Abstract
The main virulence factor of Vibrio cholerae, the cholera toxin, is encoded by the ctxAB operon, which is contained in the genome of the lysogenic filamentous phage CTXφ. This phage transmits ctxAB genes between V. cholerae bacterial populations that express toxin-coregulated pilus (TCP), the CTXφ receptor. In investigating new forms of ctxAB transmission, we found that V. cholerae filamentous phage VGJφ, which uses the mannose-sensitive hemagglutinin (MSHA) pilus as a receptor, transmits CTXφ or its satellite phage RS1 by an efficient and highly specific TCP-independent mechanism. This is a novel type of specialized transduction consisting in the site-specific cointegration of VGJφ and CTXφ (or RS1) replicative forms to produce a single hybrid molecule, which generates a single-stranded DNA hybrid genome that is packaged into hybrid viral particles designated HybPφ (for the VGJφ/CTXφ hybrid) and HybRSφ (for the VGJφ/RS1 hybrid). The hybrid phages replicate by using the VGJφ replicating functions and use the VGJφ capsid, retaining the ability to infect via MSHA. The hybrid phages infect most tested strains more efficiently than CTXφ, even under in vitro optimal conditions for TCP expression. Infection and lysogenization with HybPφ revert the V. cholerae live attenuated vaccine strain 1333 to virulence. Our results reinforce that TCP is not indispensable for the acquisition of CTXφ. Thus, we discuss an alternative to the current accepted evolutionary model for the emergence of new toxigenic strains of V. cholerae and the importance of our findings for the development of an environmentally safer live attenuated cholera vaccine.
The filamentous phage CTXφ contains the ctxAB genes encoding cholera toxin (CT), the main virulence factor of the pathogenic gram-negative bacterium Vibrio cholerae (49). In toxigenic El Tor and O139 strains of V. cholerae CTXφ is integrated at the dif site in the bacterial genome arrayed in different tandem structures along with the related satellite phage RS1 (11, 39). The genome of RS1 is a short version of the genome of CTXφ, which contains genes encoding proteins needed for replication (RstA), integration (RstB), and regulation of gene expression (RstR and RstC) but lacks the genes encoding proteins needed for assembling and secretion of viral particles (Psh, Cep, pIIICTX, Ace, and Zot), as well as CT, which is not necessary for phage morphogenesis (11). Thus, satellite phage RS1 can replicate autonomously but depends on its helper phage CTXφ for assembly and secretion and thereby for transmission of RS1 viral particles (11). Conversely, RS1 encodes the antirepressor RstC, which is not present in CTXφ (9). This protein promotes transcription of CTXφ and RS1 genes by counteracting the activity of the phage repressor RstR (9). Thus, RS1 enhances transmission of both CTXφ and itself by means of RstC antirepressor activity (9).
Classical strains of V. cholerae contain nonfunctional CTXφ prophages, whereas El Tor and O139 strains contain fully active prophages that produce infective CTXφ viral particles (10). CTXφ site specifically integrates into the host chromosome by a process dependent on the host recombinases XerC and XerD, which ordinarily catalyze the resolution of chromosome dimers at the dif recombination site (25). Other filamentous phages such as f237 of Vibrio parahaemolyticus, φLf of Xanthomonas campestris, Xfφf1 of Xilella fastidiosa, CUSφ-2 of Yersinia pestis, and VGJφ of V. cholerae seem to exploit the XerCD recombination system to integrate into the chromosome of their hosts, suggesting that lysogenic filamentous phages are more common than initially thought (5, 25, 26).
CTXφ infects V. cholerae through toxin-coregulated pilus (TCP) (49), a type IV pilus essential for intestinal colonization (47) that is encoded by a gene cluster contained in the V. cholerae pathogenicity island (VPI) (33). Although VPI seems to move horizontally between bacterial populations of V. cholerae, the transfer mechanism is still controversial. Karaolis et al. presented data suggesting that VPI is the prophage state of another filamentous phage, VPIφ, which they thought to use TcpA as major capsid protein (34); however, this hypothesis has raised several unanswered questions (11), and the results from that study could not be reproduced by other researchers (16). Perhaps VPI transmission is mediated by several mechanisms that follow different pathways; for example, O'Shea and Boyd have found that VPI can be mobilized by the generalized transducing phage CP-T1 (43); however, the main mechanism accounting for VPI transmission probably has not been discovered yet. Whatever the mechanism, VPI, carrying the CTXφ receptor, also has the ability to move horizontally between bacterial populations, providing CTXφ with the advantage of amplifying its host range when VPI moves toward new TCP-negative strains.
An evolutionary model for the origin of pathogenic V. cholerae has been proposed in which this bacterium first acquired the tcp operon and then TCP-producing strains were infected and lysogenized by CTXφ (2, 11, 17, 18, 49). However, this model has been impugned by the isolation of several strains of V. cholerae (both O1 and non-O1) that lack TCP genes but contain the CTXφ prophage (20, 45). It has been suggested that such strains arose by the mentioned model with a subsequent loss of the VPI (17, 18). Another possibility is that such strains have alternative CTXφ receptors (2, 3), but filamentous phages that use more than one receptor have not been described. However, CTXφ can infect V. cholerae in a TCP-independent fashion that requires the TolQ, TolR, and TolA proteins, but the efficiency of infection by this mechanism is quite low and needs direct cell-cell contact (23). Boyd and Waldor demonstrated that a specialized receptor such as TCP is not always essential for acquisition of CTXφ, since these authors showed that V. cholerae generalized transducing bacteriophage CP-T1 can transfer the whole CTXφ genome toward TCP-negative strains of V. cholerae (3). However, the presence of TCP-devoid, non-O1, non-O139 strains of V. cholerae that contain CTXφ prophages cannot be explained by this mechanism, since CP-T1 only infects strains belonging to the O1 serogroup (21).
We recently described a V. cholerae-specific filamentous phage, VGJφ, which infects host cells through the mannose-sensitive hemagglutinin (MSHA) pilus and that is able to integrate its genome at the same attB chromosomal site as CTXφ (5). We describe here a new type of specialized transduction mediated by VGJφ through which CTXφ and its satellite phage RS1 are transmitted to new hosts in a TCP-independent fashion. We also discuss the potential implications of this alternative mechanism for the emergence of new toxigenic serotypes of V. cholerae and for the development of an environmentally safer live attenuated cholera vaccine.
MATERIALS AND METHODS
Phages, strains, and media.
Bacterial strains and phages used in the present study are described in Table 1. Ordinarily, strains were grown in Luria-Bertani (LB) medium at 37°C. Kanamycin was added, when necessary, at 50 μg/ml. The expression of MshA and TcpA proteins was analyzed in the following media: LB medium (pH 6.5), LB medium (pH 7.0), AKI medium (29), tryptone soy broth (TSB), TSB plus 2.5 g of glucose/liter (TSBG), Dulbecco modified Eagle medium (DMEM; Sigma), and protein-free hybridoma medium (PFHM; Gibco-BRL).
TABLE 1.
