Abstract
The barA and sirA genes of Salmonella enterica serovar Typhimurium encode a two-component sensor kinase and a response regulator, respectively. This system increases the expression of virulence genes and decreases the expression of motility genes. In this study, we examined the pathways by which SirA affects these genes. We found that the master regulator of flagellar genes, flhDC, had a positive regulatory effect on the primary regulator of intestinal virulence determinants, hilA, but that hilA had no effect on flhDC. SirA was able to repress flhDC in a hilA mutant and activate hilA in an flhDC mutant. Therefore, although the flhDC and hilA regulatory cascades interact, sirA affects each of them independently. A form of BarA lacking the two N-terminal membrane-spanning domains, BarA198, autophosphorylates in the presence of ATP and transfers the phosphate to purified SirA. Phosphorylated SirA was found to directly bind the hilA and hilC promoters in gel mobility shift assays but not the flhD, fliA, hilD, and invF promoters. Given that the CsrA/csrB system is known to directly affect flagellar gene expression, we tested the hypothesis that SirA affects flagellar gene expression indirectly by regulating csrA or csrB. The sirA gene did not regulate csrA but did activate csrB expression. Consistent with these results, phosphorylated SirA was found to directly bind the csrB promoter but not the csrA promoter. We propose a model in which SirA directly activates virulence expression via hilA and hilC while repressing the flagellar regulon indirectly via csrB.
Salmonella enterica serovar Typhimurium is a motile gram-negative bacterium that causes gastroenteritis in calves and humans and a typhoid-like systemic infection in mice (68). A systemic infection requires bacterial penetration of the intestinal epithelium, a process which occurs primarily through the M cells of Peyer's patches (34). The invasiveness of Salmonella is attributed to a type III secretion system, TTSS-1, encoded by Salmonella pathogenicity island 1 (SPI1) (21, 46, 69). Injection of effector proteins by TTSS-1 directly into host cells leads to uptake of the bacterium via macropinocytosis (ruffling) (20). TTSS-1 effectors also elicit the inflammation and fluid secretion associated with gastroenteritis in susceptible host species (23, 43, 63, 64, 68).
The focus of this report is the SirA/BarA two-component regulatory system, which is one of several evolutionarily conserved housekeeping systems that regulate SPI1 (2, 5, 33). SirA orthologs are present, with different names, throughout the γ-proteobacteria, e.g., SirA in S. enterica serovar Typhimurium, GacA in Pseudomonas species, VarA in Vibrio cholerae, ExpA in Erwinia carotovora, LetA in Legionella pneumophila, and UvrY in Escherichia coli (29). In each species, sirA is located directly upstream of uvrC but is not involved in UvrC functions (DNA repair). A sensor kinase for SirA is present in each of these organisms as well. The sensor kinase is known as BarA in E. coli and Salmonella but has different names in other genera (GacS, ExpS, and LetS). In S. enterica serovar Typhimurium, SirA positively regulates the invasion of tissue culture cells and bovine gastroenteritis while negatively affecting the expression of flagellar genes (2, 5, 24, 33). In other species, sirA/barA orthologs are required for virulence gene expression, exoenzyme and antibiotic production, motility, and biofilm formation (29).
SirA is a response regulator of the FixJ family (29). It is predicted to have a C-terminal DNA binding domain (helix-turn-helix) and an N-terminal phosphoacceptor domain. BarA is the sensor kinase for SirA, as determined on the basis of biochemical data generated with E. coli (55) and genetic data generated with Salmonella, Erwinia, and Pseudomonas (5, 17, 29, 70). BarA has a predicted secondary structure similar to those of the BvgS family of hybrid sensor kinases, which contain both receiver and transmitter domains. BarA is predicted to have two transmembrane alpha helices at the N terminus followed by a cytosolic transmitter domain containing a conserved histidine residue, a central receiver domain containing a conserved aspartate residue and, finally, a phosphotransfer domain containing a conserved histidine residue (29, 70).
SPI1 is a Salmonella-specific horizontal acquisition, the expression of which is controlled by numerous housekeeping regulators located throughout the Salmonella chromosome. The primary “adaptor” regulator that links the endogenous sensory capabilities of the cell to SPI1 gene expression is hilA (7). HilA is a member of the OmpR/ToxR family that is encoded within SPI1. However, HilA lacks the phosphoryl acceptor domain typical of other OmpR family members, and its activity is probably not modulated by posttranslational modification. Instead, HilA appears to be a constitutive activator that is controlled primarily at the level of transcription (7).
Numerous genes have been identified as having either positive or negative effects on hilA expression. Positive influences include sirA/barA, the posttranscriptional regulatory system consisting of csrA and csrB, and other genes, including fis, fadD, fliZ, ompR/envZ, hilC (sirC or sprA), and hilD (2-4, 16, 18, 33, 48, 53, 56, 58, 66). Negative influences include hilE, the two-component regulatory systems phoP/phoQ and phoB/phoR, the lon protease, ams (RNase E), hupB (HU), and the histone-like protein Hha (8-10, 18, 19, 48, 62). The hilC and hilD genes both encode AraC-type regulators. These two genes differ from the others in that they are Salmonella specific and are located within SPI1. The determination of which genes encode direct regulators of hilA and which have indirect effects has begun only recently. To date, it is known that the positive regulators HilC and HilD and the negative regulators Hha and Fis can bind directly the hilA promoter (8, 19, 53, 59).
Once HilA is produced, it directly activates two operons encoding the type III secretory apparatus, the prgH operon and the invF operon. The prgH operon includes prgHIJK-orgABC, and the invF operon includes invFGEABC-spaMNOPQRS-sicA-sipBCDA-iacP-sicP-sptP. InvF is a transcription factor that activates genes encoding secreted effectors (12-14). The secreted effectors are expressed at higher levels than the structural apparatus, so InvF appears to act as an “amplifier” of HilA activity. Within SPI1, InvF activates sipBCDA and downstream genes by binding to an internal promoter upstream of sicA. The sipBCD genes encode secreted effectors that are themselves required for the translocation of other effectors into the host cell and probably form a translocase complex in the target cell membrane (31). At least six effectors are encoded outside of SPI1. They are encoded by sopA, sopD, slrP, sopE, sopE2, and sopB (sigD) (68). The sopB and sopE genes are known to be regulated by invF (12, 14).
