Abstract
Dengue virus (DENV) infections are vectored by mosquitoes and constitute one of the most prevalent infectious diseases in many parts of the world, affecting millions of people annually. Current treatments for DENV infections are nonspecific and largely ineffective. In this study, we describe the adaptation of a high-content cell-based assay for screening against DENV-infected cells to identify inhibitors and modulators of DENV infection. Using this high-content approach, we monitored the inhibition of test compounds on DENV protein production by means of immunofluorescence staining of DENV glycoprotein envelope, simultaneously evaluating cytotoxicity in HEK293 cells. The adapted 384-well microtiter-based assay was validated using a small panel of compounds previously reported as having inhibitory activity against DENV infections of cell cultures, including compounds with antiviral activity (ribavirin), inhibitors of cellular signaling pathways (U0126), and polysaccharides that are presumed to interfere with virus attachment (carrageenan). A screen was performed against a collection of 5,632 well-characterized bioactives, including U.S. Food and Drug Administration–approved drugs. Assay control statistics show an average Z' of 0.63, indicative of a robust assay in this cell-based format. Using a threshold of >80% DENV inhibition with <20% cellular cytotoxicity, 79 compounds were initially scored as positive hits. A follow-up screen confirmed 73 compounds with IC50 potencies ranging from 60 nM to 9 μM and yielding a hit rate of 1.3%. Over half of the confirmed hits are known to target transporters, receptors, and protein kinases, providing potential opportunity for drug repurposing to treat DENV infections. In summary, this assay offers the opportunity to screen libraries of chemical compounds, in an effort to identify and develop novel drug candidates against DENV infections.
Introduction
The dengue viruses (DENV) constitute a single species within the genus Flavivirus of the Flaviviridae family and are transmitted to humans through infected mosquitoes. DENV causes dengue fever, an often severe flu-like illness, and the more serious dengue hemorrhagic fever. DENV contains a single-stranded, positive-sense RNA genome of ∼10 kb in length, which encodes three structural proteins involved in particle formation (capsid [C], membrane [prM], and envelope [E]) and seven nonstructural proteins (NS) involved in viral replication (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5).1 The myriad strains of DENV can differ from each other by as much as 35% at the nucleotide level, but all strains belong to one of four serotypes (DENV-1–4),2 based on antigenic criteria. DENV enters cells by receptor-mediated endocytosis and replicates in the cytoplasm.
Over the past 30 years, infection with DENV has emerged as one of the most significant health risks in tropical and subtropical regions worldwide, with upward of 50 million people infected yearly.3 Further, according to the World Health Organization, two-fifths of the world's population is at risk for infection with DENV.4 Presently, no vaccine or specific antiviral agent is approved for use and the need to develop novel interventional strategies is pressing. Several recent reports have identified small-molecule compounds with anti-DENV activity, including ribavirin,5 mycophenolic acid,6 N-nonyl-deoxynojirimycin,7 7-deaza-2'-C-methyl-adenosine,8 6-O-butanoyl castanospermine,9 Geneticin,10 and an adenosine analog NITD008.11 However, only few of these compounds appear to have the pharmacologic properties appropriate for a commercial drug and no drug against DENV has yet shown efficacy in a clinical trial.
To date, most antiviral drug candidates target viral gene products and their use typically results in the generation of mutant drug-resistant virus. In contrast, the targeting of host factors may reduce or avoid the generation of resistance and allow the development of more durable therapies. Recently, considerable investigation has helped elucidate host–virus interactions, which occur during DENV infection and pathogenesis,12–14 although understanding of the cellular and molecular mechanisms involved remains far from complete.
Traditionally, antiviral drug development has employed cytopathic and plaque reduction assays to screen and evaluate compound efficacy.15 Plaque reduction assays are labor intensive and not amenable to screen large numbers of compounds, because of their low-throughput nature. Various methods to evaluate antiflaviviral efficacy and cytotoxicity have been developed, and these methods have been adapted to 96-well and, in some cases, 384-well microtiter plate formats from low- to high-throughput applications.16–19 To rapidly identify new antiviral drugs against DENV, screening strategies must be robust and able to evaluate large libraries of compounds for both antiviral efficacy and cytotoxicity. One of the most powerful tools available for drug discovery is high-content screening, which combines high-throughput screening (HTS) with the ability to collect cellular images of biological processes. Automated microscope technology has now advanced to the point where images from microtiter plates can be rapidly visualized and recorded with clarity.20–22 Changes in virus quantity or cell viability can be extracted from the acquired images and quantified using sophisticated analysis tools. When performed in parallel at a primary screening level, compounds can be rapidly evaluated for antiviral activity and cytotoxicity in living cells. Once successfully integrated, high-content technology can minimize both lead times for candidate generation and the probability of failures in eventual clinical trials.
In this article, we describe adaptation, validation, and screening of a high-content assay against a chemical library of known drugs and bioactives. Our assay utilizes a multiparametric readout to directly monitor viral replication in host cells, simultaneously evaluating cell viability to rapidly score compounds for activity. In the screening of a well-characterized collection of bioactives with our high-content assay, we identified hits that target several prominent classes of cellular factors including transporters, receptors, and enzymes, potentially allowing for repurposing of use as a new indication for antiviral therapies against DENV infections.
Materials and Methods
Cells, Virus, and Antibodies
The cell line human embryonic kidney HEK293 (Microbix) was grown at 37°C in an atmosphere containing 5% CO2. HEK293 cells were propagated in complete growth medium containing minimal essential Eagle's medium supplemented with 10% heat inactivated fetal bovine serum, l-glutamine, nonessential amino acids, 100 unit/mL penicillin, and 100 g/mL streptomycin. All cell culture supplies were from Fisher Scientific and Invitrogen. DENV-2 strain New Guinea C was purchased from American Type Culture Collection (ATCC, Catalog No. VR-222). DENV-2 virus stocks were propagated in C6/36 mosquito cells. Virus stock was concentrated and purified by centrifugation through a 20% sorbitol cushion at 28,000 rpm for 2 h in a SW28 rotor (Beckman). Following centrifugation, virus was resuspended in 1/100 of the original volume in Dulbecco's modified Eagle's medium with 0.1% bovine serum albumin, and aliquots were stored at −80°C. Titers of viral stocks were determined on Vero cells by immunostaining of foci. Briefly, serial dilutions of viral stocks were used to infect confluent monolayers of Vero cells. At 5 days postinoculation, cells were fixed with 4% paraformaldehyde (w/v) for 30 min. For immunostaining, cells were washed twice with 1 × phosphate-buffered saline (PBS) and then blocked for 30 min with 2% normal goat serum and 0.4% Triton X-100 (v/v). Mouse monoclonal α-E (4G2)23 purchased from ATCC (Catalog No. HB-112) was added to the assay plates at 0.4 μg/mL for 1 h, followed by three washes with 1 × PBS. To detect α-E, horse radish peroxidase–conjugated goat α-mouse IgG at 0.4 μg/mL (Santa Cruz Biotechnology) was added and incubated for 1 h, followed by 1 × PBS wash. Immunoreactive foci were stained with Vector VIP peroxidase substrate kit (Vector Labs).