V. cholerae strains and phages used in this study and susceptibility of V. cholerae strains to phages VGJ-Knφ, HybP-Knφ, and CTX-Knφ
| Straina or phage | Description and remarks | Source, yr or referencec | Susceptibility to phageb:
|
||
|---|---|---|---|---|---|
| VGJ-Knφ | HybP-Knφ | CTX-Knφ | |||
| Strains | |||||
| 569B | O1, classical, Inaba | Calcutta, India, 1945 | +++ | +++ | +++ |
| O395 | O1, classical, Ogawa | Calcutta, India, 1964 | +++ | +++ | ++ |
| 1395 | O1, classical, Inaba | CIEI | +++ | +++ | ++ |
| CA385 | O1, classical, Ogawa | Calcutta, India, 1953 | − | − | + |
| CA401 | O1, classical, Inaba | Calcutta, India, 1953 | +++ | +++ | + |
| CA-EM1 | CA401, ΔmshA | CNIC | − | − | + |
| NIH35A3 | O1, classical, Inaba | India, 1941 | ++ | ++ | ++ |
| C6706 | O1, El Tor, Inaba | Peru, 1992 | +++ | +++ | − |
| KHT46 | C6706 ΔmshA1 | 48 | − | − | − |
| KHT52 | C6706 ΔtcpA10 | 48 | +++ | +++ | − |
| 1333 | C6706 Δ(CTXφ prophage), hap::celA, remnant RS1 | CNIC | +++ | +++ | − |
| C7258 | O1, El Tor, Ogawa: prophage structure: CTXφ/RS1 | Peru, 1991 | +++ | +++ | − |
| C72K7 | C7258, CTXφ/RS1::Knr (inserted in NotI site) | CNIC | NP | NP | NP |
| N16961 | O1, El Tor, Inaba, prophage structure: CTXφ/RS1 | Bangladesh, 1975 | ++ | ++ | − |
| N16K38 | N16961, RS1/CTXφ::Knr (inserted in NotI site)/RS1 | CNIC | NP | NP | NP |
| Peru-15 | C6709 Δ(CTXφ/RS1 prophages, attRS1), rec4::ctxB | 35 | +++ | +++ | +++ |
| E7946 | O1, El Tor, Ogawa | Bahrain, 1978 | +++ | +++ | − |
| Lima | O1, El Tor, Inaba | Lima, Peru, 1991 | +++ | +++ | − |
| 3083 | O1, El Tor, Ogawa | Viet Nam, 1964 | +++ | +++ | − |
| CRC262 | O139 | Bangladesh, 2000 | +++ | +++ | − |
| CRC266 | O139 | Bangladesh, 2000 | +++ | +++ | − |
| Phages | |||||
| VGJφ | Filamentous phage of V. cholerae | 5 | NA | NA | NA |
| VGJ-Knφ | VGJφ carrying a Knr gene inserted into its unique XbaI site | 5 | NA | NA | NA |
| CTX-Knφ | CTXφ carrying a Knr gene inserted into its unique NotI site (produced by strain C72K7) | CNIC | NA | NA | NA |
| RS1-Knφ | RS1 carrying a Knr gene inserted into its unique NotI site (produced by strain N16K38) | CNIC | NA | NA | NA |
| HybP-Knφ | Recombinant hybrid phage produced by site-specific recombination of CTX-Knφ and VGJφ RFs | This study | NA | NA | NA |
| HybRS-Knφ | Recombinant hybrid phage produced by site-specific recombination of RS1-Knφ and VGJφ RFs | This study | NA | NA | NA |
Receptor strains were grown under optimal conditions for expression of TCP, which was checked in parallel by Western blotting.
569B was used as an indicator strain to adjust the titer of each phage suspension to 106 Knr transducing particles (TP)/ml, and then 100 μl of the adjusted suspension (105 Knr TP) was used to infect 108 cells of the receptor strains (multiplicity of infection of 10−3). +++, Appearance of >104 Knr transductants of the infected strain; ++, appearance of 102 to 104 Knr transductants; +, appearance of <102 Knr transductants; −, no Knr CFU detected. NP, not performed; NA, not applicable. These results are from three independent experiments.
CIEI, Centro de Investigación de Enfermedades Infecciosas, Cuernavaca, Mexico; CNIC, Centro Nacional de Investigaciones Cientificas, Havana, Cuba.
Phage methods.
Phage particles of VGJφ, CTX-Knφ, RS1-Knφ, HybP-Knφ, or HybRS-Knφ were purified and concentrated as described in (5). For infection assays, phage donor strains were grown until they reached an optical density (OD) at 600 nm of 1.5. One-milliliter portions of the cultures were filtered through a 0.22-μm-pore-size filter (Sartorius), and 50-μl portions of the filtrates were plated on solid LB medium to check for sterility. Then, 100 μl of pure and diluted cell-free culture supernatants of the donor strains were mixed with 20 μl (∼108 cells) of a fresh culture of the receptor strains grown under optimal conditions for TCP expression. The mixture was incubated 20 min at room temperature to allow infection and plated on solid LB medium supplemented with kanamycin to count kanamycin-resistant (Knr) transductants. Alternatively, infected cells were grown in LB broth to purify phage particles or single-stranded DNA (ssDNA) according to Sambrook et al. (46).
DNA methods.
Total DNA was prepared according to the method of Ausubel et al. (1). Plasmid DNA was prepared by using the WizardPlus SV System (Promega). DNA restriction and modification enzymes were used according to the manufacturer's instructions (Promega). Southern blot analyses were performed with the following digoxigenin (DIG)-labeled probes: a 643-bp fragment containing part of ctxAB genes amplified by PCR with the primer pairs 5′-ATGATCATGCAAGAGGAACTC-3′ and 5′-AGGTGTTCCATGTGCATATGC-3′ was used as the ctxAB-specific probe; the 954-bp SacI-EcoRI fragment of VGJφ RF, containing open reading frame 81 (ORF81), ORF44, ORF29, and part of ORF493, was used as the VGJφ-specific probe (see the genome sequence of VGJφ; GenBank no. AY242528); the 564-bp SacI-SphI fragment containing the rstC gene from plasmid pURS1 (4) was used as the rstC-specific probe; and, finally, the 2.9-kb EcoRI-PstI fragment containing the RS1 element from pURS1 (4) was used as the RS-specific probe.
Strand-specific probes were generated by asymmetric PCR with HybP-Knφ RF as the template. Primer CTB1 (5′-GCGATTGAAAGGATGAAGG-3′), hybridizing with the negative strand of CTXφ and inside ctxB, was used to generate strand-specific HybP-A probe and primer NJ2 (5′-TAGAACGTGTCATTGCATCG-3′), hybridizing with the negative strand of VGJφ and inside ORF136 (AY242528), was used to generate the complementary strand HybP-B probe. Nucleotides were added at the following final concentrations: 100 μM dATP, 100 μM dCTP, 100 μM dGTP, 65 μM dTTP, and 35 μM DIG-dUTP. Amplification reactions were performed with the Taq bead hot-start polymerase system (Promega).