Interestingly, in addition to binding the hilA promoter, both the HilC and the HilD proteins can directly bind the invF promoter in vitro, providing the ability to bypass the hilA requirement (3). While the hilD gene is essential for hilA expression, the hilC gene is not (16, 47, 56). Although a hilC mutation had very little impact on invasion gene expression under the conditions tested, hilC is known to be regulated by SirA, and the plasmid-based expression of hilC can lead to invasion gene expression in the absence of hilA (3, 56).
In addition to regulating virulence gene expression, sirA orthologs affect flagellar gene expression and/or motility in E. coli, S. enterica serovar Typhimurium, Pseudomonas fluorescens, Pseudomonas aeruginosa, and Vibrio cholerae (24). Swarming motility in Pseudomonas syringae also requires sirA/barA orthologs (36). Flagellar biosynthesis has been divided into three levels (11). The class 1 operon encodes two proteins, FlhD and FlhC. These form a heterotetramer that is required for the transcriptional activation of class 2 genes, which encode hook-basal body complexes and the alternative sigma factor FliA. The FliA sigma factor allows the expression of class 3 genes, which encode the filament protein, hook-associated proteins, motor proteins, and chemotaxis proteins (38, 39).
In both E. coli and S. enterica serovar Typhimurium, the flhDC promoter is a major point of signal integration. Transcription from this promoter is regulated by numerous conditions and regulators, including DNA supercoiling, temperature, growth phase, cell cycle, cyclic AMP receptor protein, OmpR, H-NS, HdfR, and LrhA (11, 37, 44). Posttranscriptionally, the flhDC transcript is stabilized by CsrA (65).
CsrA is a small RNA binding protein that controls bacterial gene expression at the level of transcript stabilization (57). Depending on the target gene, CsrA either can stabilize transcripts and promote translation or can bind near the ribosome binding site to block translation and stimulate message decay. The csrB gene encodes a regulatory RNA that can bind up to 18 CsrA proteins and antagonize CsrA function (57). The csrB gene was recently discovered to be regulated by the SirA ortholog of E. coli, UvrY (61). In S. enterica serovar Typhimurium, the CsrA/csrB system is capable of both positively and negatively regulating the virulence genes located within SPI1 (4, 5). The effects are partially mediated through hilA, but this mediation may be indirect and may involve the regulation of hilC or hilD. Additionally, csrA can continue to affect invF and sipC, but not prgH, in the absence of hilA, indicating that CsrA may directly affect the posttranscriptional regulation of multiple invasion genes (4).
In this report, we focus on the pathways leading to SirA-dependent regulation of Salmonella virulence (the HilA regulon, which includes SPI1, and the sopB gene within SPI5) and flagellar genes (the FlhDC regulon). Using both genetic and biochemical approaches, we demonstrate that SirA regulates motility and virulence through independent pathways.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37°C in Luria-Bertani (LB) broth or on LB agar (1.2% [wt/vol] agar) (EM Science, Gibbstown, N.J.) unless otherwise stated. M9 minimal glucose medium was made as described previously (52). When necessary, media were supplemented with appropriate antibiotics at the following concentrations (micrograms per milliter): ampicillin, 100; kanamycin, 50; tetracycline, 20; and chloramphenicol, 30. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at a final concentration of 40 μg/ml. Motility assays were performed with TS medium (1% tryptone [Difco, Sparks, Md.]), 1% NaCl) or LB medium supplemented with 0.3% agar (EM Science).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or description | Source, construction, or reference |
|---|---|---|
| Strains | ||
| 14028 | Wild-type S. enterica serovar Typhimurium | American Type Culture Collection |
| AT351 | 14028 flhD::Tn10 | 14028 × P22/KK2040 (40) |
| BA460 | 14028 hilA1550::MudJ flhD::Tn10 | BA1550 × P22/AT351 |
| BA461 | 14028 hilA1550::MudJ flhD::Tn10 sirA3::Cam | BA1750 × P22/AT351 |
| BA736 | 14028 sirA2::Kan | 2 |
| BA743 | SL1344 sipC11-6::Tn5 lacZYA sirA3::Cam | EE638 × P22/BA705 (2) |
| BA746 | 14028 sirA3::Cam | 2 |
| BA790 | SL1344 ΔhilA520 sirA3::Cam | 2 |
| BA1550 | 14028 hilA1550::MudJ | 2 |
| BA1750 | 14028 hilA1550::MudJ sirA3::Cam | 2 |
| BL21λDE3 | E. coli ompT hsdSB (rB− mB−) gal dcm λDE3 | Novagen |
| BW20767 | E. coli leu-63::IS10 recA1 creC510 hsdR17 endA1 zbf-5uidA(ΔMluI)::pir+thi RP4-2-tet::Mu-1kan::Tn7 | 50 |
| EE638 | SL1344 sipC11-6::Tn5 lacZYA | 7 |
| RG206 | 14028 flhD::Tn10 sirA2::Kan | AT351 × P22/BA736 |
| RG235 | 14028 csrB+/csrB::lacZYA integrant | BW20767/pRG84 mated with 14028, with selection on M9 minimal glucose plates with kanamycin |
| SL1344 | S. enterica serovar Typhimurium hisG rpsL xyl | 30 |
| TIM25 | 14028 csrA+/csrA::lacZYA integrant | BW20767/pMT6 mated with 14028, with selection on M9 minimal glucose plates with kanamycin |
| TIM26 | 14028 sirA3::cam csrA+/csrA::lacZYA integrant | TIM25 × P22/BA746 |
| TIM27 | 14028 sirA3::cam csrB+/csrB::lacZYA integrant | RG235 × P22/BA746 |
| TOP10 | E. coli mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| XL1-Blue | E. coli recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10] | Stratagene |
| Plasmids | ||
| pBA322 | pBAD18-kn sirA+ | This study (see Materials and Methods) |
| pBA416 | pET24a sirA expression vector | This study (see Materials and Methods) |
| pBAD18-kn | Arabinose-conditional expression vector; Kanr ColE1 | 26 |
| pBAD33 | Arabinose-conditional expression vector; Camr p15A | 26 |
| pCR2.1-TOPO | Cloning vector; Kanr ColE1 | Invitrogen |
| pCR-Blunt II-TOPO | Cloning vector; Kanr ColE1 | Invitrogen |
| pCR-Script AMP | Cloning vector; Ampr ColE1 | Stratagene |
| pET24a | Expression vector; Kanr ColE1 | Novagen |
| pMT6 | csrA-lacZYA fusion in pVIK112 | This study (see Materials and Methods) |
| pQE30 | Expression vector; Ampr ColE1 | Qiagen |
| pRG19 | motA::luxCDABE fusion in pSB401 | 24 |
| pRG38 | flhD::luxCDABE fusion in pSB401 | 24 |
| pRG61 | sopB::luxCDABE fusion in pSB377 | This study (see Materials and Methods) |
| pRG67 | pQE30 barA198 expression vector | This study (see Materials and Methods) |
| pRG84 | csrB-lacZYA fusion in pVIK112 | This study (see Materials and Methods) |
| pSB377 | luxCDABE transcriptional fusion vector; Ampr ColE1 | 67 |
| pSB401 | luxCDABE transcriptional fusion vector; Tetr p15a | 67 |
| pJVR4 | pBAD33 sirA+ | 2 |
| pVIK112 | lacZYA transcriptional fusion vector; Kanr oriR6K | 35 |
Growth conditions and assays of β-galactosidase and luciferase activities.
