Abstract
inv encodes invasin, which is the primary invasion factor of Yersinia enterocolitica. inv expression in vitro is regulated in response to temperature, pH, and growth phase. In vitro, inv is maximally expressed at 26°C and repressed at 37°C at neutral pH but, when the pH of the media is adjusted to 5.5, levels of inv expression at 37°C are comparable to those at 26°C. A previous genetic screen for regulators of inv identified RovA, which was found to be required for activation of inv in vitro under all conditions tested as well as in vivo. Here we describe a screen that has identified a negative regulator of inv expression, ymoA. The ymoBA locus was identified by transposon mutagenesis as a repressor of inv expression in vitro at 37°C at neutral pH. This mutant shows increased inv expression at 37°C. The mutant can be fully complemented for inv expression by a plasmid expressing ymoA. These results indicate that YmoA plays a role in the negative regulation of inv.
Yersinia enterocolitica is a gram-negative human pathogen capable of colonizing the gastrointestinal tract. Once the organism establishes itself within a host, it is able to cause a variety of syndromes including enterocolitis, mesenteric lymphadenitis, and terminal ileitis. Y. enterocolitica is normally acquired through ingestion of contaminated food or water, with swine serving as a major reservoir for strains pathogenic to humans (5).
Colonization of the intestinal epithelium first requires that the organism be able to survive the gastric barrier of the stomach. Once in the small intestine, the organism is able to adhere to specialized cells in the small intestine called M cells (14, 16). Y. enterocolitica is able to transverse these cells, ending up in the underlying lymphoid tissue (Peyer's patches), where it is able to replicate and spread to the mesenteric lymph nodes (7, 38).
Y. enterocolitica contains a variety of adherence and invasion factors to aid the bacterium in establishing an infection. The major adhesion and invasion molecules identified in Y. enterocolitica are invasin, which is the primary invasion factor (38) and which initiates cell penetration by binding to a subset of β1-integrins (19) found on the surface of M cells (8), YadA, which is believed to be involved in adherence to intestinal brush border membranes as well as mucus (24, 35, 39), and Ail, which is involved in adherence and invasion as well as the promotion of serum resistance (4, 27, 28, 40, 45).
Invasin is an outer membrane protein found in Y. enterocolitica and Yersinia pseudotuberculosis (18, 19, 28, 37). When expressed in Escherichia coli, invasin is sufficient to promote invasion of tissue culture cells (18, 28). inv mutants do not colonize host tissues as well as wild-type bacteria at early time points. However, inv mutants eventually colonize to wild-type levels, presumably due to YadA and possibly Ail (38, 39). inv is maximally expressed in late exponential to early stationary phase in vitro and is regulated in response to changes in growth conditions. When cultures are grown at 26°C at neutral pH, inv expression is activated. In contrast, cultures grown at 37°C at neutral pH repress inv expression. However, when the pH of the media is adjusted to 5.5, levels of inv expression at 37°C are comparable to those observed at 26°C (36).
A previous genetic screen for regulators of Y. enterocolitica inv identified RovA (41), which was found to be required for activation of inv in vitro under all conditions tested. Subsequently, a homologue of RovA was identified as the regulator of inv in Y. pseudotuberculosis (44). Comparing inv expression in wild-type Y. enterocolitica to inv expression in a rovA mutant showed robust expression of inv in the wild type and little to no expression in the rovA mutant. Additionally, Western blot analysis of invasin in Peyer's patches from mice 2 days postinfection demonstrated that RovA is also required for in vivo expression of inv (41). However, preliminary analysis of the inv promoter suggested there may also be a negative regulator of inv. In this study we describe a screen that has identified YmoA as a negative regulator of inv expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The “v” designation refers to strains harboring the virulence plasmid; “c” refers to strains which have been cured of the virulence plasmid. Bacterial cultures were grown in Luria-Bertani (LB) broth at 26 or 37°C. When appropriate, antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 25 (for growth of E. coli) and 12.5 μg ml−1 (for growth of Y. enterocolitica); kanamycin, 50 μg ml−1; nalidixic acid, 20 μg ml−1; spectinomycin, 50 μg ml−1; streptomycin, 50 μg ml−1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Y. enterocolitica strainsa | ||