Sequences of the novel junctions between CTXφ and VGJφ in HybP-Knφ were obtained by sequencing its replicative form (RF) with primers CTB1 (see above) for the junction att-CTX/VGJ and NJ4 (5′-CCTGTAGAAATTCCGTCTCC-3′), hybridizing inside the intergenic region I of CTXφ for the junction att-VGJ/CTX. Similarly, the sequence of the novel junctions between VGJφ and RS1 in HybRS-Knφ was obtained by sequencing its RF with the primers NJ5 (5′-CGCTCATCAGGTTCAAAACC-3′) for the att-RS1/VGJ junction and NJ4 for the att-VGJ/RS1 junction. Sequencing reactions were performed with the Thermo Sequenase CyS dye terminator kit and an ALFexpress DNA sequencer (Amersham Pharmacia Biotech).
Stability of HybP-Knφ in the bacterial host.
To evaluate the in vitro stability of HybP-Knφ, V. cholerae 1333 infected with this phage [1333(HybP-Knφ)] was inoculated into LB medium and grown without kanamycin selection until late stationary phase. The ratio of infected cells (Knr) to the total number of bacteria was determined every 2 h for 24 h. At this time, the integrity of HybP-Knφ RF was studied by restriction analysis of plasmid DNA preparations from 12 independent Knr clones.
The in vivo stability of replicating or integrated HybP-Knφ was evaluated in BALB/c suckling mice. Ten mice were orogastrically inoculated with 106 cells of 1333(HybP-Knφ) or 1333-HybI2 (an integrant of HybP-Knφ in 1333) and then incubated 24 h at 30°C without their mothers. The mice were then sacrificed; the small intestines were homogenized to recover colonizing vibrios, and the ratio of Knr cells to the total number of bacteria was determined.
Quantification of CT production.
The ability of 1333(HybP-Knφ) and 1333-HybI2 to produce CT was determined in AKI (29) and LB media by the GM1 ganglioside-dependent enzyme linked immunosorbent assay (GM1-ELISA) (24) by using a standard curve of purified CT and the anti-CTA monoclonal antibody (MAb) 1G10G5.
Virulence evaluation of the HybP-Knφ lysogen 1333-HybI2.
Toxicity was evaluated by orogastric inoculation of groups of 15 BALB/c suckling mice with 106 cells of the strains 1333-HybI2, 1333 (negative control), and C6706 (positive control) diluted in 50 μl of phosphate-buffered saline. Mice were fasted 4 h before and after inoculation and incubated for 6 days with their mothers. The survival was monitored daily during this lapse.
Protein methods.
To determine the protein composition of the hybrid phage capsids, samples of HybP-Knφ (108 particles), HybRS-Knφ (108 particles), or VGJφ as control (109 particles) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (1) with the MAb 2H1H9 specific for the major capsid protein of VGJφ phage.
Expression of MshA and TcpA protein subunits was studied by growing the strains of interest in LB medium (pH 6.5, 30°C), LB medium (pH 7.0, 37°C), TSB (37°C), or TSBG (37°C) with overnight shaking (240 rpm) and by the AKI procedure (29). For growth in DMEM or PFHM, starting suspensions of 106 bacteria/ml in each medium were incubated for 8 h static at 37°C in an atmosphere of 5% CO2. Whole-cell lysates containing ∼2 × 107 bacteria grown in each condition were fractionated by SDS-PAGE and analyzed by Western blot with the anti-TcpA MAb 10E10E1 and the anti-MshA MAb 2F12F1 (15).
The nucleotide sequences of the novel junctions attCTX/VGJ, attVGJ/CTX, attRS1/VGJ, and attVGJ/RS1 have been deposited in the GenBank database under the accession numbers AY268047, AY268048, AY349613, and AY349614, respectively.
RESULTS
CTXφ transmission mediated by VGJφ.
The filamentous phage VGJφ, recently isolated from the O139 V. cholerae strain SG25-1, enters the cell using as receptor the MSHA (5), a type IV pilus that also confers hemagglutinating capacity to cholera vibrios (22). VGJφ integrates at the same chromosomal attB site as CTXφ, and the 17-bp functional core within the attP sites of both phages are almost identical (5). Thus, we hypothesized that since both phage RFs integrate at the same attB site, they could potentially integrate into each other by their respective attP sites if they coexisted in the same cell and that, if such a cointegrate were formed, it could be transduced into a new host cell and disseminated thereafter.
To test this hypothesis, 569B was infected with VGJφ, CTX-Knφ, or coinfected with both phages. The coinfected cells were grown under kanamycin selection to warrant the permanency of CTX-Knφ, whereas VGJφ was self-maintained because it is a very prolific phage and its receptor, MSHA, is constitutively expressed in 569B (see below). Infection assays were done with cell-free culture supernatants of the classical biotype strain 569B infected with VGJφ or CTX-Knφ or coinfected with both phages and by using as receptor strains the TCP mutant KHT52 (48), the isogenic MSHA mutant KHT46 (48), their parental strain C6706, the vaccine strain Peru-15, and the classical biotype strain 569B; the last three strains express both TCP and MSHA (Table 2). As expected, 569B infected with CTX-Knφ transmitted this phage only to TCP-expressing strains not restricted by phage immunity (only to 569B and Peru-15 receptors in Table 2). Although KHT46 and C6706 strains expressed TCP, they were resistant to CTXφ because they contain CTXφ and RS1 prophages expressing RstREl Tor repressor, which confers immunity to CTXφEl Tor infection (36). 569B infected with VGJφ was able to transmit this phage to all strains, except the MSHA mutant KHT46, but this event of transfer never provided Knr to the transductants (Table 2). However, 569B coinfected with CTX-Knφ and VGJφ transduced the Knr gene to all MSHA-expressing strains (KHT52, C6706, Peru-15, and 569B in Table 2). Thus, the Knr gene originally carried by CTX-Knφ was transduced to the TCP mutant KHT52 with great efficiency, acquiring the pattern of transmission of VGJφ. Since KHT52 cannot produce TCP (the receptor of CTX-Knφ), but normally assembles MSHA pilus (the receptor of VGJφ), the appearance of KHT52 Knr transductants could only be explained by some kind of interaction between VGJφ and CTX-Knφ.
TABLE 2.
Transduction of CTX-Knφ and RS1-Knφ to different V. cholerae strains
| Donor strain | No. of Knr transductants of the receptor strainsa:
|
||||
|---|---|---|---|---|---|
| KHT52 (ΔtcpA10) | KHT46 (ΔmshA1) | C6706 (MSHA+ TCP+) | Peru-15 (MSHA+ TCP+) | 569B (MSHA+ TCP+) | |
| 569B (CTX-Knφ) | ND | NDb | NDb | 9.85 × 104 | 2.45 × 105 |
| 569B (VGJφ) | NDc | ND | NDc | NDc | NDc |
| 569B (CTXKnφ/VGJφ) | 1.22 × 105 | NDb | 1.08 × 105 | 8.36 × 104 | 2.07 × 105 |
| 569B (RS1-Knφ) | ND | NDb | NDb | 1.83 × 105 | 2.15 × 105 |
| 569B (RS1-Knφ/VGJφ) | 5.04 × 104 | NDb | 1.02 × 105 | 2.35 × 105 | 2.68 × 105 |
| C72K7 | ND | NDb | NDb | 8.90 × 103 | 9.15 × 103 |
| C72K7 (VGJφ) | 5d | NDb | 21d | 8.90 × 103 | 6.02 × 104 |
| N16K38 | ND | NDb | NDb | 7.85 × 102 | 7.64 × 102 |
| N16K38 (VGJφ) | 3d | NDb | 9d | 1.06 × 103 | 8.04 × 102 |
That is, the number of Knr transductants appearing after exposure of 108 receptor cells to 100 μl of cell-free culture supernatants of the donor. All numbers are average from at least three independent experiments. Receptor strains were grown under optimal conditions for TCP expression. ND, no Knr transductants detected.
