Taken together, these results suggest that MtrR is involved in positive regulation of farAB expression by downregulating expression of farR.
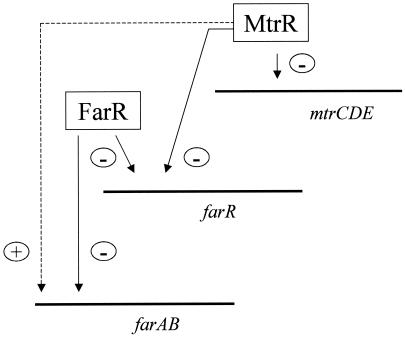
N. gonorrhoeae possesses far- and mtr-encoded efflux pumps to independently meditate gonococcal resistance to host-derived HAs. FarAB belongs to the major facilitator superfamily, while MtrCDE belongs to the resistance/nodulation/division family, of drug efflux pumps. Despite the structural dissimilarities between FarAB and MtrCDE, their expression is related in that both of the efflux operons are regulated by the same transcriptional regulatory protein (MtrR). Previously, Lee and Shafer (15) observed that MtrR was indirectly involved in the positive regulation of farAB whereas MtrR repressed mtrCDE. The results presented herein strongly suggest that MtrR modulates farAB indirectly via regulation of a second gene, farR, which encodes a repressor of farAB (Fig. 5). It is important that, because farAB expression is less than that of mtrCDE, the decreased amount of MtrE, which is shared by both efflux pumps in an MtrR-positive strain (e.g., FA19), is likely to be sufficient for maximal FarAB activity (15).
FIG. 5.
A model for MtrR regulation of farAB and mtrCDE efflux pump operons in N. gonorrhoeae. This model describes the ability of MtrR to positively regulate (+) farAB expression by repressing (−) farR and mtrCDE expression. This MtrR regulatory circuit is most likely to be important in preventing the excess expression of these efflux pumps in gonococci.
The mechanism by which MtrR regulates expression of the far and mtr systems emphasizes two important features. First, the farAB operon can be directly regulated in a negative manner by FarR. Our data also support the notion that this operon can be upregulated by MtrR and by FarR repression of farR (Fig. 5). This is different from many other efflux pump operons, which are directly regulated by an activator (e.g., the MexT activator of mexEF-oprN in P. aeruginosa [13]) or a repressor (e.g., the EmrR repressor of the emrAB operon in E. coli [18]).
The ability of MtrR to regulate mtr and far efflux operons in opposite ways highlights an important feature of gene control in gonococci (Fig. 5) because loss of MtrR repressor activity resulted in increased expression of mtrCDE but decreased expression of farAB. A similar regulatory scheme has been reported for the production of two major porins, OmpF and OmpC, which determine the permeability of the outer membrane in E. coli. The production of OmpF and OmpC is under the control of EnvZ and OmpR, a two-component signal transduction system encoded by the ompB locus. The level of OmpF, which forms a larger pore, relative to that of OmpC was modulated by the status of OmpF phosphorylation in response to environmental conditions (7, 25). Switching between OmpF and OmpC seems to be an important part of bacterial adaptation and survival under stress conditions (26).
N. gonorrhoeae uses the mtr and far efflux pump systems to resist the antimicrobial agents that bathe certain mucosal sites which this organism infects (15, 22, 34). However, overproduction of an efflux pump seems to be detrimental, as gonococcal growth was slowed when the mtr system was overproduced (9). In dealing with this problem, gonococci seem to use MtrR to adjust the total activity of efflux pumps. Our results may also explain why McFarland et al. (22) observed an Mtr-independent mechanism by which gonococci resist fecal lipids since their test strains did not express resistance to HAs such as erythromycin or Triton X-100, which would have required mtrR mutations to cause overexpression of mtrCDE. During rectal infections where gonococci would be confronted with toxic fecal lipids, those strains producing an active MtrR repressor would increase farAB expression due to the ability of MtrR to reduce farR expression. This hypothesis is in keeping with the model described in Fig. 5.
We observed that a DNA sequence upstream of farR resembles the mtrR-CDE intervening region encompassing an inverted repeat sequence. Conventional and competitive EMSA experiments that used a PCR product encompassing the sequence upstream of farR revealed that MtrR could bind to this region. This observation, coupled with the results from β-Gal fusion assays (Table 3), demonstrates that MtrR is a multigene regulator in gonococci. We are now addressing this hypothesis and are attempting to identify other MtrR-regulated genes through a combination of proteomic and genomic approaches. Because an MtrR-like protein was identified as a potential virulence factor in P. aeruginosa (38), it may be that MtrR in gonococci and similar proteins in other bacteria regulate genes involved in virulence.