Liquid Dispensing and Automation
Several different liquid-dispensing devices were used in this study. Compounds were plated and transferred using a custom-designed 384-head on a PP-384-M Personal Pipettor (Apricot Designs). The addition of cell and virus suspensions was performed using the Multidrop 384 (Thermo), and assay plates (Corning) were incubated in the Steri-Cult (Thermo) under controlled humidity and an atmosphere of 5% CO2 at 37°C. Cell fixation and immunostaining were performed using the Biotek ELx405 washer (Biotek).
Assay Miniaturization into 384-Well Microtiter Plate Format
The DENV infection assay was adapted for automated HTS. First, the assay was miniaturized from 96-well to 384-well microtiter plate format by determining optimal cell growth kinetics. HEK293 cell suspensions were dispensed into 384-well microtiter plates containing vehicle-only control with 5 μL of 10% dimethyl sulfoxide (DMSO; v/v) to measure compound carrier effect. The final concentration of DMSO vehicle was 1% (v/v). Cells were seeded at cell densities ranging from 125, 250, 500, 1,000, 2,000, 4,000 to 6,000 cells per well in 45 μL complete growth medium. At 48, 72, and 96 h postseeding, cells were fixed with 4% paraformaldehyde (w/v) for 20 min and stained for nuclei with solution containing 0.05% Triton X-100 (v/v) with 10 μM Hoechst for 10 min without any wash steps. Plates were imaged on the IN CELL Analyzer 1000 (INCA1000; GE Healthcare) using a 10 × magnifying objective allowing for four images per well, covering 70% of the well, and analyzed using Developer 1.7 for Hoechst-stained nuclei. To complete assay development, the DENV infection assay was run in an automated mode against a panel of compounds with reported antiviral activity. HEK293 cells were seeded at 4,000 cells per well in 35 μL of inoculation medium containing minimal essential Eagle's medium with 2% fetal bovine serum, l-glutamine, nonessential amino acids, 100 unit/mL penicillin, and 100 g/mL streptomycin. Compounds were tested in dose response with doubling dilution series in 10% DMSO (v/v) made in an intermediate 384-well polypropylene plate (ABgene) and 5 μL was transferred to the assay plates. Immediately after compound addition, 10 μL of inoculation medium containing DENV-2 at a multiplicity of infection of 0.5 was dispensed onto the assay plates. After 48-h incubation, cells were fixed as described previously. For immunostaining, cells were washed twice in 1 × PBS and then blocked for 1 h with blocking solution containing 2% normal goat serum (v/v) and 0.4% Triton X-100 (v/v). Mouse monoclonal antibody 4G2 against DENV glycoprotein envelope (DENV E) (α-E) was added to the assay plates for 1 h, followed by three washes with wash buffer containing 0.2% bovine serum albumin (v/v) and 0.2% Triton X-100 (v/v). To detect α-E, AlexaFluor-488 antibody (Invitrogen; A11029) at 1:1,000 dilution was added and incubated for 1 h, followed by three washes with wash buffer. Cells were then nuclei stained with Hoechst as described previously. Plates were imaged on the IN CELL Analyzer 3000 (INCA3000; GE Healthcare) using a 40 × magnifying objective allowing for four images per well, covering 45% of the well, and analyzed using Raven 1.0 software for DENV E and Hoechst-stained nuclei. As a quality control, several control wells were imaged on the INCA3000 using a 40 × magnifying objective allowing for nine images per well, covering 90% of the well, to assess mechanical stress as a result of automated wash cycles.
Chemical Libraries
The library used for the screen combines 5,632 chemicals obtained from MicroSource, Prestwick, Tocris, and other commercial sources. The MicroSource Library contains 2,000 biologically active and structurally diverse compounds from known drugs, experimental bioactives, and pure natural products. The library includes a reference collection of 160 synthetic and natural toxic substances (previously characterized as being inhibitors of DNA/RNA synthesis, protein synthesis, cellular respiration, and membrane integrity), a collection of 80 compounds representing classical and experimental pesticides, herbicides, and endocrine disruptors, and a singular collection of 720 natural products and their derivatives. The latter collection includes simple and complex oxygen heterocycles, alkaloids, sequiterpenes, diterpenes, pentercyclic triterpenes, sterols, and many other diverse representatives. The Prestwick Chemical Library is a unique collection of 1,119 high-purity chemical compounds, all off patent and selected for structural diversity and broad spectrum, with established activities in therapeutic areas including neuropsychiatry, cardiology, immunology, anti-inflammatory, and analgesia, with known safety and bioavailability in humans. Approximately 90% of the library consists of marketed drugs and 10% of bioactive alkaloids or related substances.24
Screen of Chemical Library with DENV Infection Assay
HEK293 cells were seeded at 4,000 cells per well in 35 μL of inoculation medium into a 384-well microtiter plate. Compounds were preplated in an intermediate 384-well polypropylene plate at 100 μM in 10% DMSO (v/v) and 5 μL was transferred to the assay plates. Immediately after compound addition, 10 μL of inoculation media containing DENV-2 at a multiplicity of infection of 0.5 was dispensed onto the assay plates and compounds were tested at a final concentration of 10 μM in 1% DMSO (v/v). After 48-h incubation, cells were fixed and immunostained for DENV E using the procedure described above. Images were acquired using INCA3000 and analyzed using Raven 1.0 software for DENV E and Hoechst. The chemical library was plated in thirty-two 384-well assay plates and required 48 h for imaging on the INCA3000.
Dose–Response Studies
Cell seeding, immunostaining, and plate imaging were performed using a similar procedure as described above. Serial doubling dilutions of compounds were preplated in an intermediate 384-well polypropylene plate and 5 μL was transferred to the assay plates. Compounds were tested in duplicate at a final concentration between 5 nM and 10 μM in 1% DMSO (v/v). Dose–response curves for each dataset were fitted separately using a logistic four-parameter equation as part of HTS Core's Oncology Research Informatics System or Sigmaplot (SYSTAT). The IC50 and EC50 values obtained were averaged from the duplicates. For compounds having an IC50 value of <1 μM, the dose–response study was repeated using dilutions starting at 1 μM for a more accurate determination.
Z′ Factor
The Z′ factor was used to assess assay performance. The Z′ factor constitutes a dimensionless parameter that ranges from 1 (infinite separation) to <0. It is defined as follows: Z′ = 1− (3σc+ + 3σc−)/|μc+ − μc−|, where σc+, σc−, μc+, and μc− are the standard deviations (σ) and averages (μ) of the high (c+) and low (c−) control.25 Z′ factor between 0.5 and 1 indicates an excellent assay with good separation between controls. Z′ factor between 0 and 0.5 indicates a marginal assay and <0 signifies a poor assay with overlap between controls.