It was previously shown that the maximal effects of SirA are observed when cells are actively involved in chemotaxis through motility agar (24). Growth on 1.5% agar plates or in shaking liquid broth cultures provides moderate or low, respectively, levels of sirA-dependent gene regulation (24). Therefore, whenever possible, we used motility agar and chromosomal lacZYA fusions to assess the effects of sirA, but plasmid-based luxCDABE fusions with different antibiotic resistance markers also were used when necessary. All of the results were scored qualitatively due to the technical difficulties of recovering bacteria from motility agar without denaturing β-galactosidase. For the epistasis experiments described here, this plus/minus scoring system works well. Some strains used in this study are not motile, so they were studied by streaking for isolation on 1.2% agar plates. Additionally, we have determined that SirA affects flagellar fusions more dramatically on TS plates than on LB plates. Conversely, SirA affects virulence fusions (SPI1 and SPI5) more markedly on LB plates than on TS plates. The plate type used for each experiment is listed in the appropriate figure legend. The expression of luciferase activity by reporter strains growing on agar plates or in motility agar was visualized with a Hamamatsu C2400-32 intensified charge-coupled device (CCD) camera attached to an Argus 20 image processor as described previously (24). β-Galactosidase activity was assessed qualitatively by photographing bacteria grown in the presence of X-Gal either on 1.2% agar plates or in motility agar.
DNA manipulations and genetic techniques.
DNA manipulations were performed by standard techniques (49a),typically with TOP10 as the recipient E. coli strain (Invitrogen, Carlsbad, Calif.). Restriction enzymes were purchased from Life Technologies (Rockville, Md.). Oligonucleotides were synthesized by IDT Technologies (Coralville, Iowa). Plasmid DNAs were prepared and DNA fragments were isolated from agarose gels by using the appropriate Qiagen (Valencia, Calif.) kits (QIAprep spin miniprep kit and QIAquick gel extraction kit, respectively) according to the protocols of the supplier. Electroporation of S. enterica serovar Typhimurium was achieved by using a Gene Pulser II system (Bio-Rad, Hercules, Calif.). Transduction performed with phage P22 HTint was followed by streaking for isolation on LB-EGTA agar plates containing appropriate antibiotics and by confirming smooth lipopolysaccharide and lack of pseudolysogeny on Evans blue-uranine plates as described previously (49). Mobilization of suicide vectors from E. coli to Salmonella was performed by mixing 50 μl each of stationary-phase cultures of the donor and the recipient and plating the samples on LB agar overnight at 37°C. On the following day, the bacteria were scraped from the LB plate, resuspended in M9 minimal glucose medium, and plated on M9 minimal glucose plates containing kanamycin. We have determined that the pir-dependent suicide vector pVIK112 (35), which creates lacZYA transcriptional fusions, can persist in Salmonella in the absence of a promoter fragment cloned upstream of lacZYA. Presumably, the plasmid is integrated in a region of the genome that is homologous to the vector, but this notion has not been investigated. However, all chromosomal merodiploid fusions constructed with pVIK112 were confirmed to have been integrated into the correct sites by using PCR with a primer designed to bind the Salmonella chromosome upstream of the cloned promoter region and another primer that binds the lacZ gene of the vector. Regulatory mutations were then transduced into the proper fusion strain with phage P22 HTint.
Plasmid constructions.
Plasmid pBA322 was constructed by removing the sirA fragment of pJVR4 with XbaI and SacI and cloning the fragment into pBAD18-kn digested with XbaI and SacI. Plasmid pMT6 was constructed by amplifying the csrA promoter region (nucleotides [nt] 1398 to 1704 of a sequence with GenBank accession number AE008829) with Taq DNA polymerase (GeneChoice, Frederick, Md.), cloning the fragment into pCR2.1-TOPO (Invitrogen), removing the fragment with EcoRI, and inserting it into the EcoRI site of pVIK112. Plasmid pRG61 was constructed by amplifying the sopB promoter region (nt 4 to 657 of a sequence with accession number AF021817) with Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.), cloning the fragment into pCR-Blunt II-TOPO (Invitrogen), removing the fragment with EcoRI, and inserting it into the EcoRI site of pSB377. Plasmid pRG84 was constructed by amplifying the csrB promoter region (nt 1738 to 1984 of a sequence with accession number AF076153) with Pfu Turbo DNA polymerase, cloning the fragment into pCR-Blunt II-TOPO, removing the fragment with EcoRI, and inserting it into the EcoRI site of pVIK112. Plasmid pBA416 was constructed by amplifying sirA (nt 1911 to 2602 of a sequence with accession number U88651; the 3′ primer also incorporated an XhoI site) with Pfu DNA polymerase (Stratagene), cloning the fragment into the SrfI site of pCR-Script AMP (Stratagene), removing the fragment with EcoRI and XhoI, and inserting it into pET24a (Novagen, Madison, Wis.) digested with EcoRI and XhoI. Plasmid pRG67 was constructed by amplifying a fragment of barA lacking the first 198 codons (nt 697 to 2889 of a sequence with accession number AF171069) with Pfu DNA polymerase (Stratagene). The 5′ primer included a BamHI site, and the 3′ primer included a PstI site. The resulting PCR product was cloned into pCR-Blunt II-TOPO, removed with BamHI and PstI, and cloned into pQE30 (Qiagen) digested with BamHI and PstI.