| JB580v | Serogroup O:8 Nalr ΔyenR(r− m+) | 20 |
| JB580c | Serogroup O:8 Nalr ΔyenR(r− m+) | 20 |
| JB41v | inv′-′phoA inv+ Nalr Cmr | 1 |
| YVM567c | inv′-′phoA inv+ymoB::mTn5Kn2 Nalr Cmr | This work |
| YVM923c | ΔymoB Nalr | This work |
| YVM967c | ΔymoB inv′-′phoA inv+ Nalr Cmr | This work |
| YVM976c | ymoB::mTn5Kn2 Nalr | This work |
| YVM1009c | rovA-lacZ rovA+ Nalr Cmr | This work |
| YVM1010c | ymoA-lacZ ymoB+ Nalr Cmr | This work |
| YVM1012c | ymoB::mTn5Kn2 ymoA-lacZ Nalr Cmr | This work |
| YVM1013c | ymoB::mTn5Kn2 rovA-lacZ rovA+ Nalr Cmr | This work |
| Plasmid | ||
| pBAD33 | Expression vector controlled by araBAD promoter, Cmr | 15 |
| pCR2.1 | TA cloning vector, Knr Ampr | Invitrogen |
| pHG329 | pUC19 polylinker, pBR322 ori, Ampr | 43 |
| pKENmut2 | gfp vector, Ampr | 9 |
| pSR47s | Suicide plasmid with sacB as a counter-selectable marker, Knr | 25 |
| pUTminiTn5Kn2 | Transposon vector, Kmr | 17 |
| pWKS30:StrSpec | pWKS30, which has a Strr-Spcr cassette in the HindIII site of the polylinker, Ampr Strr Spcr | Laboratory collection |
| pWKS30 | cloning vector, Ampr | 46 |
| pBY1 | pCR2.1 with gfp mut2 from pKENmut2, Ampr | This work |
| pBY8 | pWKS30::Strr-Spcr with gfp mut2 from pBY1, Ampr Strr Spcr | This work |
| pBY9 | pGEM5 with 600-bp inv promoter (−415 to +170 of coding region), Ampr | This work |
| pBY10 | pBY8 with 600-bp inv promoter from pBY9, Ampr Strr Spcr | This work |
| pBY11 | pCR2.1 with 215-bp inv promoter (−69 to +170 of coding region), Ampr | This work |
| pBY12 | pBY8 with 215-bp inv promoter from pBY11, Ampr Strr Spcr | This work |
| pBY34 | pHG329 containing ymoBA from 11F3, Ampr Knr | This work |
| pELL13 | pBAD33 containing ΔymoB and ymoA and upstream promoter region, Cmr | This work |
| pELL16 | pFuse carrying sequence just upstream of rovA, Cmr | This work |
| pELL21 | pWKS30::Strr-Spcr carrying ΔymoB and ymoA and upstream promoter region, Cmr | This work |
| pKN8 | Suicide lacZYA reporter vector pFUSE with BglII linker cloned into smaI site, Cmr | Laboratory collection |
| pKN38 | pSR47S carrying upstream and downstream regions of ymoB, Knr | This work |
| pKN46 | pKN8 carrying sequence just upstream of ymoA, Cmr | This work |
| pKN47 | pWKS30:Strr-Spcr carrying ymoBA and upstream promoter region, Cmr | This work |
| pREV2 | pCR2.1 carrying inv promoter region −415 to +170 | Laboratory collection |
All strains listed are derivatives of strain JB580.
Strain and plasmid construction.
To construct the inv-gfp transcriptional fusions, gfp was amplified from pKENmut2 (9) with primer GFP#1, which has a SacII linker (5′-TCC CCG CGG GGA AAG AAG GAG ATA TAC ATA TGA GT-3′), and primer GFP#2, which has a SacI linker (5′-CGA GCT CGT ATT TGT ATA GTT CAT CCA TGC C-3′). The amplified fragment contained a ribosome binding site and the coding region of the gfp gene. The PCR product was initially subcloned into the TA cloning vector pCR2.1 (Invitrogen) to generate pBY1. Subsequently, the SacII/SacI gfp fragment was subcloned into the SacII/SacI sites of pWKS30:StrSpec (46) to generate pBY8. To generate the inv-gfp transcriptional fusion, the full-length inv promoter region −415 to +170 (36) was subcloned from pREV2 via the 5′ BamHI site and the 3′ NotI site into pBY8 to generate pBY10. Primer inv#3 (5′-GCA TTT CAT TTG TCA TTG C-3′) and primer inv#1 (5′-CCG ATC GAT AAT ATT AGC C-3′) were used to amplify the 215-bp (−69 to +170) inv promoter fragment, which was cloned into pCR2.1 to generate pBY11. The 215-bp fragment was then subcloned into pBY8 via 5′ BamHI and 3′ NotI sites within the polylinker of pCR2.1, generating pBY12. Plasmids were confirmed by sequencing.