Although the TCP is intact in KHT46 and C6706, these strains are not efficiently infected by CTX-Knφ or RS1-Knφ due to phage immunity provided by RstREl Tor repressor present in these strains. Notice that Peru-15 and 569B lack immunity to CTX-Knφ or RS1-Knφ because Peru-15 does not contain any copy of the RstR repressor gene and 569B expresses RstRCl repressor, which belongs to a phage immunity group (classical) different from the immunity group of the CTX-Knφ used in this study (El Tor).
No Knr transductants were detected, but RFs were isolated from the same recipient strains grown without Kanamycin selection.
These colonies were determined to be transductants of HybP-Knφ by Southern blot. A total of 108 cells of the receptor strains plated in LBK did not produce any Knr spontaneous mutant.
Plasmid DNA preparations from 24 Knr transductants of KHT52 from three independent experiments digested with EcoRI, which cuts once into VGJφ RF but does not cut into CTX-Knφ RF, produced two bands of ca. 15.7 and 7.5 kb, respectively (not shown). Analysis by Southern blot with two DIG-labeled probes specific for VGJφ and CTXφ showed that the smaller 7.5-kb band hybridized only with the VGJφ-specific probe while the larger 15.7-kb band hybridized with both the VGJφ- and the CTXφ-specific probes (not shown). These results indicated the simultaneous presence of two plasmids: the VGJφ RF of 7.5 kb and a larger plasmid whose size (15.7 kb) was compatible with a cointegrate structure comprising both VGJφ and CTX-Knφ RFs fused into a single recombinant molecule, thus supporting our initial hypothesis. We designated the larger new plasmid pHybP-Knφ (from hybrid phage). Electroporation of these plasmid DNA preparations (containing both plasmids) into KHT46 allowed us to propagate and purify clones containing pHybP-Knφ alone, giving final confirmation of its existence (VGJφ was washed out due to lack of its receptor, MSHA, in this strain).
Restriction analysis of pHybP-Knφ produced bands characteristic of CTXφ or VGJφ and new bands corresponding to the novel junctions between both phage DNAs (Fig. 1AI). In addition, pHybP-Knφ hybridized with VGJφ- and CTXφ-specific probes in Southern blots (Fig. 1AII and III), indicating that this molecule contained sequences from both phages. These results were absolutely compatible with the theoretic cointegrate structure of pHybP-Knφ derived from our initial hypothesis in which the RFs of VGJφ and CTX-Knφ recombined site specifically through their respective attP sites (Fig. 1B). Finally, DNA sequencing of the novel junctions of pHybP-Knφ confirmed our hypothesis and showed that the genome of both phages were opposite in the hybrid RF (Fig. 1C).
FIG. 1.
Analysis of HybP-Knφ and HybRS-Knφ RF structure. (A) Restriction and Southern analysis of HybP-Knφ RF. In subpanel I, DNA samples of HybP-Knφ RF digested with HindIII (lane 1), EcoRI (lane 2), XhoI (lane 3), EcoRI-BglII (lane 4), and appropriate molecular size markers (lane 5) were electrophoresed in 0.8% agarose gels. In subpanels II and III are shown Southern blot analyses with the ctxAB- and VGJφ-specific probes represented in panel B. The lanes are the same as in subpanel I, except for lane 5, which was loaded with VGJφ RF DNA in subpanel II or with CTX-Knφ RF DNA in subpanel III as negative controls. (B) Structure of pHybP-Knφ. The genes of CTXφ are represented in white, the genes of VGJφ are represented in gray, and the Knr gene is represented in black. The enzymes and probes used to determine pHybP-Knφ structure by restriction and Southern in panel A are shown. The novel junctions between phage DNAs, att-VGJ/CTX and att-CTX/VGJ, are also represented. Notice that the VGJφ and CTXφ genomes are opposite. (C) Nucleotide sequences of attP sites of CTXφ and VGJφ before site-specific recombination and sequences of the novel junctions after recombination, as determined by DNA sequencing. The core sequences of homology are shown in uppercase, with identities boxed. DNA sequences of CTXφ or VGJφ origin are shown in gray or black, respectively, or in white over black for sequences of undetermined origin at the moment of recombination (5). A double-headed arrow delimits the region where the cut and rejoining of the recombination partners took place. (D, E, and F) The same analyses as in panels A, B, and C, respectively, but for HybRS-Knφ. In DIII (lane 5) the negative control loaded was RS1-Knφ RF DNA.
KHT46 carrying pHybP-Knφ transmitted this hybrid molecule to several MSHA-expressing V. cholerae strains, indicating that pHybP-Knφ was generating an ssDNA genome that was being packaged and exported in a new phage particle designated HybP-Knφ. KHT46 produced ca. 107 HybP-Knφ particles/ml/OD unit of culture when the titers were measured by using 569B as receptor strain. In general, the titers of HybP-Knφ ranged from 106 to 108 phage particles/ml/OD unit of cultures, depending on the donor and receptor strain used to measure phage titer (not shown). This value was intermediate between titers of CTXφ (105 to 106) and VGJφ (1010 to 1011) when measured under the same conditions.
RS1 transmission mediated by VGJφ.
The RF of the satellite phage RS1 contains an attP site nearly identical to that of CTXφ; we therefore evaluated whether VGJφ could also transmit the satellite phage RS1 according to the same procedure described above to assess CTXφ transmission. Thus, RS1-Knφ was used in place of CTX-Knφ to coinfect strain 569B, together with VGJφ (Table 2). As occurred with CTX-Knφ, RS1-Knφ was efficiently transmitted to the TCP mutant KHT52 (Table 2), suggesting that a hybrid phage was also formed by recombination between RS1 and VGJφ RFs. Here again, the analysis of several KHT52 transductants by restriction and Southern blotting (not shown) confirmed that the RFs of RS1-Knφ and VGJφ had recombined site specifically to form a hybrid RF designated pHybRS-Knφ, which generated an ssDNA hybrid genome that was packaged and exported in a phage particle designated HybRS-Knφ. The analysis of one representative clone of pHybRS-Knφ purified from KHT46 (as described above for pHybP-Knφ) is shown in Fig. 1D. Sequencing of the novel junctions between RS1 and VGJφ DNAs (Fig. 1F) confirmed the structure of HybRS-Knφ RF and that RS1 and VGJφ DNAs were opposite in the hybrid molecule (Fig. 1E).
VGJφ-dependent transmission of CTXφ and RS1 from their lysogenic state.