Purification of His6-tagged proteins.
E. coli XL1-Blue/pRG67 (His6-BarA198 [BarA198 is a form of BarA lacking the two N-terminal membrane-spanning domains]) and E. coli BL21λDE3/pBA416 (SirA-His6) were grown in 1 liter of broth containing 16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl with appropriate antibiotics (55). E. coli BL21λDE3/pBA416 was grown on a rotary shaker at 37°C. At an optical density at 550 nm of 0.5, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 1 mM, followed by 4 h of shaking incubation at 37°C. E. coli XL1-Blue/pRG67 was grown in a 3-liter jacketed bioreactor (Z61103CT04; Applikon Dependable Instruments DV, Schiedam, The Netherlands) at 37°C with 500 ml of air per liter of medium per min of aeration and with agitation at 200 rpm. When the culture reached an optical density at 550 nm of 0.5, IPTG was added to 1 mM, and incubation was continued overnight at 17°C. Cells from both strains were harvested by centrifugation and suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) containing 1 mg of lysozyme/ml and 1 mM protease inhibitor phenylmethylsulfonyl fluoride. Cells were then disrupted with a French press and mild sonication. The His6-tagged proteins were purified by nickel affinity chromatography with nickel resin from Qiagen according to the manufacturer's instructions. Elution was performed with lysis buffer containing increasing concentrations of imidazole. The His6-tagged proteins eluted at between 50 and 200 mM imidazole. The preparation of His6-BarA198 was dialyzed in a Slyde-A-Lyzer 7K cassette (Pierce, Rockford, Ill.) overnight in dialysis buffer (0.1 M Tris-HCl, 30 mM potassium glutamate, 1 mM dithiothreitol [DTT], 0.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 20% [vol/vol] glycerol [pH 8.0]) and then concentrated in a spin dialysis unit with a 30-kDa molecular-mass cutoff (Vivascience, Carlsbad, Calif.). SirA-His6 was spin dialyzed in a Vivascience spin dialysis unit (10-kDa molecular-mass cutoff) and then mixed 2:1 with 3× phosphorylation buffer containing 14% glycerol (55). Protein concentrations were estimated by using a modified Lowryassay (DC protein assay kit; Bio-Rad) or a bicinchoninic acid protein assay (Pierce) with serial dilutions of bovine serum albumin (BSA) as a standard. The protein preparations were stored at −80°C.
Phosphorylation and transphosphorylation assays.
Phosphorylation and transphosphorylation reactions were carried out by using phosphorylation buffer (55) with 40 μM [γ-32P]ATP at room temperature (specific activity, 3,000 Ci/mmol; Perkin-Elmer Life Sciences, Wellesley, Mass.). The phosphorylation reaction was initiated by the addition of [γ-32P]ATP to 15 μg of His6-BarA198 in phosphorylation buffer (total volume, 150 μl). Aliquots (15 μl) were removed at various times and added to an equal volume of 2× LSB (0.09 M Tris-HCl [pH 6.8], 20% [vol/vol] glycerol, 2% [wt/vol] sodium dodecyl sulfate [SDS], 0.02% [wt/vol] bromophenol blue, 0.1% [wt/vol] DTT) to stop the reaction. The transphosphorylation reaction was initiated by the addition of 30 μg of SirA-His6 to a 150-μl phosphorylation reaction mixture with or without His6-BarA198. Aliquots (15 μl) were removed at various times and added to an equal volume of 2× LSB to stop the reaction. Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) with 12.5% acrylamide gels. Radioactive regions of gels were detected with a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and quantitated with ImageQuant 5.2 software (Molecular Dynamics).
Gel mobility shift assays.
Promoter regions upstream of csrA, csrB, flhD, fliA, hilA, hilC, hilD, and invF were amplified by PCR with S. enterica serovar Typhimurium 14028 as a template. The DNA fragments spanned nucleotides of sequences with the following GenBank accession numbers: hilA is nt 550 to 850 of U25352, flhD is nt 548 to 850 of AF029300, hilC is nt 2146 to 2472 of AE008831, hilD is nt 6729 to 7027 of AE008831, csrA is nt 1398 to 1704 of AE008829, invF is nt 12500 to 12800 of AE008832, fliA is nt 250 to 550 of AB010947, and csrB is nt 1738 to 1984 of AF076153. The DNA fragments were purified by agarose gel electrophoresis followed by gel extraction with a QIAquick gel extraction kit. A 250-ng quantity of each fragment was end labeled for 1 h at 37°C with [γ-32P]ATP and T4 polynucleotide kinase according to the manufacturer's instructions (Boehringer Mannheim, Indianapolis, Ind.). The labeled fragment was then separated from unincorporated nucleotides with ProbeQuant G-50 microcolumns (Amersham Life Sciences, Piscataway, N.J.).
DNA binding reactions were carried out with a total volume of 20 μl containing 5 μl of 3× DNA binding buffer (129 mM Tris-HCl, 90 mM potassium acetate, 24 mM MgSO4, 81 mM ammonium acetate, 3 mM DTT, 240 mM KCl, 30% glycerol; this is a modification of a buffer from reference 22), 5 μl of His6-BarA198-SirA-His6 transphosphorylation reaction mixture (as described above except with nonradioactive ATP), 2 μl of labeled DNA fragment (≈5 ng; 54,000 cpm), 2 μl of poly(dI-dC) (1 μg/μl), 1 μl of BSA (1 μg/μl), and 0.5 μl of 0.5 M EDTA. DNA binding reactions were carried out at room temperature for 25 min, and then samples were subjected to native PAGE with 5% polyacrylamide gels and a buffer containing 90 mM Tris, 90 mM H3BO3, and 2 mM EDTA. Radioactive regions of gels were detected with a Storm PhosphorImager and quantitated with ImageQuant 5.2 software.