To complement the mutation in YVM567c, plasmids pKN43 and pELL21 were constructed. To generate pKN43, ymoB was amplified from JB580v with primer ybaJ-5.2, which contains an EcoRI linker (5′-GGA ATT CCG GTG AGC GGA GAA ATG ATT TAT ATT AAT A-3′), and primer ybaJ-3.2, which contains an EcoRI linker (5′-GGA ATT CCT CAA TAT AAA AAT AAT ATA GGG AAA CTA TCC-3′). The resulting product was cloned into the EcoRI site of pWKS30:StrSpec. pELL21 was constructed by digesting pELL13 with EcoRI/XbaI; the resulting fragment contained a deletion of ymoB, leavingymoA under the control of its native promoter. The fragment was subcloned into the EcoRI/XbaI sites of pWKS30:StrSpec. Plasmids were confirmed by sequencing.
lacZ transcriptional fusions were generated by cloning PCR fragments into pKN8 (3) and mating recipient (Y. enterocolitica) and donor (E. coli) strains of bacteria. To construct pKN46, primer ymoA-5L, which contains an XbaI linker (5′-GCT CTA GAC ACA TAT ACT CTG TTT AGT AGT TAC GGA ATC-3′), and primer ymoA-3L, which contains a BglII linker (5′-GAA GAT CTG TTG CCA TAC AGT AGG TGG AAT TTTA TCA T-3′), were used to amplify a 360-bp product that contained most of the ymoA gene. The resulting product was digested with XbaI/BglII and cloned into the same sites in pKN8. To generate pELL16, the promoter region (500 bp upstream of the start codon) of rovA was amplified from JB580v with primer rovAFusexba, which contains an XbaI linker (5′-CGT CTA GAT TCC ACA TCC ACC AAC-3′), and primer rovA-2, which has a BamHI linker (5′-CGC GGA TCC TGC TAA ATC AGA TCC TAA TGT CGA TTC CAA-3′). After amplification the products were digested with XbaI/BamHI and cloned into the XbaI/BglII sites in pKN8. Plasmids were confirmed by sequencing.
Strains JB580c (20) and YVM976c were mated with S17-λpir (29) containing either pKN46 or pELL16 to generate chromosomal lacZ fusions. Briefly, 400 μl of overnight cultures was mixed and resuspended in 200 μl of LB broth and spotted on LB agar. The plate was incubated overnight at 26°C, and the bacteria were harvested and resuspended in 1 ml of LB broth. Dilutions of 10−1 and 10−2 were spread on LB agar containing nalidixic acid (to select against the donor E. coli strain), and chloramphenicol (to select for the recipient Y. enterocolitica strain). Integration was confirmed by PCR with a primer within lacZ and a primer 5′ of the homologous region used for recombination.
Green fluorescent protein assays.
Cultures of JB580v containing inv-gfp transcriptional fusions were grown in 4 ml of LB broth overnight at 26°C with selection and then subcultured to an optical density at 600 nm (OD600) of 0.2 in 3 ml of LB broth. Cultures were grown at 26 or 37°C for 24 h, and levels of fluorescence intensity were compared. Fluorescence intensity was determined with a Beacon 2000 variable-fluorescence polarization system (Panvera). The fluorescence was calculated by dividing the average fluorescence intensity by the culture OD600. In all cases, JB580v with pBY8 was used to determine background fluorescence.
Screen for negative regulators of inv.
Strain JB41v (1) was mutagenized with mTn5Kn2 by mating with S17-λpir containing pUTmTn5Kn2 as previously described (17). To select for mutants that showed increased inv expression at 37°C and neutral pH, as shown by their dark blue appearance, plates containing nalidixic acid (to select against the donor E. coli strain), chloramphenicol (to select for the recipient Y. enterocolitica strain), kanamycin (to select for transposon insertions), and 5-bromo-4-chloro-3-indolylphosphate (XP; 40 μg ml−1; to screen for alkaline phosphatase [AP] activity indicating alterations in inv promoter activity) were incubated at 37°C for 2 days. Colonies were then screened for inv-phoA expression. Of 300 mutants selected from plates 2, 6B4 and 11F3, showed increased expression of inv-phoA at 37°C when tested by a quantitative AP assay.