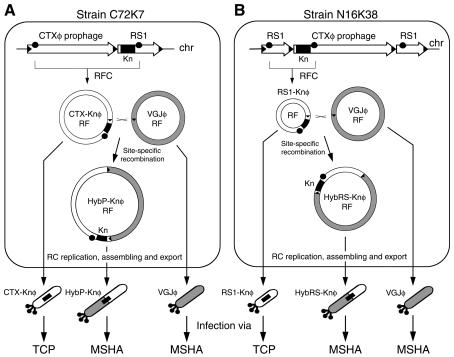
Since the initial source of HybP-Knφ (and HybRS-Knφ) was 569B coinfected with both CTX-Knφ (or RS1-Knφ) and VGJφ, where both phages were replicating, we studied HybP-Knφ and HybRS-Knφ production from El Tor strains with resident Knr-labeled CTXφ or RS1 prophages after infection with VGJφ. Thus, we used strains C72K7 and N16K38, which normally produce CTX-Knφ and RS1-Knφ particles, respectively, from the resident prophages (Fig. 2). When these strains were infected with VGJφ, they detectably produced HybP-Knφ or HybRS-Knφ, albeit with significantly reduced titers (Table 2). However, once the few HybP-Knφ or HybRS-Knφ particles produced by these strains infected another receptor like KHT52, they could replicate and produce large amounts of hybrid phage particles (∼107 particles/ml/OD unit) from the new host.
FIG. 2.
Schematic representation of hybrid phages HybP-Knφ and HybRS-Knφ formation in V. cholerae El Tor strains. (A) Production of the hybrid phage HybP-Knφ in the El Tor strain C72K7 infected with VGJφ. Extrachromosomal CTXφ RF is generated by replication from the chromosome (RFC) by a process dependent on the presence of replication origins (dots) in tandem (40), and then CTXφ and VGJφ RFs recombined site specifically through their respective attP sites (solid arrowheads) to produce HybP-Knφ RF, which generates hybrid phage HybP-Knφ particles. Only hybrid particles can transduce the Knr trait (solid box) by using the MSHA receptor. (B) Production of the hybrid phage HybRS-Knφ in the El Tor strain N16K38 infected with VGJφ by the same mechanism described in panel A for HybP-Knφ. Again, only HybRS-Knφ hybrid particles transduce the Knr trait through the MSHA.
DNA and protein composition of HybP-Knφ and HybRS-Knφ particles.
Genomic ssDNA preparations of HybP-Knφ particles obtained from KHT46(HybP-Knφ) could only be sequenced with oligonucleotides hybridizing with the positive DNA strand of VGJφ or the negative DNA strand of CTXφ; also, this genomic DNA hybridized only with the HybP-A probe, which is specific for the positive strand of VGJφ and the negative strand of CTXφ but not with the reverse probe HybP-B (Fig. 3A to C). These results were identical for HybRS-Knφ (not shown), clearly indicating that the replication machinery of VGJφ prevailed by far over that of CTXφ during replication of the hybrid phages.
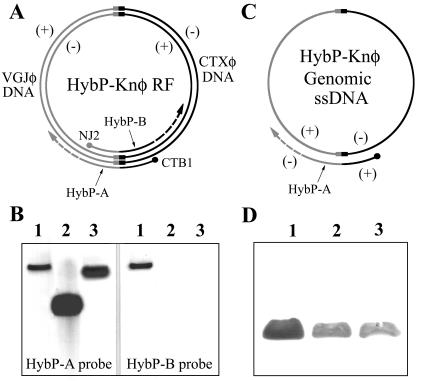
FIG. 3.
Analysis of DNA and protein composition of HybP-Knφ. (A) Schematic representation of the construction of HybP-A and HybP-B strand-specific probes by asymmetric PCR with HybP-Knφ RF as a template and the primers CTB1 and NJ2 (represented by dots). VGJφ and CTXφ DNAs are represented in gray and black, respectively, connected by their respective attP sites. (B) Southern blot analysis of genomic DNA from HybP-Knφ particles with strand-specific probes. Lanes: 1, HybP-Knφ RF linearized with SphI; 2, genomic DNA (positive strand) of VGJφ as a control; 3, genomic ssDNA of phage HybP-Knφ. As expected, both strand-specific probes hybridized with the RF of HybP-Knφ, from where they were generated, but only HybP-A probe hybridized with the ssDNA genome extracted from VGJφ or HybP-Knφ particles. (C) Structure of the ssDNA genome of HybP-Knφ deduced from the Southern results, showing that it is composed of the positive strand of VGJφ and the negative strand of CTXφ. (D) Western blot analysis with MAb 2H1H9 specific for the major capsid protein of VGJφ. Lanes: 1, VGJφ (109 particles); 2, HybP-Knφ (108 particles); 3, HybRS-Knφ (108 particles).
HybP-Knφ and HybRS-Knφ particles profiled by SDS-PAGE to study their protein contents revealed a protein profile identical to that of VGJφ (5). The major capsid protein of VGJφ was evident in silver-stained gels (not shown) when HybP-Knφ or HybRS-Knφ particles (108) were electrophoresed, and its identity was confirmed by reaction with a MAb specific for this protein (Fig. 3D), indicating that the hybrid phage capsid is mostly (if not totally) composed by the same proteins of VGJφ capsid.
Stability of HybP-Knφ.
The time course stability and structural integrity of HybP-Knφ studied during in vitro growth of 1333(HybP-Knφ), in the absence of kanamycin selection, demonstrated that HybP-Knφ preserved the same recombinant structure and remained present in about one-third of the bacterial population until the late stationary phase (24 h) (not shown).
Colonization of 1333(HybP-Knφ) in the suckling mice intestines also supported the permanency of HybP-Knφ in a similar fraction of the bacterial population at 24 h of colonization but, in contrast to the in vitro condition, the in vivo conditions promoted site-specific integration of pHybP-Knφ into an as-yet-unidentified chromosomal site different from the integration site of VGJφ, as revealed by the unique Southern blot banding pattern in a significant number of clones tested (see the analysis of one representative clone in Fig. 4). One of these in vivo isolated Knr clones, 1333-HybI2, extensively cultured in vitro and in vivo showed that all vibrios in the bacterial population retained the Knr trait. Southern blot analysis of 10 clones isolated from both culture conditions revealed the same original prophage structure of 1333-HybI2 (not shown).
FIG. 4.
Site-specific integration of HybP-Knφ into a chromosomal site different from the attRS1. Southern blot analysis of total DNAs of 1333 (lane 1), 1333-HybI2 (lane 2), and HybP-Knφ RF DNA (lane 3) digested with BglII and hybridized with ctxAB (left)- and RS1 (right)-specific probes. The “J” denotes bands containing the novel junctions of HybP-Knφ with the chromosome, and “L” denotes the linearized RF. Note that in the right panel the RS1-specific bands, belonging to the RS1 prophage contained in 1333's chromosome (lane 1) remain identical in 1333-HybI2 after integration of HybP-Knφ (lane 2), indicating that integration occurred by a different locus.