RESULTS
SirA activates the HilA regulon independently of flhD.
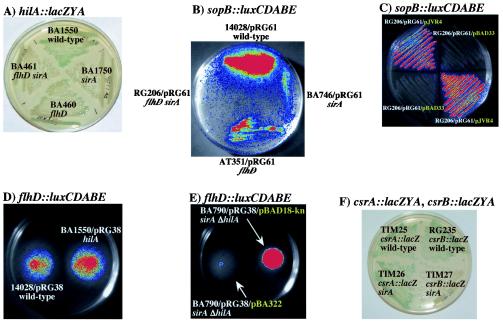
The HilA regulon is known to be affected by mutations in flhC and fliA (15, 32, 48). Complementation studies revealed that fliZ, a gene downstream of fliA and expressed from the same transcript, could restore SPI1 gene expression in flhC or fliA mutants (48). We confirmed that SPI1 gene expression is decreased by an flhD::Tn10 mutant. flhD mutant strains showed less hilA::lacZYA expression and sopB::luxCDABE expression than flhD+ strains (Fig. 1A and B).
FIG. 1.
Interactions among SirA, the FlhD regulon, and the HilA regulon. (A) HilA expression is positively affected by flhD, and sirA regulates hilA in the absence of flhD. A lacZYA fusion to hilA produces more blue pigment in the wild-type background, BA1550, than in the flhD mutant, BA460, or the sirA mutant, BA1750. Pigment production decreases further in an sirA flhD double mutant, BA461. The plate shown is an LB plate that contained X-Gal and was incubated at 37°C. (B) A representative member of the HilA regulon, sopB, is positively affected by flhD, and sirA regulates sopB in the absence of flhD. A sopB::luxCDABE fusion (pRG61) yields more light in the wild-type background, 14028, than in the flhD mutant, AT351, or the sirA mutant, BA746 (intensity scale goes from blue [low luminescence] to red [high luminescence]). Light production decreases further in an sirA flhD double mutant, RG206. The plate shown is an LB plate that was incubated at 37°C. (C) SirA can regulate sopB in an flhD mutant. RG206/pRG61 is an sirA flhD double mutant carrying a sopB::luxCDABE fusion on pRG61. pJVR4 carries sirA under arabinose control, while pBAD33 is the vector control. sopB::luxCDABE expression is higher in the strain carrying pJVR4. The plate shown is an LB plate that was incubated at 37°C. (D) HilA has no effect on an flhD::luxCDABE fusion. 14028/pRG38 is an hilA+ strain carrying an flhD::luxCDABE fusion, while BA1550/pRG38 is an isogenic hilA mutant. The strains show equal levels of luminescence, indicating that hilA does not regulate flhD. The plate shown is a TS motility agar plate that was incubated at 37°C. (E) SirA represses flhD in an hilA mutant. BA790 is an hilA sirA double mutant. The presence of sirA on a plasmid, pBA322, decreases the expression of the flhD::luxCDABE fusion on plasmid pRG38 compared to the vector control, pBAD18-kn. The plate shown is a TS motility agar plate that was incubated at 37°C. (F) A csrB::lacZYA fusion, but not a csrA::lacZYA fusion, is expressed at lower levels in an sirA mutant (TIM25 versus TIM26 for csrA::lacZYA; RG235 versus TIM27 for csrB::lacZYA). The plate shown is an LB plate that contained X-Gal and that was incubated at 37°C.
Because both sirA and flhD positively affect the HilA regulon, we tested the hypothesis that SirA affects the HilA regulon only by regulating flhDC. If this hypothesis were true, then SirA would not be able to affect the HilA regulon in the absence of flhD. This hypothesis was examined in three ways. First, it can be seen in Fig. 1A that an sirA flhD double mutant has lower hilA::lacZYA expression than an flhD single mutant (BA460 versus BA461). This finding indicates that sirA regulates hilA in the absence of flhD. Second, we compared the expression of a sopB::luxCDABE fusion in an flhD mutant and an flhD sirA double mutant (AT351/pRG61 versus RG206/pRG61) (Fig. 1B). The presence of sirA has a positive effect on the sopB fusion, even though flhD is mutated (Fig. 1B). Third, we constructed an arabinose-conditional sirA strain that lacks flhD and carries a sopB-luxCDABE fusion (RG206/pRG61/pJVR4). A comparison of the luciferase activities of this strain and an isogenic vector control strain (RG206/pRG61/pBAD33) clearly demonstrated that sirA is able to activate sopB expression in the absence of flhD (Fig. 1C). The basal level of sirA expression from pJVR4 is sufficient to achieve this effect without the addition of arabinose to the media. This methodology also served as a sirA complementation test to demonstrate that the regulatory effects are due to sirA and not to a secondary mutation.
HilA does not affect the FlhD regulon, and SirA represses flhD independently of hilA.
Wild-type S. enterica serovar Typhimurium and an isogenic hilA deletion mutant, both carrying a plasmid-based flhD::luxCDABE fusion, were compared during growth in motility agar (14028/pRG38 versus BA1550/pRG38) (Fig. 1D). Both strains were found to express similar levels of luciferase activity, indicating that hilA does not affect the expression of flhD under these conditions (Fig. 1D). Similar results were obtained with transcriptional fusions to motA (14028/pRG19 versus BA1550/pRG19) (data not shown). These results demonstrate that hilA does not affect the flagellar regulon and strongly suggest that SirA could not regulate the flagellar regulon via hilA. Confirmation of this hypothesis was obtained by examining a plasmid-based flhD::luxCDABE fusion, pRG38, in an arabinose-conditional sirA strain with a deletion in hilA (BA790/pRG38/pBA322). In TS motility agar, this strain produced less light than the vector control strain (BA790/pRG38/pBAD18-kn), demonstrating that SirA is able to repress the flagellar regulon in the absence of hilA (Fig. 1E). Similar results were obtained with an motA::luxCDABE fusion (BA790/pRG19/pBA322 versus BA790/pRG38/pBAD18-kn) (data not shown). The basal level of sirA expression from pBA322 was sufficient to achieve this effect without the addition of arabinose to the media. This methodology also served as a sirA complementation test to demonstrate that the regulatory effects are due to sirA and not to a secondary mutation.