Southern blotting was performed to determine the sizes of the Knr-encoding fragments from mutants 6B4 and 11F3. The blots were probed with the EcoRI Knr fragment from the pUTminiTn5Kn2 plasmid. The 11F3 fragment was cloned into pHG329 (43) by making a subgenomic library and selecting for Knr to generate pBY34. However, the 6B4 mutation could not be cloned by this method. Since mutant 6B4 was not malleable to cloning via a subgenomic library, the location of the mutation was determined by Southern blotting using the Knr-encoding fragment and ymoA as probes and PCR with primers ymo1 (5′-GAA GAT CTT GCT ATT TCA CAT GTT GCC-3′), which anneals to the 3′ end of ymoA and reads into ymoA, P6 (5′-CCT AGG CGG CCA GAT CTG AT-3′), which anneals to the Tn5 I repeat and reads away from Tn5, and P7 (5′-GCA CTT GTG TAT AAG AGT CAG-3′), which anneals to the Tn5 O repeat to read away from Tn5.
β-Galactosidase assays.
Cultures were grown in triplicate overnight in 2 ml of LB broth at 37 or 26°C. β-Galactosidase activity was measured according to previously described methods (26).
AP assays.
Cultures were grown in triplicate overnight in 2 ml of LB broth at 37 or 26°C. AP activity was measured according to previously described methods (23).
Nucleotide sequence accession number.
The GenBank accession number assigned to the ymoBA locus is AY387659.
RESULTS
Truncation of the inv promoter results in constitutive expression at 37 and 26°C.
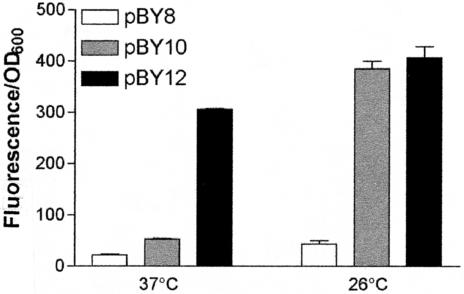
To determine if sequences upstream of the −35 region of the inv promoter were important for promoter function, a full-length promoter and a truncated promoter were fused to gfp (Fig. 1). The plasmids were transformed into JB580v, and levels of expression of the inv-gfp fusions at 37 (temperature where the inv promoter is repressed) and 26°C (temperature where the inv promoter is activated) were compared (Fig. 2). pBY10 (−415 to +170) shows normal regulation at 37 and 26°C, with inv expression elevated at 26°C compared to that at 37°C. pBY12 (−69 to +170) shows derepression of the inv promoter at 37°C, with levels of promoter activity comparable to those at 26°C. These results suggest the presence of a repressor binding site between positions −415 and −69 on the inv promoter.
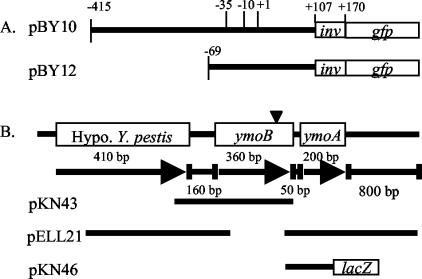
FIG. 1.
Schematic of inv-gfp promoter fusions and ymoBA locus. (A) The predicted −35, −10, and +1 sites, along with the start codon (+107) and fusion junction (+170) are marked on pBY10. (B) The ymoBA locus contains a hypothetical Y. pestis gene just upstream of ymoB and no open reading frames in the first 800 bp downstream of ymoA. Arrows indicate the direction of transcription, and numbers of base pairs between the genes are also indicated. Inverted triangle, point of transposon insertion; solid black lines below the locus, regions of the locus cloned into plasmids pKN43 (ymoB), pELL21 (ymoA), and pKN46 (ymoA-lacZ).
FIG. 2.
Effect of promoter truncation on inv-gfp expression. Overnight cultures of Y. enterocolitica strain JB580v carrying the indicated plasmids were grown in triplicate at 26°C, subcultured to an OD600 of 0.2, and grown for 24 h. Fluorescence from the inv-gfp fusions was calculated by dividing average fluorescence by the OD600. pBY10 (−415 to +170), which has the full-length inv promoter, and pBY12 (−69 to +170), which has a truncated inv promoter, were compared. pBY8 is the vector with promoterless gfp and was used to determine background levels of fluorescence.
Identification of a negative regulator of inv.