Image Acquisition, Data Analysis, and Screening Data Management
Cell titration studies were performed on the INCA1000 epifluorescent microscope system. The INCA1000 is a rapid imaging platform built around the Nikon automated microscope and integrated with a high-resolution charge-coupled device camera. Images were captured at 364 nm excitation/450 nm emission in the blue channel for Hoechst-stained nuclei, with an exposure time of 100 ms. Four images per well were collected using a 10 × magnifying objective and required 3 s per well, with a total imaging time of 20 min for a complete 384-well microtiter plate. Images were analyzed using the Developer 1.7 software's built in segmentation algorithm. The segmentation algorithm determines nuclei count based on the number of objects with Hoechst pixel intensities above background and required a total analysis time of 15 min for a complete 384-well microtiter plate. Image acquisition for assay development and screening was performed on the automated laser confocal INCA3000 microscope system. This laser scanning confocal imager comprises two laser light sources, three excitation lines, and three highly sensitive 12-bit charge-coupled device cameras allowing simultaneous imaging of three fluorophores with continuous laser-based autofocus. Image acquisition was captured at the following wavelengths: 364 nm excitation/450 nm emission in the blue channel for Hoechst-stained nuclei and 488 nm excitation/535 nm emission in the green channel for DENV E with an exposure time of 1.5 ms. Four images per well were collected using a 40 × magnifying objective and required 14 s per well, with a total imaging time of 90 min for a complete 384-well microtiter plate. Images were analyzed using the Raven 1.0 software's built-in object intensity analysis module. The object intensity analysis module uses a segmentation algorithm to count nuclei based on number of objects with Hoechst pixel intensities above background. For DENV E intensity per well, a cell masked overlay was generated using nuclei as a marker and intensity was measured within the defined boundaries. Analysis of Hoechst-stained nuclei and DENV E required 20 min for a complete 384-well microtiter plate. Analysis data files were loaded into Oncology Research Informatics System, a custom-built suite of modules for compound registration, plating, and data management and powered by ChemAxon Cheminformatic tools (ChemAxon). Data were analyzed and the summary of the identified positives was exported as structure data files for further reporting. The DENV E and nuclei inhibition was expressed as percentage compared with high and low controls, defined as % Inhibition = (high control average – read value)/(high control average – low control average) × 100. Compounds that resulted in >20% reduction of nuclei count when compared with controls were defined as cytotoxic.
Classification and Pathway Analysis
To classify the hits, an extensive literature search was performed to find relevant publications and other supporting information on those compounds. Ingenuity's Pathways Analysis software was also used to extract information about the interaction of these compounds with major canonical pathways and search for its potential targets.
Results
High-Content Assay for Inhibitors of DENV Infection
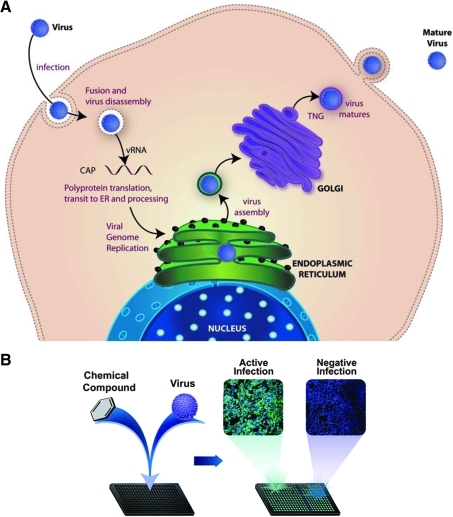
To identify inhibitors of DENV infection, we adapted a cell-based assay focusing on a high-content platform for screening of small-molecule libraries. We infected human HEK293 cells with DENV-2 immediately after compound addition, and 2 days later measured the amount of viral protein produced in the cells. Inoculation of HEK293 cells with DENV-2 results in the production of large amounts of viral DENV E, localized to intracellular membrane compartments, which can be detected by immunofluorescence. Previous studies have confirmed that the level of DENV E is reflective of the level of infectious virus production and that input virus does not produce a detectable level of signal at 2 days postinoculation (data not shown). A reduction in DENV E protein production could potentially be an indication of reduced viral entry, genome replication, and processing (Fig. 1A) and have been shown in similar studies on flaviviruses.19 Compounds that inhibit infection can therefore be determined by immunofluorescence of DENV E, compared with controls (Fig. 1B). Immunofluorescence detection of DENV E allows for direct imaging and quantification on an automated microscope.
Fig. 1.
High-content assay workflow. (A) Life cycle of DENV in host cells. (B) Design schematic of DENV infection assay. HEK293 cells (4,000 cells per well) are treated with compound, followed by DENV-2 inoculation at an MOI of 0.5, with antiviral activity and cytotoxicity evaluated at 48 h postinfection. DENV is detected by fluorescent antibody–based detection of the DENV-2 structural glycoprotein envelope (α-E). Compounds that inhibit viral replication show little or no green florescent signal. Cytotoxicity of compounds is evaluated by measuring a reduction in number of nuclei per well. Plates were fixed and stained as described in the Materials and Methods section. Images were acquired using the INCA3000, with green channel for DENV E and blue channel for detection of Hoechst-stained nuclei. DENV, dengue virus; DENV E, Dengue virus glycoprotein envelope; INCA3000, IN CELL Analyzer 3000; MOI, multiplicity of infection.
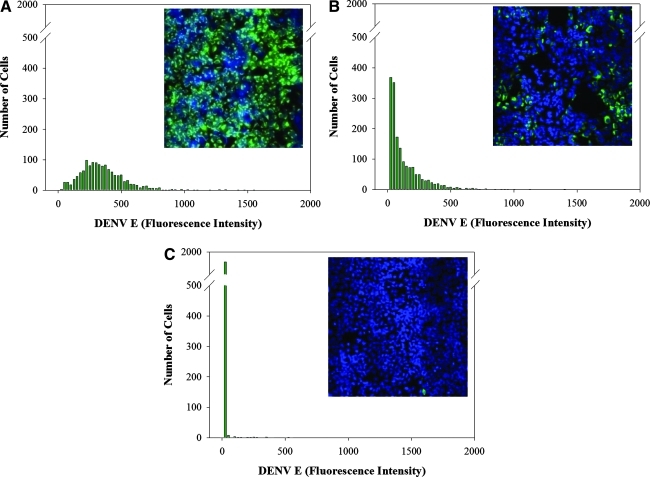
An advantage of this high-content approach is that it allows for compounds to be scored on the fly using a multiparameter readout for DENV inhibition by the amount of viral protein detected, and cell cytotoxicity by the relative number of nuclei present after treatment. Using the INCA3000 built-in analysis module, we first identified the nuclei of each cell using an object recognition tool based on pixel gradient in the blue Hoechst channel. To assess DENV infection, a cell mask was applied over the nuclei and total pixel intensity in the green channel was assessed. A cell-by-cell distribution plot was generated for each well and green pixel intensity was calculated to determine the level of DENV E present at 48 h after treatment with the compound. Cell-by-cell distribution plots are distinct depending on the amount of antiviral activity as depicted by vehicle-only control with 1% DMSO (v/v) (Fig. 2A), a moderately effective inhibitor (Fig. 2B), and the complete inhibitor carrageenan (Fig. 2C). Images of these cells are shown in the insets of Figure 2 to illustrate varying levels of DENV inhibition. To evaluate cytotoxicity, total nuclei count per well in test sample was compared with vehicle-only control (1% DMSO, v/v) samples. A reading of >20% loss in nuclei was scored as cytotoxic.