SirA affects a regulatory gene above flhD in the flagellar regulon, csrB.
For E. coli, it is known that UvrY (SirA) positively regulates csrB (61). The csrB gene encodes a regulatory RNA that binds to and antagonizes CsrA (45, 57). In E. coli and Salmonella, the CsrA protein is known to positively affect FlhDC expression posttranscriptionally (41, 65). In Salmonella, CsrA also affects the HilA regulon (4, 41). Therefore, we hypothesized that the effect of SirA on flagella could be indirect and due to the regulation of csrB by SirA. Chromosomal merodiploid lacZYA transcriptional fusions to csrA and csrB were constructed and tested for regulation by sirA. The csrA fusion showed no regulation by sirA (TIM25 versus TIM26) (Fig. 1F), but the csrB fusion was activated by sirA (RG235 versus TIM27) (Fig. 1F). These results are consistent with recent findings for E. coli (61).
Purification and phosphorylation of SirA and BarA198.
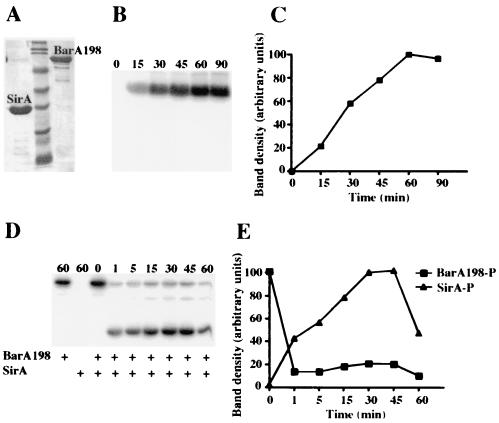
Given the complexity of the genetic networks controlled by SirA, it was imperative to determine which promoters are bound by SirA and which are only affected indirectly. To do this, we purified SirA and a soluble portion of BarA to be used in phosphorylating SirA. Both proteins were tagged with six histidine residues to facilitate purification by nickel affinity chromatography essentially as described by Pernestig et al. (55). The sirA construct uses the full-length gene and incorporates the six histidine residues at the C terminus of the protein (pBA416). The barA construct lacks residues 1 to 198 and contains an N-terminal His6 tag to form BarA198 (pRG67). Previously, an E. coli variant of this construct was found to be capable of transferring phosphate from ATP to SirA and was found to be more suitable than the wild type for purification because it lacks the two transmembrane helices found at the N terminus (55). To confirm that our SirA-His6 construct was functional in vivo, we electroporated pBA416 or the vector control pET24a into an sirA mutant S. enterica serovar Typhimurium strain carrying an sipC::lacZYA fusion. His6-tagged sirA fully complemented the chromosomal sirA mutation and activated the sipC::lacZYA fusion (BA743/pBA416 versus BA743/pET24a) (data not shown). Purified BarA198 autophosphorylated in the presence of [γ-32P]ATP and transferred phosphate to SirA (Fig. 2B to E). The amount of autophosphorylated BarA198 in the reaction steadily increased for up to 60 min, at which point it maintained a steady-state level (Fig. 2B and C). The presence of SDS in the time-zero sample prevented the autophosphorylation of BarA198 (Fig. 2B and C). The addition of SirA-His6 to a BarA198 autophosphorylation reaction allowed BarA198 to transfer phosphate to SirA. Transphosphorylation between BarA198 and SirA was detectable within 1 min and continued to increase for 30 min (Fig. 2D and E). The pool of phosphorylated SirA appeared to be stable for at least 30 min. SirA did not accept phosphate directly from [γ-32P]ATP in the absence of BarA198, and the presence of SDS in the time-zero sample prevented transphosphorylation between SirA and BarA198 (Fig. 2D and E). These results are consistent with those observed for E. coli BarA198 and UvrY (SirA) (55).
FIG. 2.
Purified BarA198 can autophosphorylate and transfer phosphate to SirA in vitro. (A) Purified SirA and BarA198 after nickel affinity chromatography, SDS-PAGE, and Coomassie blue staining. Both proteins are His6 tagged. Molecular mass markers (Bio-Rad prestained broad-range standards) are in the center lane (from bottom to top: 7.1, 21, 29, 35, 49, 80, 124, and 209 kDa). SirA-His6 is predicted to be 24.95 kDa; His6-BarA198 is predicted to be 81.60 kDa. (B) Time course of BarA198 autophosphorylation in the presence of [γ-32P]ATP. Reactions were allowed to proceed for the times indicated above each lane in minutes before aliquots were removed and the reactions were stopped by the addition of sample buffer containing SDS. Samples were resolved by SDS-PAGE and detected with a PhosphorImager. (C) Plot of band densities as determined by ImageQuant software from panel B. (D) Transphosphorylation of SirA by BarA198-P. BarA198 that had been preincubated with [γ-32P]ATP for 25 min was mixed with SirA as indicated. One reaction contained SirA with [γ-32P]ATP and no BarA198. Reactions were stopped at the times indicated above each lane in minutes by the addition of sample buffer containing SDS. Samples were resolved by SDS-PAGE and detected with a PhosphorImager. (E) Plot of band densities as determined by ImageQuant software from panel D. P, phosphorylated.
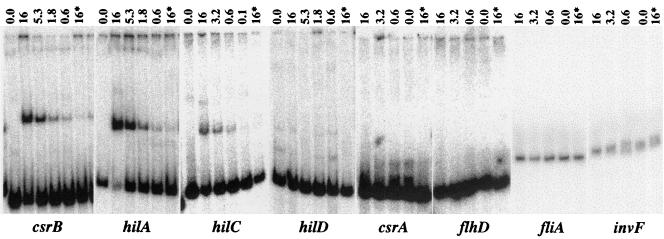
SirA binds the csrB, hilA, and hilC promoters.