Strain JB41v, which contains a translational fusion of inv-phoA integrated on the chromosome, was mutated with mTn5Kn2. To identify mutants that had altered regulation at 37°C, colonies were screened on indicator plates containing XP, which turns blue in the presence of AP. Colonies that showed normal repression of inv appeared light blue at 37°C. Colonies with a mutation in a potential repressor appeared dark blue on the indicator plates due to the increased expression of inv-phoA. Eleven independent matings, yielding approximately 28,000 mutants, were performed. Of the 28,000 mutants 300 appeared to have increased inv-phoA expression at 37°C. These were further characterized by performing AP assays at 26 and 37°C. Of the 300 mutants, 6B4 and 11F3 showed increased expression of inv-phoA at 37°C (data not shown). These two mutants were from independent matings.
To determine the sequences of the mutated genes in mutants 6B4 and 11F3, Southern blotting was performed to identify the size of the Knr-encoding fragments in both mutants (data not shown). The fragment from 11F3 was cloned into pHG329, generating pBY34. Sequence analysis of the transposon-chromosome junction in pBY34 indicated that the transposon had inserted just upstream of a previously identified gene, ymoA. Because mutant 6B4 was not malleable to cloning via a subgenomic library, the location of the mutation was determined by Southern blot and PCR analysis (data not shown). The results indicated that the transposon insertion was in the same general location as, but in the opposite orientation to, that in 11F3. The PCR product for 6B4 obtained with primers P6 and ymo1 was sequenced directly with primer ymo1, and this confirmed that the transposon was inserted 2 bp upstream of, but in the opposite orientation as, the insertion in 11F3.
It was determined by sequencing that both 6B4 and 11F3 contained a mutation in a gene we named ymoB. YmoB has homology to a hypothetical protein YbaJ from E. coli, which has no known function (64% identity and 82% similarity). Examination of the genetic locus revealed differences between E. coli and Y. enterocolitica. The sequence upstream of ybaJ in E. coli contains acrB, while the upstream sequence in Y. enterocolitica contains a hypothetical Yersinia pestis gene of unknown function. hha is located just downstream of ybaJ in E. coli, and further sequencing revealed that ymoA, a gene that has been previously shown to influence regulation of Y. enterocolitica virulence factors (10), is just downstream of ymoB. YmoA and Hha are members of a new class of proteins that regulate genes in response to different environmental conditions (21). This raised the possibility that the insertion in ymoB has a polar effect on the expression of ymoA, reducing the level of ymoA transcription, leading to increased inv-phoA expression at 37°C. 6B4 was chosen from the two mutants for further study and was designated strain YVM567c.
ymoA is able to restore wild-type inv-phoA expression in YVM567c.
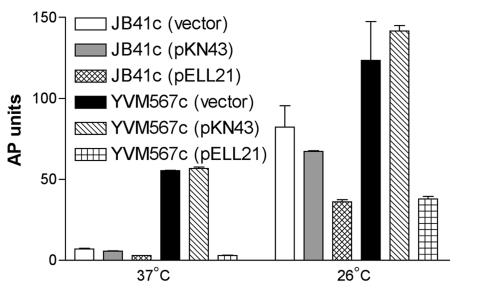
To test the ability of ymoB and ymoA to restore wild-type inv-phoA expression in YVM567c, plasmids pKN43(ymoB) and pELL21 (ymoA) were transformed into YVM567c, generating strains YVM567c(pKN43) and YVM567c(pELL21). Levels of inv-phoA expression from these strains were compared to those from wild-type strains JB41c(pKN43) and JB41c(pELL21). All cultures were grown in LB broth overnight at 26 or 37°C. Strain YVM567c(pKN43) showed levels of inv-phoA expression comparable to those of YVM567c, indicating that ymoB is not able to complement the mutation in YVM567c. Strain YVM567c(pELL21) showed levels of inv-phoA expression comparable to those seen in strain JB41c(pELL21), indicating that ymoA alone was able to complement the mutation in YVM567c (Fig. 3). JB41c and YVM567c containing the vector showed no difference in regulation compared to JB41c and YVM567c with no plasmids (data not shown). To further test whether or not ymoB was involved in the repression of inv, an in-frame ymoB deletion mutant was constructed and tested for inv expression. The ΔymoB mutant, YVM967c, showed normal levels of inv expression compared to JB41c (data not shown). Together these data suggested that the phenotype of loss of inv repression observed with the transposon insertion mutation in ymoB was probably due to a polar effect on ymoA expression rather than loss of ymoB. We first tried to make an in-frame deletion of ymoA using two different suicide plasmids, but both strategies failed. Generation of an insertional ymoA mutant was also unsuccessful. All attempts to construct a mutation in ymoA were unsuccessful unless ymoA was supplied in trans on a plasmid, indicating that ymoA is probably an essential gene for this particular strain of Y. enterocolitica.