Fig. 2.
High-content image acquisition and data analysis. Analysis of images acquired from INCA3000. HEK293 cells (4,000 cells per well) treated with compound and inoculated with DENV-2 (MOI of 0.5) were evaluated for antiviral activity at 48 h postinfection. (A) Distribution plot and image of DENV E fluorescence intensity in a well treated with vehicle-only 1% dimethyl sulfoxide (v/v). (B) Distribution plot and image of DENV E fluorescence intensity in a well treated with 10 μM of moderately effective inhibitor. (C) Distribution plot and image of DENV E fluorescence intensity in a well treated with 4 μg/mL of carrageenan. Plates were fixed and stained as described in the Materials and Methods section. Images were acquired using INCA3000, with green channel for DENV E and blue channel for detection of Hoechst-stained nuclei.
Assay Adaptation to 384-Well Microtiter Plate Format
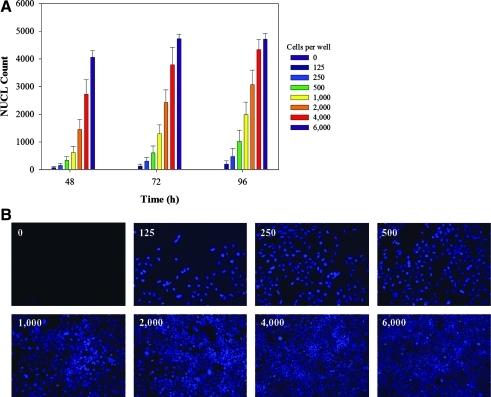
As a first step toward automated HTS, the assay was formatted for use in 384-well microtiter plates. We investigated the growth kinetics of HEK293 cells to determine optimal seeding conditions for the miniaturized 384-well microtiter plate format. Cells were seeded at densities between 125 and 6,000 cells per well and monitored by Hoechst-stained nuclei imaging at 48, 72, and 96 h postseeding. At seeding densities between 125 and 500 cells per well, cells did not grow or showed slow recovery, indicating subpar cultivation conditions (Fig. 3A). Between 1,000 and 6,000 cells per well were recovered, which showed linear growth kinetics indicating optimal seeding conditions. Seeding density was selected at 4,000 cells per well for overall consistency of cell distribution (Fig. 3B).
Fig. 3.
Assay adaptation to 384-well microtiter plate format. Assay miniaturization studies in 384-well microtiter plate format. (A) HEK293 cells were seeded at cell densities ranging from 0 ( ), 125 (
), 125 ( ), 250 (
), 250 ( ), 500 (
), 500 ( ), 1,000 (
), 1,000 ( ), 2,000 (
), 2,000 ( ), 4,000 (
), 4,000 ( ) to 6,000 (
) to 6,000 ( ) cells per well. For each density range, average and standard deviation are calculated from 48 data points. (B) Images of Hoechst-stained nuclei at 72 h postseeding of cells ranging from 0 to 6,000 per well. Images were acquired using IN CELL Analyzer 1000, with blue channel for detection of Hoechst-stained nuclei (NUCL).
) cells per well. For each density range, average and standard deviation are calculated from 48 data points. (B) Images of Hoechst-stained nuclei at 72 h postseeding of cells ranging from 0 to 6,000 per well. Images were acquired using IN CELL Analyzer 1000, with blue channel for detection of Hoechst-stained nuclei (NUCL).
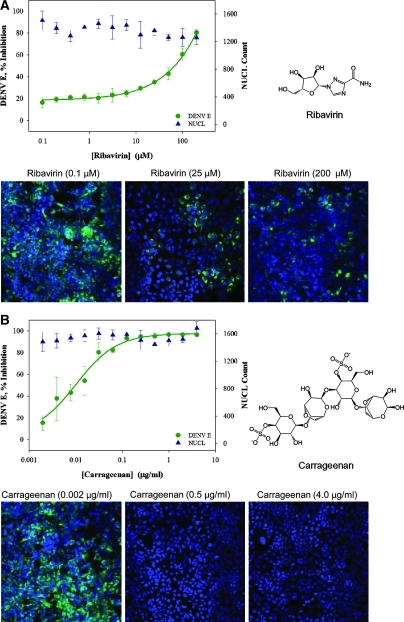
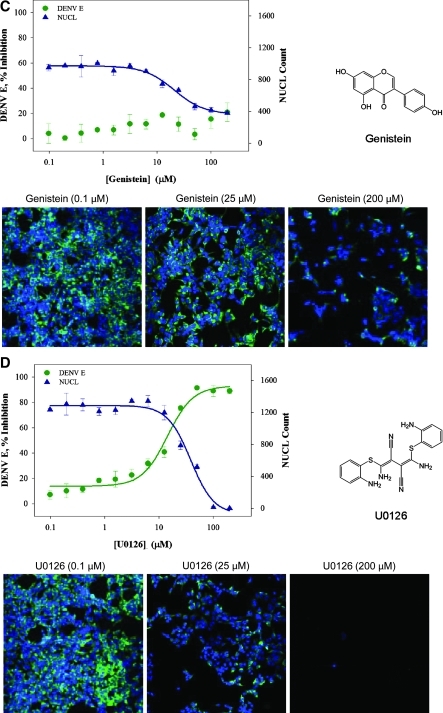
To evaluate the assay in a 384-well format, a panel of four compounds with reported antiviral activity was selected for testing. Genistein, a soybean isoflavone and tyrosine kinase inhibitor, modulates cellular tyrosine phosphorylation, which has been implicated as having a role in viral replication.26 The guanosine analog ribavirin can disrupt viral infection, likely by competitively inhibiting inosine-5'-monophosphate dehydrogenase, an essential enzyme involved in the synthesis of guanine nucleotides. Depletion of guanosine-5'-triphosphate pools by ribavirin prevents RNA capping by viral polymerases, impacting viral replication.27 U0126, a mitogen-activated protein kinase 1 and 2 inhibitor, affects viral replication by interfering with host extracellular signal-regulated kinase pathways.28 Carrageenans, polysaccharides made of sulfated galactose subunits, are thought to produce antiviral activity through interference with virus adsorption and internalization into the host cell.29 Genistein, ribavirin, and U0126 were diluted from stock for final testing in a 12-point doubling concentration series between 100 nM and 200 μM. Carrageenan was tested in a 12-point doubling concentration series between 2 ng/mL and 4 μg/mL. HEK293 cells were treated with compound plus DENV-2 for 48 h and virus quantity was measured by immunostaining. Compound cytotoxicity was also evaluated, as described above. The assay range was established using controls with 1% DMSO (v/v) plus DENV-2 for high signal and only 1% DMSO (v/v) for low signal. Ribavirin and carrageenan exhibited an IC50 at 75 ± 5 μM and 0.01 ± 0.004 μg/mL, respectively, with no detected cytotoxicity (Fig. 4A, B). Genistein showed no inhibition of DENV infection at any concentration tested and cytotoxicity EC50 at 21 ± 3 μM (Fig. 4C). The IC50 for U0126 was 14 ± 2 μM, with pronounced cytotoxicity at an EC50 of 38 ± 5 μM (Fig. 4D). Based on these observations, carrageenan at 4 μg/mL was selected for use as a positive antiviral (low) control for each assay plate to monitor screening reliability.