Therefore, the replication and packaging processes used by HybP-Knφ (which are the same used by VGJφ) preserved the structure of the hybrid recombinant genome during cell growth and sustained the inheritance of the hybrid phage by a significant fraction of the cell population in vitro. In vivo, the inheritance of the recombinant phage was guaranteed by the site-specific integration of HybP-Knφ into the bacterial chromosome also in a significant fraction (about one-third) of the bacterial population. Once HybP-Knφ integrated it was stably maintained in the bacterial chromosome.
Expression of CT genes transduced by HybP-Knφ.
1333(HybP-Knφ) and 1333-HybI2 cultured in AKI medium produced 80 to 90 ng of CT/ml as measured by GM1-ELISA. This amount is similar to the CT levels produced by its toxigenic progenitor C6706 in similar condition but contrasted with the null production of CT by the attenuated vaccine strain 1333. Thus, ctxAB genes are actively expressing CT from the replicating and lysogenic state of HybP-Knφ in the AKI condition. Curiously, when CT production was measured in LB medium, 1333(HybP-Knφ) produced 60 to 70 ng of CT/ml; however, 1333-HybI2, as well as the control parental strain C6706, did not produce CT under this condition. Thus, CT expression from HybP-Knφ behaved in a different manner when HybP-Knφ DNA was in the replicative or integrated state showing the same pattern of CT expression from CTXφ (38). These results suggest that ctxAB genes are not under the control of the ToxR regulon when HybP-Knφ is in the replicative state but that they are regulated by this regulatory system when HybP-Knφ is integrated.
HybP-Knφ mediates virulence conversion.
As expected, attenuated strain 1333 was not lethal at all to suckling mice; however, strain 1333-HybI2 inoculated orogastrically into suckling mice at a dose of 106 CFU reproduced the toxigenic behavior of the pathogenic parental C6706 in the kinetics of mouse killing (Fig. 5). At day 6 no mice survived inoculation with 1333-HybI2 or C6706 (Fig. 5), indicating that infection and lysogenization with HybP-Knφ have full potential for virulence reversion of attenuated vaccine strains such as 1333.
FIG. 5.
Time course survival of suckling mice inoculated with 1333, 1333-HybI2, or their parental strain C6706.
HybP-Knφ host range.
The host range of HybP-Knφ was the same of VGJφ (5) and was unaffected by the expression or not of TCP, indicating that HybP-Knφ entry into host cells depended only on the MSHA (Table 1). In addition, HybP-Knφ infected most strains tested more efficiently than CTXφ, even under optimal conditions for TCP expression (Table 1).
MshA versus TcpA expression.
To compare the in vitro relative expression of HybP-Knφ and CTXφ receptors, we analyzed the expression of their major pilus subunits, MshA and TcpA, in classical, El Tor, and O139 strains under different culture conditions by Western blotting. Although in vitro TcpA expression was detected only in restricted conditions and not in all strains, MshA was expressed at similar levels in all strains and conditions tested (Table 3). These results indicated that MSHA is very likely a more ubiquitously expressed receptor than TCP.
TABLE 3.
Comparative expression of TcpA and MshA protein subunits under different growth conditions
| Protein subunit and conditiona | Expression in strainsb
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O1, classical
|
O1, El Tor
|
O139
|
||||||||||||
| 569B | O395 | CA401 | CA385 | NIH35A3 | 1395 | C6706 | C7258 | N16961 | E7946 | 3083 | Lima | CRC262 | CRC266 | |
| MshA | ||||||||||||||
| LB (pH 7, 37°C) | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| LB (pH 6.5, 30°C) | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AKI | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| TSB or TSBG | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| DMEM | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| PFHM | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| TcpA | ||||||||||||||
| LB (pH 7, 37°C) | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LB (pH 6.5, 30°C) | + | + | − | − | − | + | − | − | − | − | − | − | − | − |
| AKI | + | − | − | − | − | − | + | + | + | + | + | + | + | + |
| TSB or TSBG | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| DMEM | + | NP | NP | NP | NP | NP | − | − | − | − | − | − | − | − |
| PFHM | + | NP | NP | NP | NP | NP | + | + | + | + | + | + | + | + |
LB, LB medium.
+, Clearly visible band by Western blotting; −, no band detected. NP, not performed.
DISCUSSION
The present study describes a novel type of specialized transduction for transmission of CTXφ or its satellite phage RS1 between V. cholerae strains, which is mediated by the filamentous phage VGJφ. Our results strongly suggest that this mechanism consists in the site-specific recombination of the RF of VGJφ and CTXφ (or RS1) to form a single-hybrid molecule in a donor V. cholerae cell. This hybrid RF generates an ssDNA genome that is packaged and exported in hybrid phage particles designated HybPφ (for VGJφ/CTXφ hybrid) or HybRSφ (for VGJφ/RS1 hybrid). HybPφ and HybRSφ are recombinant and stable phages whose genomes consist of the positive strand of VGJφ and the negative strand of CTXφ or RS1, respectively, linked by their respective attP sites. Although it is possible that the phages first integrate in tandem in the bacterial chromosome and then excise to form the hybrid phage, this mechanism is very unlikely because (i) integration of VGJφ has not been detected in vitro, a condition in which the hybrid phages are obtained, and (ii) strains in which VGJφ and CTXφ/RS1 are integrated in tandem (5) do not produce hybrid phage particles (data not shown). In addition, the hybrid phages are efficiently produced by classical strains in which VGJφ and CTXφ (or RS1) are replicating in the same host cell (Table 2). However, the hybrid phages are produced at very low levels by El Tor strains of V. cholerae (in which CTXφ and RS1 are integrated) when these strains are infected with VGJφ (Table 2), a finding which suggests that the extrachromosomal RFs of VGJφ and CTXφ (or RS1) are needed for production of the hybrid phages by site-specific recombination.
This mechanism of specialized transduction constitutes a novel TCP-independent pathway for CTXφ or RS1 transmission and dissemination. The processes of replication and assembly of HybPφ and HybRSφ, as well as their capsid, are essentially those of VGJφ; thus, HybPφ and HybRSφ also use MSHA as receptor. The MSHA is probably a more available phage receptor than TCP since expression of MshA protein is not restricted to specific growth conditions, as TcpA is, and the MSHA has been found also in non-O1 non-O139 strains of V. cholerae (14). Thus, a hybrid phage such as HybPφ could have advantage over CTXφ for transmitting the CT genes under certain conditions, such as those found in the environment. Interestingly, HybPφ infected all El Tor and O139 strains tested in the present study more efficiently than CTXφ, even under optimal conditions for TCP expression (Table 1); therefore, phage HybPφ (containing a CTXφEl Tor genome) circumvents phage immunity (36), supporting the idea that the replication machinery of VGJφ is leading replication of HybPφ. Although the differences between the efficiency of infection of classical strains with HybPφ and CTXφ are less pronounced because these strains are not immune to CTXφEl Tor (36), these strains were also infected more efficiently by HybPφ than by CTX-Knφ (Table 1), which likely indicates that HybPφ tranduces CT genes more efficiently than CTXφ, the ordinary vehicle of these genes, in all conditions tested.