FIG. 3.
Effect of complementing YVM567c with ymoA and ymoB. Cultures were grown in triplicate overnight at 26 or 37°C and assayed for AP activity. All strains have a chromosomal inv-phoA fusion to monitor promoter activity. JB41c is used as the wild-type control, YVM567c contains a ymoB::mTn5Kn2 mutation, YVM567c(pKN43) contains ymoB::mTn5Kn2 plus ymoB complementing clone, and YVM567c(pELL21) contains ymoB::Tn5Kn2 plus ymoA complementing clone.
The data in Fig. 3 also suggested that YVM567c has increased inv-phoA expression at 26°C compared to JB41c. These results are consistent with previously published reports that show a mutation in ymoA derepressed virF and the yop regulon of the pYV virulence plasmid of Y. enterocolitica strain W22711, which is a derivative of W22703, a wild-type O:9 strain, leading to higher levels of expression at both 26 and 37°C (10). Our data show the same type of derepression of inv in strain YVM567c, with higher expression of inv-phoA at both 37 and 26°C.
mTn5Kn2:ymoB has a polar effect on the expression of ymoA.
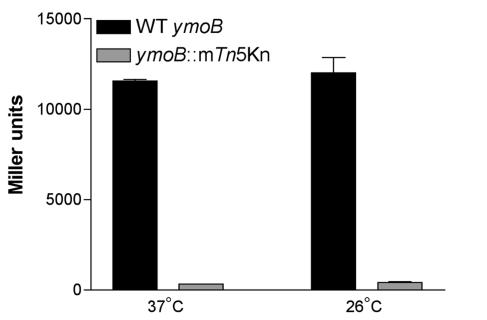
To determine if the mutation in strain YVM567c has a polar effect on the expression of ymoA, chromosomal lacZ-ymoA fusions were constructed, generating a merodiploid. The fusion was constructed in strains JB580c and YVM976c, generating strains YVM1010c and YVM1012c, respectively. Cultures were grown in LB broth overnight at 26 or 37°C, and promoter activity was monitored by measuring β-galactosidase activity (Fig. 4). There was greatly reduced β-galactosidase activity in strain YVM1012c, which contains the ymoB:mTn5Kn2 mutation, compared to that in the wild-type strain YVM1010c. To rule out the possibility that YmoB is required for the expression of the ymoBA locus, we complemented YVM1012c with plasmid pKN43 and saw no difference in the expression of lacZ compared to that of YVM1012c (data not shown). The vector alone had no effect on expression of lacZ in YVM1012c (data not shown). These results reveal that the mutation in strain YVM567c exerts a polar effect on the transcription of ymoA. Taken together with previous data, YmoA appears responsible for the derepression of inv observed in strain YVM567c.
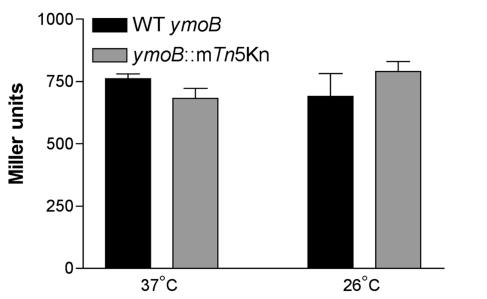
FIG. 4.
Effect of ymoB::mTn5Kn2 on the expression of ymoA. Cultures were grown in triplicate overnight at 26 and 37°C and assayed for β-galactosidase activity. Both strains contain a chromosomal ymoA-lacZ fusion to monitor promoter activity. YVM1010c (black bars) is used as the wild-type (WT) control and is compared to YVM1012c (gray bars), which contains the ymoB::mTn5Kn2 mutation.
Increased levels of rovA are not responsible for the increase in inv-phoA expression seen in strain YVM567c.
RovA has previously been shown to be required for the expression of inv in Y. enterocolitica and Y. pseudotuberculosis (30, 41). To rule out the possibility that increased rovA expression in strain YVM567c was responsible for the increased levels of inv expression, chromosomal lacZ fusions to the rovA promoter were constructed in strains JB580c and YVM976c, yielding strains YVM1009c and YVM1013c, respectively. Cultures were grown overnight in LB broth at 26 and 37°C, and levels of β-galactosidase activity for the two strains were compared (Fig. 5). The data show no difference in the level of rovA-lacZ expression between strains YVM1009c and YVM1013c. These results indicate that the increased inv expression observed in YVM567c was not due to increased levels of rovA transcription in strain YVM567c.