Fig. 4.
DENV infection assay against a panel of reported antivirals. HEK293 cells (4,000 cells per well) were treated with compounds and inoculated with DENV-2 (MOI 0.5) and then evaluated 48 h later for antiviral activity and cytotoxicity. (A) Treatment with ribavirin from 100 nM to 200 μM. Ribavirin IC50 against DENV was 75 ± 5 μM, with no detected cytotoxicity. (B) Treatment with carrageenan concentrations ranging from 2 ng/mL to 4 μg/mL. Carrageenan IC50 against DENV was 0.01 ± 0.004 μg/mL, with no detected cytotoxicity. (C) Treatment with genistein concentrations ranging from 100 nM to 200 μM resulted in no inhibition of DENV, with cytotoxicity EC50 of 21 ± 3 μM. (D) Treatment with U0126 concentrations ranging from 100 nM to 200 μM. U0126 IC50 against DENV was 14 ± 2 μM and cytotoxicity EC50 of 38 ± 5 μM. For each data point, average and standard deviation are calculated from triplicate data points. DENV E ( ) denotes detection of DENV-2 structural DENV E with α-E. NUCL (
) denotes detection of DENV-2 structural DENV E with α-E. NUCL ( ) denotes detection of Hoechst-stained nuclei. Plates were fixed and stained as described in the Materials and Methods section. Images were acquired using INCA3000, with green channel for DENV E and blue channel for detection of Hoechst-stained nuclei.
) denotes detection of Hoechst-stained nuclei. Plates were fixed and stained as described in the Materials and Methods section. Images were acquired using INCA3000, with green channel for DENV E and blue channel for detection of Hoechst-stained nuclei.
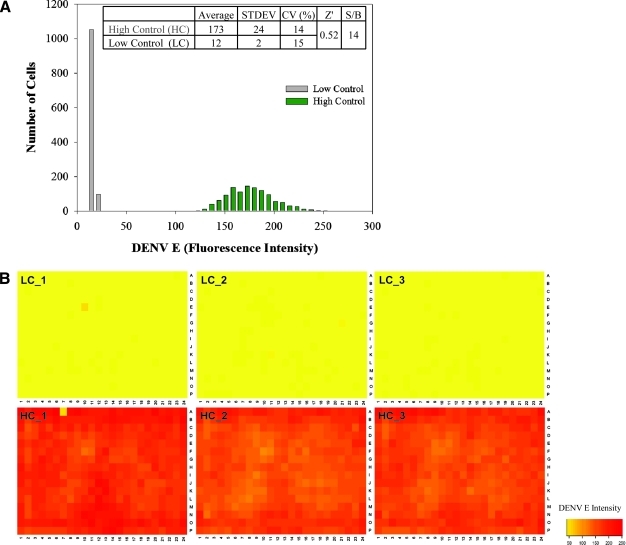
Next, we tested assay performance in a control run using three 384-well microtiter plates for negative (high) control (1% DMSO, v/v) and three 384-well microtiter plates for low control (4 μg/mL carrageenan in 1% DMSO, v/v). The assay had an average high signal of 173 ± 24 intensity and average low signal of 12 ± 2 intensity with percentage of coefficient of variation (CV%) at 14% and 15%, respectively. The Z′ factor was calculated to be acceptable for cell-based assays at 0.52, with a signal-to-background ratio (S/B) of 14 (Fig. 5A). A plate heat map analysis was also performed to assess inter- and intrawell variability in the microtiter plates and we observed no systematic plate effects (Fig. 5B). A Z′ of 0.52 and CVs <15% indicated excellent performance considering the heterogeneity of cellular response typical of cell-based assays. Further, as predicted, nuclei count indicated no cytotoxicity in both high and low controls. These data gave us confidence in moving forward with automated screening with this high-content assay.
Fig. 5.
Assessment of control wells. HEK293 cells (4,000 cells per well) were treated with either 4 μg/mL carrageenan as the low control (no glycoprotein E production) or 1% dimethyl sulfoxide (v/v) as the high control (high glycoprotein E production) for the infection assay and inoculated with DENV-2 (MOI of 0.5) and evaluated 48 h postinfection. High and low control plates were fixed and stained as described in the Materials and Methods section for evaluation. (A) Frequency distribution plot of the control wells. A total of 1,152 data points were used for each control, yielding a Z' value of 0.52 and a signal-to-background ratio of 14. (B) Plate heat map of resulting three high and three low control plates, showing no major deviations and no observed edge effects. LC, low control; HL, high control.
Chemical Screen for Inhibitors of DENV Infection
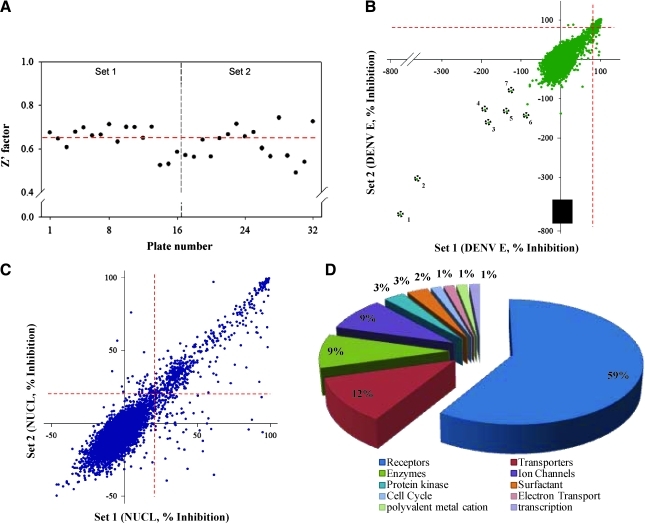
To identify small-molecule inhibitors of DENV infection, the newly adapted assay was optimized for HTS use as depicted in Table 1 and tested against 5,632 compounds comprised of several collections including U.S. Food and Drug Administration–approved drugs, known bioactives, and experimental substances. The screen was performed in duplicate to assess the assay's variability and reproducibility. The library was plated in sixteen 384-well microtiter plates with columns 13 and 14 empty for high and low control wells. To monitor the assay's performance throughout the screen, 1% DMSO (v/v) was added into column 13 for high control and 4 μg/mL carrageenan into column 14 for low control. Plate-by-plate Z′ values for screen set 1 and set 2 averaged at 0.63, indicating excellent performance and consistency with previous assay development data (Fig. 6A). To measure reproducibility, the datasets were plotted in a scatter graph to examine correlation for each compound. A scatter plot of DENV E % inhibition and nuclei % inhibition showed high overall correlation as depicted by the slope of the pattern (Fig. 6B, C). In both readouts, a large majority of compounds clustered around the center axis, indicating no activity, and only few outliers were present.
Table 1.