Acknowledgments
We are grateful to J. Pohl and O. Stuchlik of the Emory Microchemical Facility for their help in FarR purification and N-terminal sequencing of FarR. We are thankful to the Gonococcal Genome Sequencing Project (supported by NIH grant AI-38399) of the University of Oklahoma (B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, T. Dulcey, L. Lewis, and D. W. Dyer) for providing the sequence of the FA1090 genome online. We are also thankful to L. Pucko, S. Katzif, and S. Satola for helping with manuscript preparation and careful reading.
The protein purification and sequencing work performed at the Microchemical Facility was supported by NIH-NCRR grants 02878, 12878, and 13948. Work in our laboratories was supported by NIH grants. AI-21150-17 (W.M.S.) and AI-37945 (R. Lehrer, UCLA Health Sciences Center). J.P.F. was supported by NIH training grant 5T32 AI-07470. W.M.S. is the recipient of a Senior Research Career Scientist Award from the VA Medical Research Service.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. [DOI] [PubMed] [Google Scholar]
- 3.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delahay, R. M., B. D. Robertson, J. T. Balthazar, W. M. Shafer, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic compounds. Microbiology 143:2127-2133. [DOI] [PubMed] [Google Scholar]
- 6.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forst, S., and M. Inouye. 1988. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu. Rev. Cell Biol. 4:21-42. [DOI] [PubMed] [Google Scholar]
- 8.Guymon, L. F., and P. F. Sparling. 1975. Altered crystal violet permeability and lytic behavior in antibiotic-resistant and -sensitive strains of Neisseria gonorrhoeae. J. Bacteriol. 124:757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guymon, L. F., D. L. Walstad, and P. F. Sparling. 1978. Cell envelope alterations in antibiotic-sensitive and -resistant strains of Neisseria gonorrhoeae. J. Bacteriol. 136:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hächler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 12.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhler, T., S. F. Epp, L. K. Curty, and J.-C. Pechère. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181:6300-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lee, E.-H., and W. M. Shafer. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839-845. [DOI] [PubMed] [Google Scholar]
- 16.Lim, D., K. Poole, and N. Strynadka. 2002. Crystal structure of the MexR repressor of the mexAB-oprM multi-drug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 277:29253-29259. [DOI] [PubMed] [Google Scholar]
- 17.Lomovskaya, O., and K. Lewis. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomovskaya, O., K. Lewis, and A. Martin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 21.Martin, R. G., and J. L. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFarland, L., T. A. Mietzner, J. S. Knapp, E. Sandstrom, K. K. Homes, and S. A. Morse. 1983. Gonococcal susceptibility to fecal lipids can be mediated by an mtr-independent mechanism. J. Clin. Microbiol. 18:121-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, P. F., and M. C. Sulavik. 1996. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol. Microbiol. 21:441-448. [DOI] [PubMed] [Google Scholar]
- 25.Mizuno, T., and S. Mizushima. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4:1077-1082. [DOI] [PubMed] [Google Scholar]
- 26.Nikaido, H., and M. Vaara. 1987. Outer membrane, p. 7-22. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.). Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C.
- 27.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 29.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouquette-Loughlin, C., I. Stojiljkovic, T. Hrobowski, J. T. Balthazar, and W. M. Shafer. 2002. Inducible, but not constitutive, resistance of gonococci to hydrophobic agents due to the MtrC-MtrD-MtrE efflux pump requires TonB-ExbB-ExbD proteins. Antimicrob. Agents Chemother. 46:561-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 32.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon of Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafer, W. M., L. F. Guymon, I. Lind, and P. F. Sparling. 1984. Identification of an envelope mutation (env-10) resulting in increased antibiotic susceptibility and pyocin resistance in a clinical isolate of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 25:767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafer, W. M., J. T. Balthazar, K. E. Hagman, and S. A. Morse. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141:904-911. [DOI] [PubMed] [Google Scholar]
- 35.Shafer, W. M., X.-D. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silver, L. E., and V. L. Clark. 1995. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene 166:101-104. [DOI] [PubMed] [Google Scholar]
- 37.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-oprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong, A., A. Gottman, C. Park, M. Baetens, S. Pandza, and A. Martin. 2000. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 44:2905-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziha-Zarifi, I., C. Llanes, T. Kohler, J. C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]