CTXφ transmission mediated by a VGJφ-like phage is a plausible explanation for the occasional emergence of non-O1, non-O139 TCP-devoid toxigenic strains of V. cholerae. It is highly probable that other environmentally circulating V. cholerae filamentous phages can potentially transduce CTXφ or RS1 by a mechanism similar to the one described here. As a supporting example, phage fs-2 contains a 715-bp fragment that is highly homologous (97% identity) to part of RS1 satellite phage, flanked at one side by an attRS1 (27), strongly suggesting that fs-2 and RS1 recombined once upon their evolutionary history. In addition, it has been recently found that RS1 is transduced by the filamentous phage KSFφ by an unknown TCP-independent mechanism (19) that perhaps is the same mechanism described in the present study. More recently, we isolated another related filamentous phage, designated VEJφ, from the V. cholerae O139 strain MO45, which is also able to transmit CTXφ by the mechanism described here mediated by VGJφ (results not shown). Phages such as VGJφ and VEJφ seem to be relatively abundant in strains of the O139 serogroup of V. cholerae, since other related filamentous phages have been isolated from strains of this serogroup, such as VSK, fs1, and 493 (30, 31, 32, 42).
Consequently, in Fig. 6 we propose an evolutionary model that could explain the origin of new virulent strains from nontoxigenic environmental strains of V. cholerae or from another related bacterial species. Acquisition of CTXφ phage by a nontoxigenic V. cholerae strain does not automatically create a new pathogen since other factors besides CT are needed for manifestation of the full virulence phenotype (12, 13, 37). Among these factors are TCP, which allows virulent strains to colonize the human small intestine (47), and the lipopolysaccharide, since only two serogroups of V. cholerae (O1 and O139) are known to cause epidemic cholera (7, 28). Therefore, for a phage such as HybPφ to convert a nontoxigenic V. cholerae strain into a full pathogen it is necessary that the phage infect a host cell with adequate virulence factors. One possibility is that these hybrid phages infect V. cholerae strains (non-O1, non-O139) which do not contain CT encoding genes but which contain VPI and thereby the TCP gene cluster and toxT gene, whose product is needed to activate TCP and CT expression. Although, they are not the rule, such strains have been isolated, including some with new variant alleles of toxT and tcpA (6, 20, 41). If such strains additionally have the MSHA gene cluster, they could express this pilus in a broader number of conditions than TCP, giving advantage to a phage such as HybPφ over CTXφ to infect them. Also, the possibility that a phage such as HybPφ infects V. cholerae strains with other unknown virulence attributes, such as a colonization factor different from TCP cannot be absolutely excluded. Furthermore, another possibility for the emergence of new pathogens is that HybPφ-like phages infect other bacterial species expressing MSHA homologous pili where the hybrid phage could integrate and express CT. The homology between the putative adsorption proteins of phages VGJφ and Vf33, which infects the enteropathogen Vibrio parahaemolyticus, suggests that this bacterium species expresses a pilus homologous to MSHA; in fact, VGJφ seems to be more related to Vf33 than to other V. cholerae phages such as CTXφ or fs2 (5).
FIG. 6.
Speculative model for the emerging of new pathogens from V. cholerae (non-O1, non-O139), or another bacterium species, mediated by a phage such as VGJφ. (A) A VGJφ-like phage infects a toxigenic strain of V. cholerae through MSHA pilus. (B) Incoming RF of this phage site specifically recombines with CTXφ RF, producing a hybrid genome, which is packaged and exported in hybrid phage particles such as HybPφ. (C and D) Hybrid particles eventually encountered, infected, and integrated their hybrid genome into the chromosome of an MSHA+ nontoxigenic strain of V. cholerae or into another related host species expressing an MSHA homologous pilus (C) to produce a new genetically stable toxigenic strain (D). The nontoxigenic V. cholerae receptor could be a VPI+ strain, expressing MSHA but not TCP in the environment conditions, such as live attenuated vaccine strains. Although not represented, the inverse situation in which CTXφ infects V. cholerae with a resident phage such as VGJφ would also originate a hybrid phage such as HybPφ. CTXφ phage, its RF, and its receptor (TCP) are represented in white; VGJφ phage, its RF, and its receptor (MSHA) are represented in gray; and HybPφ phage and its RF are represented in gray and white. Chr, bacterial chromosome; RFC, replication from the chromosome; Int., integration; CTXpro, CTXφ prophage; VGJpro, VGJφ prophage.
More tangible is the fact that live attenuated vaccine strains, which contain MSHA and TCP gene clusters, can potentially reacquire ctxAB genes in the environment by infection with a phage such as HybPφ. Infection with HybPφ mediates virulence conversion of the V. cholerae vaccine attenuated strain 1333, in which this phage is able to integrate into the chromosome during in vivo growth in the small intestines of suckling mice, stabilizing the inheritance of CT genes. In contrast to VGJφ and CTXφ, which integrate preferentially at the right flanking end repeat of RS1 (attRS1) (5, 8), HybPφ integrates at an as-yet-unidentified chromosomal site of 1333 (Fig. 4), indicating that the recombinant nature of HybPφ shifts integration specificity. Therefore, previously conceived mutations, such as the deletion of the attRS1 site (44), directed to avoid stable reversion to virulence of live attenuated vaccine strains, could be ineffective to prevent integration of HybPφ-like phages. Thus, it would be desirable that live attenuated vaccine strains do not express MSHA to diminish the risk of virulence reversion in the environment after the release of these strains during massive vaccination campaigns. In addition, it would be desirable that vaccine strains lack any VGJφ-related phage (replicating or lysogenized) to avoid dissemination of CTXφ in case it is reacquired.
The present study emphasizes the importance and versatility of horizontal gene transfer mechanisms for the evolution of bacterial pathogens. To our knowledge, this is the first report of the transmission of one filamentous phage genome by another filamentous phage, which constitutes an example of how mobile genetic elements can subtly interact to expand the pathways for gene transfer. This finding should lead to future studies about the interaction between filamentous phages in particular and between mobile genetic elements in general for horizontal gene transfer in bacteria.