Dengue Virus Infection Assay Workflow
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Cell plating | 35 μL | 4,000 HEK293 cells in inoculation MEM media |
| 2 | Incubation time | 12 h | 37°C, 5% CO2 |
| 3 | Library compounds | 5 μL | 100 μM in 10% DMSO |
| 4 | Low control | 5 μL | 40 μg/mL carrageenan in water |
| 5 | High control | 5 μL | 10% DMSO |
| 6 | Virus Infection | 10 μL | Addition of DENV-2 at an MOI of 0.5 |
| 7 | Incubation time | 48 h | 37°C, 5% CO2 |
| 8 | Fix | 50 μL | 4% PFA for 20 min |
| 9 | Wash | 50 μL | Two times with 1 × PBS |
| 10 | Blocking buffer | 50 μL | 2% NGS, 0.4% TX-100 in 1 × PBS for 1 h |
| 11 | α-E primary staining | 50 μL | 1:1,000 dilution in 1 × PBS for 1 h |
| 12 | Wash buffer | 50 μL | Three times with 0.2% BSA, 0.2% TX-100 in 1 × PBS |
| 13 | Detection secondary staining | 50 μL | 1:1,000 dilution of Alexa-Fluor 488 in 1 × PBS for 1 h |
| 14 | Wash buffer | 50 μL | Three times with 0.2% BSA, 0.2% TX-100 in 1 × PBS |
| 15 | Nuclear staining | 50 μL | 10 μM Hoechst, 0.05% TX-100 in 1 × PBS for 10 min |
| 16 | Assay readout | 364 nm/450 nm and 488 nm/535 nm (ex/em) | INCA3000 microscope system |
| 17 | Image analysis | — | Multiparametric analysis using Raven 1.0 software |
Step Notes
1. Cells are prepared in media and dispensed into a 384-well assay plate with Multidrop 384
2. The 384-well assay plates are stored in the Steri-Cult, an automated, temperature-controlled incubator
3. to 5. Dispensing using a custom-designed 384-head on a PP-384-M Personal Pipettor; 30 s per 384-well microplate
6. Virus prepared in media and dispensed into 384-well assay plate with Multidrop 384
7. The 384-well assay plates are stored in the Steri-Cult
8. to 15. Dispensing and aspirating on the ELx405 washer; 1 min per 384-well microtiter plate
16. Readout performed on an automated platform; 14 s per well, with a total imaging time of 90 min per 384-well microtiter plate
17. Analysis of DENV E and Hoechst-stained nuclei; 20 min per 384-well microtiter plate
BSA, bovine serum albumin; DENV, Dengue virus; DMSO, dimethyl sulfoxide; INCA3000, IN CELL Analyzer 3000; MOI, multiplicity of infection; PBS, phosphate-buffered saline.
Fig. 6.
DENV HCA screening results. Assay sensitivity and performance were evaluated against a library containing 5,632 compounds screened in duplicates to assess reproducibility. (A) Plate-by-plate Z' factor across sets 1 and 2. Average Z' factor across both sets was 0.63, depicted with a red dashed line. (B) Scatter plot analysis of DENV E % inhibition in set 1 versus set 2. Positive threshold at 80% inhibition is depicted with red dashed lines. Compounds with high DENV E intensity are circled in black and shown in Table 2. (C) Scatter plot analysis of NUCL % inhibition in set 1 versus set 2. Cytotoxicity threshold was arbitrarily set at 20% and is depicted with red dashed lines. (D) Classification of positives identified from the screen.
The screen was run at a single compound concentration of 10 μM, and using 80% inhibition of DENV infection as a threshold cutoff, 273 compounds were identified as positive. To rapidly filter cytotoxic compounds, nuclei inhibition was set at a 20% threshold, thereby eliminating 184 compounds. Using the high-content multiparametric readout, 79 unique compounds remain as positives, yielding an initial hit rate of 1.4%. Ten compounds among the positives were identified twice as they were provided by two separate vendors (MicroSource and Prestwick) in the pilot set, further indicating assay robustness. Positives fell into several major categories (Fig. 6D) based on their previously characterized interactions, with 59% of the compounds interacting with receptors and about 12% with transporters. An additional 9% of the compounds are predicted to specifically inhibit enzymes, and another small-percentage group likely interferes with the function of ion channels, especially calcium.
As with many high-content screens, the cell-based assay suffers from artifacts generated by fluorescent compounds during the course of large chemical library screens. We have identified seven compounds with very high green pixel intensity ranging in DENV E % inhibition from −103% to −751%, indicative of an increase over high control (Table 2). An advantage of high-content screening in addition to on-the-fly scoring is the ability to recall images of wells for secondary analysis. The seven compounds were reanalyzed and found to be autofluorescent, including merbromin, which is known to possess fluorescent properties. Acriflavine and daunorubicin are DNA intercalators and represent another class of fluorescent compounds.
Table 2.
Chemical Structures and Images of Seven Identified Hits Found to Optically Interfere in Dengue Virus Infection Assay
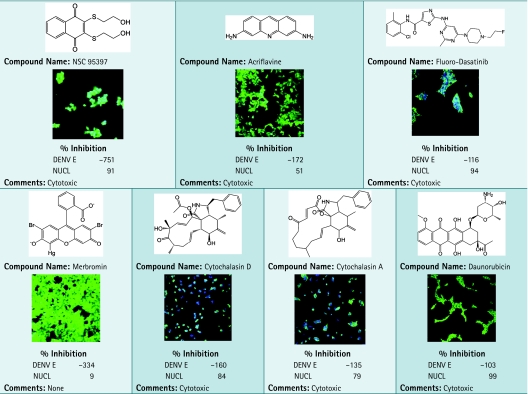 |
Compounds identified with high green pixel intensity from library screening in DENV infection assay are shown. Primary screening data as % inhibition are shown and averaged from duplicate data points. Plates were fixed and stained as described in the Materials and Methods section. Images were acquired using INCA3000, with green channel for DENV E and blue channel for detection of Hoechst-stained nuclei (NUCL).
Dose–Response Studies
Antiviral activity of the candidates was confirmed in dose–response studies on 74 resupplied compounds that have passed quality control and purity standards from vendors. The compounds were tested in dose–response studies in a 12-point doubling concentration series from 5 nM to 10 μM. Z′ factor was calculated at 0.68 for HEK293. The confirmation rate was nearly 100%, with 73 compounds showing IC50 values for DENV inhibition from nanomolar to micromolar range. A small subset of positives identified with the DENV infection assay are shown in Table 3, including kinase inhibitors BIBU 1361, W-9, and H-89, although mechanism of action in this particular assay remains unclear. Antimycin A, acetylspiramycin, benzethonium, and selamectin are in use as antimicrobials or pesticides. MPP, 3α-[(4-chlorophenyl)phenylmethoxy] tropane, niguldipine, 3α-bis-(4-fluorophenyl) methoxytropane, and LE 135 are part of a class of compounds that affect or modulate receptor activity. Interestingly, quinacrine, formerly used as an antimalarial, and bebeerines were identified as positive for antiviral activity against DENV. Of all 74 compounds resupplied for confirmation, only the antibacterial colistin had no effect on DENV in the dose–response evaluation. Altogether, these data show that the DENV infection assay was robust and reproducible and was able to identify selective nanomolar inhibitors, thus establishing this method as well suited for HTS of compound libraries.