FIG. 5.
Effect of ymoB::mTn5Kn2 on rovA expression. Cultures were grown in triplicate overnight at 26 and 37°C and assayed for β-galactosidase activity. Both strains contain a chromosomal rovA-lacZ fusion to monitor promoter activity. YVM1009c (black bars) is used as the wild-type (WT) control and is compared to YVM1013c (gray bars), which contains the ymoB::mTn5Kn2 mutation.
DISCUSSION
Invasin is an important virulence factor in Y. enterocolitica, allowing attachment and efficient translocation of the bacteria through M cells (38). Without inv the progress of Y. enterocolitica through the infection process is delayed (38). Previous work has shown that inv is positively regulated by RovA, a transcriptional regulator in the MarR/SlyA family (41). In this study we have found that YmoA plays a role in the negative regulation of inv. Promoter truncation experiments revealed the presence of a negative regulatory sequence in the inv promoter involved in temperature control of inv expression, and a subsequent genetic screen showed that the ymoBA locus was involved in the derepression of inv at 37°C. Complementing the mutation in the ymoBA locus with ymoB and ymoA revealed that ymoA and not ymoB was able to restore wild-type expression of inv. A lacZ fusion to ymoA showed that the mutation in the ymoBA locus exerted a polar effect on the transcription of ymoA. We have also shown that the mutation in the ymoBA locus does not affect the transcription of rovA. These results suggest that YmoA plays a negative regulatory role in the expression of inv.
YmoA is a member of a growing class of transcriptional regulators with homology to histone-likeproteins (21). YmoA previously was shown to be involved in the regulation of Yop proteins and YadA in Y. enterocolitica (10). It is not clear why a ymoA mutant in the strain W22711 background was viable whereas a ymoA mutation in the 8081 strain background appears to be lethal. However, the W22711 mutant was isolated in the O:9 serotype background, and different serotypes of Y. enterocolitica are known to have genetic differences (6, 13). It is also possible that the ymoA mutant in the W22711 background simultaneously picked up a suppressor mutation. Another member of this family, Hha, has been implicated in the regulation of hly, a pore-forming toxin, in E. coli (31). It has also been reported that the Hha homologue from Salmonella enterica serovar Typhimurium is involved in repressing the invasion phenotype by altering expression of the regulatory gene hilA (12). It is interesting that other examples of H-NS-like proteins that regulate virulence factors have been shown to increase expression when the temperature is shifted from 26 to 37°C. This is not the case for inv, as expression is downregulated at 37°C and upregulated at 26°C, making regulation of inv by YmoA distinct.
Recent studies have shown that Hha from S. enterica serovar Typhimurium is able to bind specifically to the hilA promoter (12). Other studies on Hha from E. coli have shown that Hha is not able to bind the hly promoter specifically (33). However, recent work has described the ability of Hha and YmoA to interact with H-NS (32). Subsequent work done on the regulation of the hly promoter showed that Hha and H-NS act together to negatively regulate expression of hemolysin, with H-NS providing the binding specificity for the hly promoter (22). It has been suggested that H-NS and Hha form a multiprotein complex on the hly promoter and repress transcription. YmoA and Hha are functionally interchangeable, suggesting similar regulatory mechanisms (2). Further evidence to support a role for H-NS in the regulation of inv comes directly from the sequence of the inv promoter, which contains an AT-rich stretch of 30 bp centered on the −35 sequence, raising the possibility that the promoter may be intrinsically bent. It is widely accepted that H-NS binds to bent DNA (11, 42). Based on the data for the regulation of hly and the ability of YmoA to interact with H-NS, it is reasonable to speculate that YmoA and H-NS may form a repression complex at the inv promoter.
RovA is a member of the MarR family of transcriptional regulators, and has significant amino acid identity with SlyA from S. enterica serovar Typhimurium (41). When the slyA homologue from E. coli is overexpressed, it is able to activate clyA (cytotoxin) in laboratory strains of E. coli (34). It was recently found that H-NS plays a role in silencing expression of this gene in E. coli and that SlyA is able to overcome this silencing and activate transcription from the clyA promoter (47). Likewise, RovA is able to activate expression of the inv promoter, and we hypothesize that it may do so by overcoming the repression by YmoA. It remains to be seen what role if any is played by H-NS in the regulation of inv. RovA has been shown to bind the inv promoter (30), and it is possible that under some conditions (e.g., 37°C) YmoA or a YmoA-H-NS complex can interfere with RovA binding or alter the interaction of RovA with the transcriptional machinery. Further experiments with purified YmoA and RovA investigating their interaction with the inv promoter will provide further evidence for the mechanism by which the inv promoter is both repressed and activated.