Purified SirA was phosphorylated in the presence of ATP and BarA198 and then used in a gel mobility shift assay to determine whether SirA directly binds particular promoter regions. Eight promoters were tested in this assay: flhD, fliA, hilA, hilC, hilD, invF, csrA, and csrB. The gel mobility of the csrB and hilA promoters was shifted strongly, while the shift of the gel mobility of hilC was reproducibly weaker (Fig. 3 and data not shown). The remaining five promoters were not shifted at all. Nonspecific competitor DNA [poly(dI-dC)] and protein (BSA) were added to all reactions to minimize nonspecific interactions of the labeled DNA fragments with the proteins. As expected for a specific protein-DNA interaction, the addition of unlabeled promoter DNA as a specific competitor diminished the shift of the labeled hilA, csrB, and hilC fragments (Fig. 3). The concentration of phosphorylated SirA required to shift each of these promoters was determined to be approximately 600 nM. To evaluate the requirement for SirA phosphorylation for the binding of these promoters, the hilA and csrB promoters were used in experiments that directly compared the abilities of phosphorylated and nonphosphorylated SirA-His6 to shift these promoters. Interestingly, both promoters were shifted regardless of the phosphorylation state of SirA. The only difference was that the band intensity of the shifted species was decreased by approximately twofold when nonphosphorylated SirA-His6 rather than phosphorylated SirA-His6 was used (data not shown).
FIG. 3.
SirA alters the gel mobilities of promoter DNA fragments. Eight promoter DNA fragments (shown below panels) were tested for their abilities to bind purified SirA in a gel mobility shift assay. SirA was phosphorylated by incubation with BarA198 and ATP for 25 min and then was added to promoter DNA labeled with [γ-32P]ATP. Each reaction was resolved by nondenaturing PAGE, and samples were detected with a PhosphorImager. The micromolar concentration of SirA in each reaction is indicated above each lane. An asterisk indicates that a 30- to 50-fold excess of unlabeled promoter DNA fragment was added to the reaction as a specific competitor. All reactions contained nonspecific competitor DNA [2 μg of poly(dI-dC)] and protein (0.2 μg of BSA).
DISCUSSION
BarA/SirA orthologs are present throughout the γ-proteobacteria and control secondary metabolism and virulence functions. However, only in S. enterica serovar Typhimurium has a systematic effort been made to identify the entire SirA regulon (2, 24). The genes identified can be grouped into two main subregulons, the FlhDC regulon, which controls motility and chemotaxis (≈55 genes), and the HilA regulon, which controls invasion-associated TTSS-1 (≈40 genes). In this report, we have elucidated the regulatory triad that exists among SirA, FlhDC, and HilA. Pathways by which their genes are affected by SirA and promoters that are directly bound by SirA have been identified.
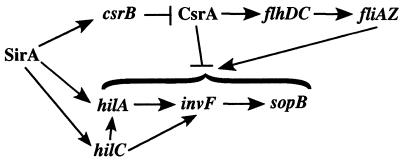
The expression of the FlhDC regulon is decreased in the presence of sirA (24). In this study, we have determined that this repressive effect likely is indirect. The purified SirA protein does not bind the flhDC promoter, but it does bind the csrB promoter. Additionally, a transcriptional fusion to csrB responds to sirA. Therefore, SirA directly activates csrB, which antagonizes the activity of CsrA, which is known to directly affect flhDC (Fig. 4). SirA does not regulate the csrA gene or bind the csrA promoter. These results match those of previous studies of E. coli in which UvrY (SirA) was demonstrated to directly activate a csrB::lacZ fusion in in vitro transcription-translation reactions (61). Therefore, the pathways by which SirA represses flagellar gene expression appear to be similar in E. coli and S. enterica serovar Typhimurium.
FIG. 4.
Model of the SirA regulatory cascade. SirA directly activates the csrB, hilA, and hilC promoters. HilA and HilC activate the invF gene, the product of which activates genes encoding secreted effectors, including sopB. HilA also activates genes encoding the structural apparatus of TTSS-1 (data not shown). The csrB RNA directly binds and antagonizes the activity of CsrA. CsrA directly increases FlhDC expression posttranscriptionally. CsrA and fliZ also affect multiple levels of the HilA regulon through undefined mechanisms.
The second subset of the SirA regulon is not present in E. coli. This is the HilA regulon, which includes TTSS-1, encoded within SPI1, along with secreted effectors that are encoded within SPI1 and elsewhere around the chromosome. In contrast to the expression of the FlhDC regulon, the expression of the HilA regulon is increased by sirA (2, 5, 33). A major point of signal integration for this regulon is the hilA promoter. Once expressed, HilA activates the structural components of TTSS-1 and another regulatory gene, invF (6, 7). InvF then activates secreted effectors (12, 14). Although HilA is the major regulator, two other regulators encoded within SPI1, HilC and HilD, can directly increase the expression of both hilA and invF independently of hilA (3, 53, 56). A link between the FlhDC and the HilA regulons was suggested by the observations that mutations in flhC and fliA resulted in decreased expression of the HilA regulon (15, 32, 48). Complementation experiments demonstrated that fliZ (part of the fliAZ operon) could restore hilA expression to both mutants, indicating that fliZ or a gene affected by fliZ is the direct regulator of hilA (48). It was reasonable to hypothesize, therefore, that activation of the HilA regulon by SirA may be indirect and mediated by the FlhDC regulon. In this study, we have eliminated this possibility both genetically and biochemically. Genetic experiments indicated that SirA can activate the HilA regulon in the absence of flhDC, and biochemical experiments indicated that SirA can directly bind the hilA and hilC promoters. SirA binding to the hilC promoter is consistent with previous genetic data indicating that SirA can bypass the hilA gene to regulate invasion determinants through hilC (56).
Phosphotransfer between the E. coli orthologs of BarA and SirA (UvrY) has been demonstrated with purified proteins in vitro (55). We have now demonstrated phosphotransfer between BarA and SirA from a second organism, S. enterica serovar Typhimurium. However, SirA did not require in vitro phosphorylation to bind these promoters. These results are very similar to the results obtained for E. coli, in which UvrY (SirA) was able, without a phosphorylation step, to activate csrB::lacZ in in vitro transcription-translation assays (61). These observations are interesting given the in vivo requirement for BarA or acetylphosphate for SirA activity (5, 42). It is possible that SirA-His6 was already phosphorylated to some extent when purified from E. coli. Alternatively, SirA may bind specific promoter sequences in the absence of phosphorylation but fail to activate transcription until it is phosphorylated. Further studies are required to determine the mechanism of promoter activation by SirA orthologs.