Table 3.
Chemical Structures and Images of 17 Positives Identified in Dengue Virus Infection Assay
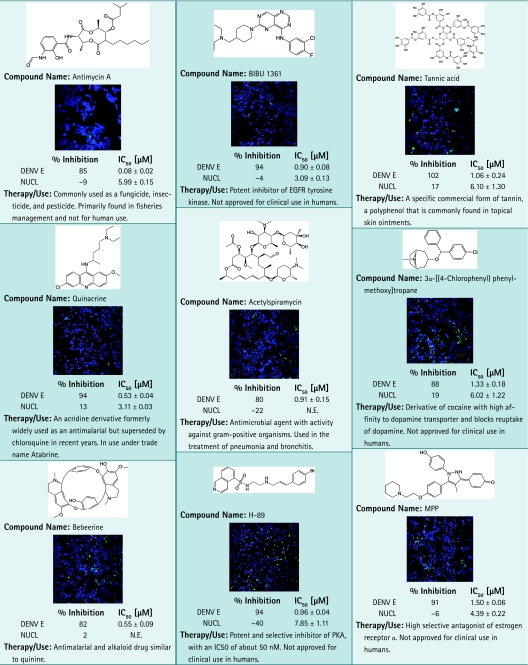 |
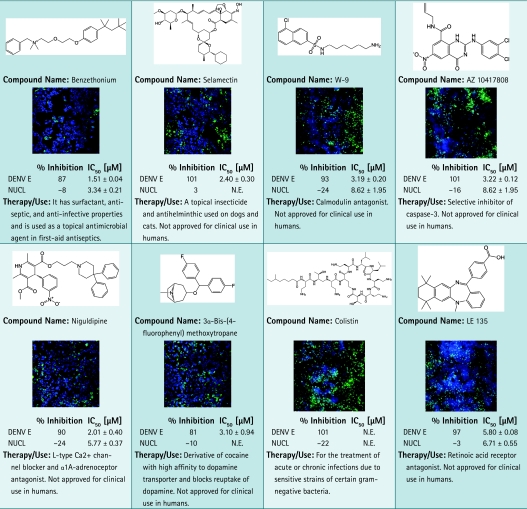 |
Images of cells tested with compounds at 2.5 μM are shown. Primary screening data as % inhibition and subsequent dose–response results are shown. For each data point, average and standard deviation calculated from duplicate data points are given. Plates were fixed and stained as described in the Materials and Methods section. Images were acquired using INCA3000, with green channel for DENV E and blue channel for detection of Hoechst-stained nuclei.
EGFR, epidermal growth factor receptor; N.E., no effect.
Discussion
The DENV infection assay described here constitutes a rapid and reliable screening method for inhibitors of DENV infection in mammalian cells. The assay evaluates two of the major criteria for drug candidate identification, namely efficacy and toxicity, and is amenable to evaluating a large number of compounds in a mechanized and consistent manner. The screening was performed in a human epithelial cell line with wild-type virus. Virus protein production and cell viability were monitored indirectly, using antibody-based detection and nuclei staining, respectively. In our screening system, compounds of interest could be rapidly identified by their ability to inhibit DENV infection without producing cytotoxicity. HEK293 cells were selected because they are of human origin, are highly infectable by DENV, grow rapidly, and can endure the mechanical challenges of HTS format. Several control wells were imaged with nine images per well covering 90% of the well and showed no loss of Hoechst-stained nuclei after many automated wash cycles (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/adt). Using a high-content approach and moderate virus inoculation, the assay was able to identify compounds that inhibit DENV replication or spread at any point in the infectious cycle. Images were acquired using the INCA3000 microscope system and compounds were scored with a multiparametric readout. Previously, we successfully identified antipoxiviral agents using this high-content approach against vaccinia virus.30
Recently, a number of other reports have appeared documenting the use of HTS formats to identify inhibitors of flavivirus infections, including DENV, in cultured cells.31 One study explored adaptation of a cytopathic effect assay on DENV-infected cells for HTS, with a library of ∼1,280 compounds and a hit rate of 0.31%.17 However, the time lapse before measurable cytopathic effect appears to be problematic for optimization of this type of assay. Target-based HTS employing protease assays have been used to evaluate compounds against the closely related flavivirus, West Nile virus.32 The assay had a hit rate of ∼0.3% and identified the 8-hydroxyquinoline family of compounds as possessing antiviral activity. Although the applicability of this assay is suggested for DENV, such results have not been published. A recent study incorporated a docking assay with cell-based flavivirus immunodetection, in which compounds predicted to bind to allosteric sites of the virus were identified virtually and then confirmed by cell-based flavivirus immunodetection.33 Although virtual screening indicated a subset of compounds as worthy of investigation, issues regarding the functional accessibility of viral protein allosteric binding sites and the cellular entry of compounds were notable. Other methods of screening potential inhibitors of DENV infections, including an RNA-dependent RNA polymerase assay,34 fluorescence quenching assay,35 virus-like particles–luciferase reporter assay,18 and replicon-base assays,36 have produced results of interest, but are typically limited to investigating compound activity against a subset of protein targets or specific events of the viral life-cycle. Most of the above assays have yet to be experimentally validated by screening larger-sized libraries and are likely to encounter considerable functional constraints upon scale-up. In most cases, cytotoxicity assays will need to be performed separately, increasing the cost and complexity of the screening process.
Chu and Yang used a similar high-content approach scored for levels of DENV E immunofluorescence, but did not include cytotoxicity evaluation of the test compounds, to screen a collection of 120 known kinase inhibitors using Vero cells.19 The authors identified several compounds as active when added preinfection and then characterized the relative level of activity of some of those compounds when added postinfection. Among the active compounds were inhibitors of the kinases Src, ABL, c-Kit, and PDGFR, which were found to block virus production at different stages of the viral life-cycle. Five of the compounds reported as active in that study were included in our screening collection, yet only one was active in our assay system (Supplementary Table S1), namely, mycophenolic acid. Notably, we found these compounds to be cytotoxic under the conditions of our assay, at a screening concentration of 10 μM. Mycophenolic acid was present in our screening collection in triplicate as it was supplied by three different vendors and all three compounds were found to inhibit DENV with some observed cytotoxic effects. The other inherent difference between the two screens is the way hits have been scored; in our case, we have set a threshold of 80% inhibition of production of protein E, whereas Chu and Yang used a 50% threshold.19
By combining automated microscopy with HTS technology, we have configured a robust and efficient high-content screening method using live virus infection, which monitors cytotoxicity of test compounds simultaneously with immunofluorescence of the viral glycoprotein E. We miniaturized this assay to a 384-well microtiter plate format for screening large chemical libraries against DENV-infected cells. We are currently investigating the mechanism of action of the identified hits with the aim of identifying some candidates for further evaluation as potential therapeutics against DENV infections.