Acknowledgments
This study was supported by a National Institutes of Health grants AI27342 and AI52167 awarded to V.L.M.
We thank Peter Dube, Scott Handley, Kim Walker, and Clemencia Maria Rojas for critical review of the manuscript.
REFERENCES
- 1.Badger, J. L., and V. L. Miller. 1998. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J. Bacteriol. 180:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balsalobre, C., A. Juarez, C. Madrid, M. Mourino, A. Prenafeta, and F. J. Munoa. 1996. Complementation of the hha mutation in Escherichia coli by the ymoA gene from Yersinia enterocolitica: dependence on the gene dosage. Microbiology 142:1841-1846. [DOI] [PubMed] [Google Scholar]
- 3.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 4.Bliska, J. B., and S. Falkow. 1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. USA 89:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, P. B. 1975. Pathogenicity of Yersinia enterocolitica for mice. Infect. Immun. 11:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, M. A., B. H. Hirst, and M. A. Jepson. 1998. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R., C. Sluiters, I. Delor, D. Geib, K. Kaniga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 11.Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83:231-234. [DOI] [PubMed] [Google Scholar]
- 12.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foultier, B., P. Troisfontaines, S. Muller, F. R. Opperdoes, and G. R. Cornelis. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55:37-51. [DOI] [PubMed] [Google Scholar]
- 14.Grutzkau, A., C. Hanski, H. Hahn, and E. O. Riecken. 1990. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut 31:1011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanski, C., U. Kutschka, H. P. Schmoranzer, M. Naumann, A. Stallmach, H. Hahn, H. Menge, and E. O. Riecken. 1989. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O8 with intestinal mucosa during experimental enteritis. Infect. Immun. 57:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 18.Isberg, R. R., and S. Falkow. 1985. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262-264. [DOI] [PubMed] [Google Scholar]
- 19.Isberg, R. R., and J. M. Leong. 1990. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60:861-871. [DOI] [PubMed] [Google Scholar]
- 20.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 21.Madrid, C., J. M. Nieto, and A. Juarez. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291:425-432. [DOI] [PubMed] [Google Scholar]
- 22.Madrid, C., J. M. Nieto, S. Paytubi, M. Falconi, C. O. Gualerzi, and A. Juarez. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184:5058-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantle, M., L. Basaraba, S. C. Peacock, and D. G. Gall. 1989. Binding of Yersinia enterocolitica to rabbit intestinal brush border membranes, mucus, and mucin. Infect. Immun. 57:3292-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Miller, V. L., K. B. Beer, G. Heusipp, B. M. Young, and M. R. Wachtel. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 41:1053-1062. [DOI] [PubMed] [Google Scholar]
- 28.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 31.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juarez. 1991. The hha gene modulates haemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285-1293. [DOI] [PubMed] [Google Scholar]
- 32.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodriguez, and A. Juarez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juarez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 34.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 35.Paerregaard, A., F. Espersen, O. M. Jensen, and M. Skurnik. 1991. Interactions between Yersinia enterocolitica and rabbit ileal mucus: growth, adhesion, penetration, and subsequent changes in surface hydrophobicity and ability to adhere to ileal brush border membrane vesicles. Infect. Immun. 59:253-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 37.Pepe, J. C., and V. L. Miller. 1990. The Yersinia enterocolitica inv gene product is an outer membrane protein that shares epitopes with Yersinia pseudotuberculosis invasin. J. Bacteriol. 172:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepe, J. C., M. R. Wachtel, E. Wagar, and V. L. Miller. 1995. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect. Immun. 63:4837-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierson, D. E., and S. Falkow. 1993. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect. Immun. 61:1846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 42.Rimsky, S., F. Zuber, M. Buckle, and H. Buc. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42:1311-1323. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, G. S., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 44.Thomson, N. R., A. Cox, B. W. Bycroft, G. S. Stewart, P. Williams, and G. P. Salmond. 1997. The rap and hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol. Microbiol. 26:531-544. [DOI] [PubMed] [Google Scholar]
- 45.Wachtel, M. R., and V. L. Miller. 1995. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect. Immun. 63:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 47.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]