Acknowledgments
We thank Richard A. Finkelstein for providing many of the V. cholerae strains used in this study, Ronald K. Taylor for providing KHT46 and KHT52 mutant strains, G. B. Nair for strains CRC262 and CRC266, Risset Silvera for technical assistance, and Aminael Sánchez for technical assistance and critical review of the manuscript.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith., and K. Struhl. 1995. Short protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Boyd, E. F., B. M. Davis, and B. Hochhut. 2001. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 9:137-144. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. F., and M. K. Waldor. 1999. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: generalized transduction of CTXφ by bacteriophage CP-T1. Infect. Immun. 67:5898-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos, J., R. Fando, A. Silva, B. L. Rodriguez, and J. A. Benitez. 1998. Replicating function of the RS1 element associated with Vibrio cholerae CTX φ prophage. FEMS Microbiol. Lett. 164:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Campos, J., E. Martínez, E. Suzarte, B. L. Rodríguez, K. Marrero, T. Ledón, R. Del Sol, and R. Fando. 2003. VGJφ, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTXφ. J. Bacteriol. 185:5685-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitnis, D. S., K. D. Sharma, and R. S. Kamat. 1982. Role of somatic antigen of Vibrio cholerae in adhesion to intestinal mucosa. J. Med. Microbiol. 15:53-61. [DOI] [PubMed] [Google Scholar]
- 8.Davis, B. M., H. H. Kimsey, W. Chang, and M. K. Waldor. 1999. The Vibrio cholerae O139 Calcutta bacteriophage CTXφ is infectious and encodes a novel repressor. J. Bacteriol. 181:6779-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, B. M., H. H. Kimsey, A. V. Kane, and M. K. Waldor. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21:4240-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, B. M., K. E. Moyer, E. F. Boyd, and M. K. Waldor. 2000. CTX prophages in classical biotype Vibrio cholerae: functional phage genes but dysfunctional phage genomes. J. Bacteriol. 182:6992-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, B. M., and M. K. Waldor. 2003. Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 6:35-42. [DOI] [PubMed] [Google Scholar]
- 12.DiRita, V. J., N. C. Engleberg, A. Heath, A. Miller, J. A. Crawford, and R. Yu. 2000. Virulence gene regulation inside and outside. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehara, M., S. Shimodori, F. Kojima, Y. Ichinose, T. Hirayama, M. J. Albert, K. Supawat, Y. Honma, M. Iwanaga, and K. Amako. 1997. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol. Lett. 154:293-301. [DOI] [PubMed] [Google Scholar]
- 15.Falero, G., B. L. Rodriguez, T. Valmaseda, M. E. Perez, J. L. Perez, R. Fando, A. Robert, J. Campos, A. Silva, G. Sierra, and J. A. Benitez. 1998. Production and characterization of a monoclonal antibody against mannose-sensitive hemagglutinin of Vibrio cholerae. Hybridoma 17:63-67. [DOI] [PubMed] [Google Scholar]
- 16.Faruque, A. S., J. Zhu, Asadulghani, M. Kamruzzaman, and J. J. Mekalanos. 2003. Examination of diverse toxin-coregulated pilus-positive Vibrio cholerae strains fails to demonstrate evidence for Vibrio pathogenicity island phage. Infect. Immun. 71:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque, S. M., Asadulghani, M. N. Saha, A. R. Alim, M. J. Albert, K. M. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXφ: molecular basis for origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faruque, S. M., M. Kamruzzaman, Asadulghani, D. A. Sack, J. J. Mekalanos, and G. B. Nair. 2003. CTXφ-independent production of the RS1 satellite phage by Vibrio cholerae. Proc. Natl. Acad. Sci. USA 100:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh, C., R. K. Nandy, S. K. Dasgupta, G. B. Nair, R. H. Hall, and A. C. Ghose. 1997. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb. Pathog. 22:199-208. [DOI] [PubMed] [Google Scholar]
- 21.Guidolin, A., and P. A. Manning. 1985. Bacteriophage CP-T1 of Vibrio cholerae: identification of the cell surface receptor. Eur. J. Biochem. 153:89-94. [DOI] [PubMed] [Google Scholar]
- 22.Hanne, L. F., and R. A. Finkelstein. 1982. Characterization and distribution of the hemagglutinins produced by Vibrio cholerae. Infect. Immun. 36:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilpern, A. J., and M. K. Waldor. 2000. CTXφ infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 182:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmgren, J. 1973. Comparison of the tissue receptors for Vibrio cholerae and Escherichia coli enterotoxins by means of gangliosides and natural cholera toxoid. Infect. Immun. 8:851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber, K. E., and M. K. Waldor. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656-659. [DOI] [PubMed] [Google Scholar]
- 26.Iida, T., K. Makino, H. Nasu, K. Yokoyama, K. Tagomori, A. Hattori, T. Okuno, H. Shinagawa, and T. Honda. 2002. Filamentous bacteriophages of vibrios are integrated into the dif-like site of the host chromosome. J. Bacteriol. 184:4933-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikema, M., and Y. Honma. 1998. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144:1901-1906. [DOI] [PubMed] [Google Scholar]
- 28.Iredell, J. R., U. H. Stroeher, H. M. Ward, and P. A. Manning. 1998. Lipopolysaccharide O-antigen expression and the effect of its absence on virulence in rfb mutants of Vibrio cholerae O1. FEMS Immunol. Med. Microbiol. 20:45-54. [DOI] [PubMed] [Google Scholar]
- 29.Iwanaga, M., and K. Yamamoto. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jouravleva, E. A., G. A. McDonald, C. F. Garon, M. Boesman-Finkelstein, and R. A. Finkelstein. 1998. Characterization and possible functions of a new filamentous bacteriophage from Vibrio cholerae O139. Microbiology 144:315-324. [DOI] [PubMed] [Google Scholar]
- 31.Jouravleva, E. A., G. A. McDonald, J. W. Marsh, R. K. Taylor, M. Boesman-Finkelstein, and R. A. Finkelstein. 1998. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect. Immun. 66:2535-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kar, S., R. K. Ghosh, A. N. Ghosh, and A. Ghosh. 1996. Integration of the DNA of a novel filamentous bacteriophage VSK from Vibrio cholerae 0139 into the host chromosomal DNA. FEMS Microbiol. Lett. 145:17-22. [DOI] [PubMed] [Google Scholar]
- 33.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 35.Kenner, J. R., T. S. Coster, D. N. Taylor, A. F. Trofa, M. Barrera-Oro, T. Hyman, J. M. Adams, D. T. Beattie, K. P. Killeen, and D. R. Spriggs. 1995. Peru-15, an improved live attenuated oral vaccine candidate for Vibrio cholerae O1. J. Infect. Dis. 172:1126-1129. [DOI] [PubMed] [Google Scholar]
- 36.Kimsey, H. H., and M. K. Waldor. 1998. CTXφ immunity: application in the development of cholera vaccines. Proc. Natl. Acad. Sci. USA 95:7035-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klose, K. E. 2001. Regulation of virulence in Vibrio cholerae. Int. J. Med. Microbiol. 291:81-88. [DOI] [PubMed] [Google Scholar]
- 38.Lazar, S., and M. K. Waldor. 1998. ToxR-independent expression of cholera toxin from the replicative form of CTXφ. Infect. Immun. 66:394-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253-263. [DOI] [PubMed] [Google Scholar]
- 40.Moyer, K. E., H. H. Kimsey, and M. K. Waldor. 2001. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXφ. Mol. Microbiol. 41:311-323. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakasone, N., Y. Honma, C. Toma, T. Yamashiro, and M. Iwanaga. 1998. Filamentous phage fs1 of Vibrio cholerae O139. Microbiol. Immunol. 42:237-239. [DOI] [PubMed] [Google Scholar]
- 43.O'Shea, Y. A., and E. F. Boyd. 2002. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol. Lett. 214:153-157. [DOI] [PubMed] [Google Scholar]
- 44.Pearson, G. D., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90:3750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Said, B., H. R. Smith, S. M. Scotland, and B. Rowe. 1995. Detection and differentiation of the gene for toxin co-regulated pili (tcpA) in Vibrio cholerae non-O1 using the polymerase chain reaction. FEMS Microbiol. Lett. 125:205-209. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, N.Y.
- 47.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]