Supplementary Material
Abbreviations
- BSA
bovine serum albumin
- DENV
Dengue virus
- DENV E
Dengue virus glycoprotein envelope
- DMSO
dimethyl sulfoxide
- EGFR
epidermal growth factor receptor
- HTS
high-throughput screening
- INCA1000
IN CELL Analyzer 1000
- INCA3000
IN CELL Analyzer 3000
- MOI
multiplicity of infection
- NS
nonstructural proteins
- NUCL
nuclei
- PBS
phosphate-buffered saline
Acknowledgments
The authors thank Dr. Toshimitsu Takagi for critically reading the manuscript, and members of the HTS Core Facility for their help during the course of this study, and Tony J. Riley (Medical Graphics, MSKCC) for his contribution to the artwork on this article. The HTS Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and an NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44. This work was supported by the Pacific Northwest Regional Center of Excellence (U54 AI 081680).
Disclosure Statement
No competing financial interests exist.
References
- 1.Chambers TJ. Hahn CS. Galler R. Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD. Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 3.Gubler DJ. Meltzer M. Impact of dengue/dengue hemorrhagic fever on the developing world. Adv Virus Res. 1999;53:35–70. doi: 10.1016/s0065-3527(08)60342-5. [DOI] [PubMed] [Google Scholar]
- 4.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takhampunya R. Ubol S. Houng HS, et al. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J Gen Virol. 2006;87:1947–1952. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- 6.Diamond MS. Zachariah M. Harris E. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology. 2002;304:211–221. doi: 10.1006/viro.2002.1685. [DOI] [PubMed] [Google Scholar]
- 7.Wu SF. Lee CJ. Liao CL, et al. Antiviral effects of an iminosugar derivative on flavivirus infections. J Virol. 2002;76:3596–3604. doi: 10.1128/JVI.76.8.3596-3604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen DB. Eldrup AB. Bartholomew L, et al. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother. 2004;48:3944–3953. doi: 10.1128/AAC.48.10.3944-3953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitby K. Pierson TC. Geiss B, et al. Castanospermine, a potent inhibitor of dengue virus infection in vitro and in vivo. J Virol. 2005;79:8698–8706. doi: 10.1128/JVI.79.14.8698-8706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang XG. Mason PW. Dubovi EJ, et al. Antiviral activity of geneticin against dengue virus. Antiviral Res. 2009;83:21–27. doi: 10.1016/j.antiviral.2009.02.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Z. Chen YL. Schul W, et al. An adenosine nucleoside inhibitor of dengue virus. Proc Natl Acad Sci USA. 2009;106:20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodenhuis-Zybert IA. Wilschut J. Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci. 2010;67:2773–2786. doi: 10.1007/s00018-010-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martina BE. Koraka P. Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev. 2009;22:564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastorino B. Nougairède A. Wurtz N, et al. Role of host cell factors in flavivirus infection: implications for pathogenesis and development of antiviral drugs. Antiviral Res. 2010;87:281–294. doi: 10.1016/j.antiviral.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Morrey JD. Smee DF. Sidwell RW. Tseng C. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antiviral Res. 2002;55:107–116. doi: 10.1016/s0166-3542(02)00013-x. [DOI] [PubMed] [Google Scholar]
- 16.Tomlinson SM. Malmstrom RD. Russo A. Mueller N, et al. Structure based discovery of dengue virus protease inhibitors. Antiviral Res. 2009;82:110–114. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Che P. Wang L. Li Q. The development, optimization and validation of an assay for high throughput antiviral drug screening against Dengue virus. Int J Clin Exp Med. 2009;2:363–373. [PMC free article] [PubMed] [Google Scholar]
- 18.Qing M. Liu W. Yuan Z, et al. A high-throughput assay using dengue-1 virus-like particles for drug discovery. Antiviral Res. 2010;86:163–171. doi: 10.1016/j.antiviral.2010.02.313. [DOI] [PubMed] [Google Scholar]
- 19.Chu JJ. Yang PL. c-Src protein kinase inhibitors block assembly and maturation of dengue virus. Proc Natl Acad Sci USA. 2007;104:3520–3525. doi: 10.1073/pnas.0611681104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antczak C. Takagi T. Ramirez CN, et al. Live-cell imaging of caspase activation for high-content screening. J Biomol Screen. 2009;14:956–969. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivnitski-Steele I. Ramirez CN. Djaballah H. Human embryonic stem cells in drug discovery: are we there yet? Int Drug Discov. 2010;5:24–28. [Google Scholar]
- 22.Desbordes SC. Placantonakis DG. Ciro A, et al. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henchal EA. Gentry MK. McCown JM. Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- 24.Shelton CC. Tian Y. Shum D, et al. A miniaturized 1536-well format gamma-secretase assay. Assay Drug Dev Technol. 2009;7:461–470. doi: 10.1089/adt.2009.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang JH. Chung TDY. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 26.Talavera D. Castillo AM. Dominguez MC, et al. IL8 release, tight junction and cytoskeleton dynamic reorganization conducive to permeability increase are induced by dengue virus infection of microvascular endothelial monolayers. J Gen Virol. 2004;85:1801–1813. doi: 10.1099/vir.0.19652-0. [DOI] [PubMed] [Google Scholar]
- 27.Leyssen P. Balzarini J. De Clercq E. Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol. 2005;79:1943–1947. doi: 10.1128/JVI.79.3.1943-1947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trujillo-Murillo K. Rincón-Sánchez AR. Martínez-Rodríguez H, et al. Acetylsalicylic acid inhibits hepatitis C virus RNA and protein expression through cyclooxygenase 2 signaling pathways. Hepatology. 2008;47:1462–1472. doi: 10.1002/hep.22215. [DOI] [PubMed] [Google Scholar]
- 29.Talarico LB. Damonte EB. Interference in dengue virus adsorption and uncoating by carrageenans. Virology. 2007;363:473–485. doi: 10.1016/j.virol.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 30.Deng L. Dai P. Ciro A, et al. Identification of novel antipoxviral agents: mitoxantrone inhibits vaccinia virus replication by blocking virion assembly. J Virol. 2007;81:13392–13402. doi: 10.1128/JVI.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble CG. Chen YL. Dong H, et al. Strategies for development of dengue virus inhibitors. Antiviral Res. 2010;85:450–462. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Mueller NH. Pattabiraman N. Ansarah-Sobrinho C, et al. Identification and biochemical characterization of small-molecule inhibitors of West Nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52:3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang QY. Patel SJ. Vangrevelinghe E, et al. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53:1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niyomrattanakit P. Chen YL. Dong H, et al. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J Virol. 2010;84:5678–5686. doi: 10.1128/JVI.02451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodenreider C. Beer D. Keller TH, et al. A fluorescence quenching assay to discriminate between specific and nonspecific inhibitors of dengue virus protease. Anal Biochem. 2009;395:195–204. doi: 10.1016/j.ab.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Massé N. Davidson A. Ferron F, et al. Dengue virus replicons: production of an interserotypic chimera and cell lines from different species, and establishment of a cell-based fluorescent assay to screen inhibitors, validated by the evaluation of ribavirin's activity. Antiviral Res. 2010;86:296–305. doi: 10.1016/j.antiviral.2010.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.