The observation that SirA can directly bind the hilA promoter is consistent with the adaptor hypothesis, in which horizontally acquired virulence genes often carry with them a regulatory locus that is in turn controlled by endogenous housekeeping sensors (25). Using this adaptor regulator, the horizontally acquired virulence genes can immediately tap the extensive sensory apparatus of the host bacterium. This notion raises the possibility that if SPI1 were to move from Salmonella into another γ-proteobacterium, TTSS-1 might be regulated correctly (unless the new host required activation at different host locations and/or under different environmental conditions). Furthermore, it is possible that the SirA binding site in the hilA promoter evolved in another organism before the acquisition of SPI1 by Salmonella.
Given that SirA controls horizontally acquired virulence genes in such a large number of pathogenic species, it is a potential therapeutic target. A ligand that could block signal detection by BarA may be sufficient to disrupt virulence gene expression in numerous pathogens. The nature of the signal detected by BarA is presently unknown, although several hypotheses have been proposed. For E. coli, it has been noted that SirA affects central carbon metabolism, suggesting that a metabolite may the signal (54). For L. pneumophila, it has been observed that ppGpp has no effect on target genes in the absence of the barA/sirA orthologs letA/letS, and it was suggested that ppGpp may be the signal (27). Interestingly, relA (encoding a ppGpp synthase) appears to be divergently transcribed from barA in the Salmonella chromosome. For P. fluorescens, a dichloromethane-extractable substance in culture supernatants affects target gene expression, but not in a mutant lacking the predicted sensor domain of gacS (barA) (28, 70). If this substance truly were the signal for BarA, then the implication is that SirA/BarA is part of a novel quorum-sensing system. Interestingly, for E. coli and Salmonella (but not for Pseudomonas), sirA is located downstream of sdiA, a luxR family member that detects the N-acylhomoserine lactone production of other microbial species (1, 51, 60). It will be very interesting if SdiA and SirA both are found to be quorum-sensing regulators. Of course, all of these potential signals may affect other regulators within the same regulatory cascade, and not BarA itself. Further experimentation is required to determine the signal(s) and mechanisms by which BarA/SirA orthologs control virulence gene expression throughout the γ-proteobacteria.
Acknowledgments
This publication was made possible by grant no. 5 RO1 AI50002-03 from the National Institute of Allergy and Infectious Diseases (to B.M.M.A.).
We are grateful to Adam Toguchi and Rasika Harshey for constructing and sharing AT351, Bill Metcalf for BW20767, and Cathy Lee for EE638. We are grateful to Irina Artsimovitch for biochemical guidance, John Gunn and Irina Artsimovitch for critical reading of the manuscript, and Jon-David Sears and the OSU fermentation facility for fermentations.
REFERENCES
- 1.Ahmer, B. M. M., J. van Reeuwijk, C. D. Timmers, P. J. Valentine, and F. Heffron. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmer, B. M. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 3.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-728. [DOI] [PubMed] [Google Scholar]
- 4.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 6.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 7.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 8.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 175:4475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 14.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichelberg, K., and J. E. Galan. 2000. The flagellar sigma factor FliA (sigma(28)) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect. Immun. 68:2735-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichelberg, K., W. D. Hardt, and J. E. Galan. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 18.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 19.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis, C. L., T. A. Ryan, B. D. Jones, S. J. Smith, and S. Falkow. 1993. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364:639-642. [DOI] [PubMed] [Google Scholar]
- 21.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galinier, A., A. M. Garnerone, J. M. Reyrat, D. Kahn, J. Batut, and P. Boistard. 1994. Phosphorylation of the Rhizobium meliloti FixJ protein induces its binding to a compound regulatory region at the fixK promoter. J. Biol. Chem. 269:23784-23789. [PubMed] [Google Scholar]
- 23.Galyov, E. E., M. W. Wood, R. Rosqvist, P. B. Mullan, P. R. Watson, S. Hedges, and T. S. Wallis. 1997. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 25:903-912. [DOI] [PubMed] [Google Scholar]
- 24.Goodier, R. I., and B. M. Ahmer. 2001. SirA orthologs affect both motility and virulence. J. Bacteriol. 183:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guiney, D. G. 1997. Regulation of bacterial virulence gene expression by the host environment. J. Clin. Investig. 99:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 28.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 30.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 31.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 33.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 34.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalogeraki, V. S., and S. C. Winans. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69-75. [DOI] [PubMed] [Google Scholar]
- 36.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko, M., and C. Park. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 182:4670-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komeda, Y. 1982. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J. Bacteriol. 150:16-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutsukake, K., Y. Ohya, S. Yamaguchi, and T. Iino. 1988. Operon structure of flagellar genes in Salmonella typhimurium. Mol. Gen. Genet. 214:11-15. [DOI] [PubMed] [Google Scholar]
- 41.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633-1645. [DOI] [PubMed] [Google Scholar]
- 42.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 43.Lee, C. A., M. Silva, A. M. Siber, A. J. Kelly, E. Galyov, and B. A. McCormick. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. USA 97:12283-12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 45.Liu, M. Y., G. Gui, B. Wei, J. F. Preston III, L. Oakford, U. Yuksel, D. P. Giedroc, and T. Romeo. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272:17502-17510. [DOI] [PubMed] [Google Scholar]
- 46.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 47.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49a.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Michael, B., J. N. Smith, S. Swift, F. Heffron, and B. M. Ahmer. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pernestig, A. K., D. Georgellis, T. Romeo, K. Suzuki, H. Tomenius, S. Normark, and O. Melefors. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pernestig, A. K., O. Melefors, and D. Georgellis. 2000. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 56.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 58.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 59.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. N., and B. M. M. Ahmer. 2003. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson, P. R., E. E. Galyov, S. M. Paulin, P. W. Jones, and T. S. Wallis. 1998. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect. Immun. 66:1432-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 66.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]
- 67.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, S., R. A. Kingsley, R. L. Santos, H. Andrews-Polymenis, M. Raffatellu, J. Figueiredo, J. Nunes, R. M. Tsolis, L. G. Adams, and A. J. Baumler. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]
- 70.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]