With the discovery of penicillin as an efficient antibacterial agent isolated from the fungus Penicillium notatum, microorganisms attracted considerable attention as a new source for pharmaceutical agents. Screening of microbial extracts revealed the large structural diversity of natural compounds with broad biological activities, such as antimicrobial, antiviral, immunosuppressive, and antitumor activities. Like penicillin, many of these products are small peptide molecules consisting of 3 to 22 residues with often unusual structural elements. These include heterocyclic elements,d-amino acids, and glycosylated and N-methylated residues, suggesting a nonribosomal origin of biosynthesis. Due to the potent pharmacological activities of these compounds, there was an overwhelming interest in exploring their mechanism of synthesis. Lipmann et al. reported as early as the 1970s that the cyclic peptides gramicidin S and tyrocidine from Bacillus spp. were produced in a nucleic acid-independent way through the use of large enzyme complexes similar to fatty acid synthases (17). Subsequently, other peptidic natural products were shown to be assembled by large enzymes, referred to as nonribosomal peptide synthetases (NRPS), which utilize the multiple-carrier thiotemplate mechanism (34). A common feature of many nonribosomally produced peptides is their constrained structure, which ensures a precise functionality important for a proper interaction with the dedicated molecular target in the cell. Nature achieves this rigidity in molecular structure through several strategies: the molecule can be oxidatively cross-linked, as in vancomycin, heterocyclized, as in penicillin or, more commonly, cyclized, as in fengycin (Fig. 1). Cyclization seems to be the predominant way of constraining nonribosomally synthesized peptides. Because peptide cyclization from the point of view of chemical synthesis is difficult to achieve without protection of all of the side chains, there has been rapidly growing interest in exploration of the enzymatic cyclization mechanism for the development of new synthesis routes.
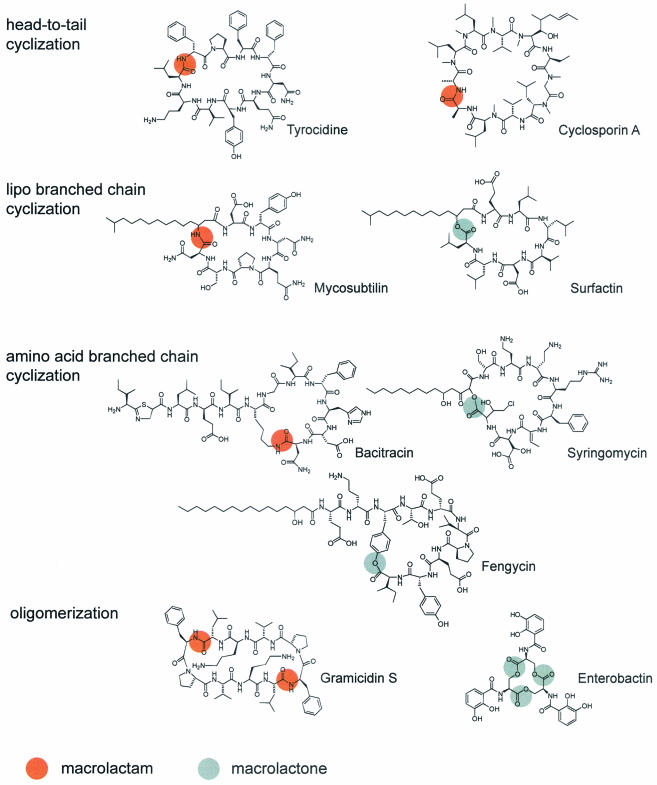
FIG. 1.
Naturally occurring macrolactones and macrolactams.
A SHORT WALK DOWN THE NRPS ASSEMBLY LINE
The nonribosomal machinery for peptide synthesis uses large multienzyme complexes as an assembly line to catalyze stepwise peptide condensation. The substrates of these multienzyme complexes are not restricted to the 20 amino acids, since hundreds of building blocks are now known to be integrated and modified by postsynthesis action. Common to this assembly line is the incorporation of nonproteinogenic amino acids, such as d-isomers, carboxy acids, and N-methylated residues, as well as the incorporation of heterocyclic rings and fatty acids (Fig. 1). Glycosylation and oxidative cross-linking are common further postsynthetic modifications by enzymes that are associated with the NRPS machinery.
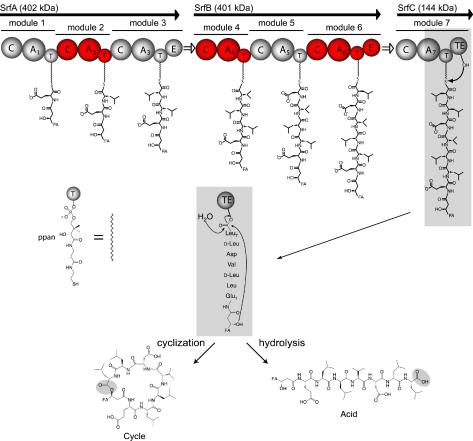
In order to understand the principles of this enzymatically directed peptide synthesis, the mechanistic features of NRPS are briefly summarized with the surfactin synthetase from Bacillus subtilis as an example (see also references 18, 20, 25, and 34). The surfactin synthetase is a large multienzyme complex consisting of three enzymatic subunits, SrfA (402 kDa), SrfB (401 kDa), and SrfC (144 kDa), which consist of seven modules that comprise 24 catalytic domains (Fig. 2). Each module is responsible for the specific incorporation of one dedicated substrate into the growing heptapeptide chain (21). The N-terminal module of an assembly line, the initiation module, specifically recognizes and activates the N-terminal amino acid of the peptide product. All chemical reactions necessary to incorporate and modify each substrate are mediated by a catalytically independent set of domains incorporated within the modules. The first step in biosynthesis is the recognition and activation of a dedicated substrate by the adenylation domain (A domain; about 550 amino acids) (4). By analogy to aminoacyl-tRNA synthetase, the A domain catalyzes the activation of a substrate as aminoacyladenylate through the Mg2+-dependent hydrolysis of ATP and the release of pyrophoshate (7). In the next step, the aminoacyladenylate intermediate is transferred to the free thio group of the cofactor phosphopantetheine, which is tethered to the thiolation domain (T domain, or peptidyl carrier protein; about 80 amino acids) located downstream of the A domain (29, 36). The phosphopantetheine arm is attached to an invariant serine residue of the apo-T domain by a dedicated 4′-phosphopantetheine (ppan) transferase that uses coenzyme A (CoA) as a substrate (15, 22). The intermediates, tethered by the reactive thioester to the flexible cofactor phosphopantetheine (in each module), can be transferred to other domains for subsequent catalytic reactions. Peptide bond formation between two adjacent substrates is catalyzed by the condensation domain (C domain; about 450 amino acids), which is located between the A and T domains of subsequent modules (11). The C domain catalyzes the nucleophilic attack of the amino acid bound to the downstream T domain with its free α-amino group on the activated thioester of the upstream T-domain-bound intermediate (1). For the surfactin initiation reaction, peptide bond formation occurs between modules 1 and 2 by the nucleophilic attack of the α-amino group of leucine on the thioester-activated carboxy group of glutamate to give a dipeptide which is then translocated to module 2.
FIG. 2.
NRPS assembly of surfactin. The surfactin synthetase consists of 24 individual domains responsible for the catalysis of 24 chemical reactions. These domains catalyze activation (A domain), covalent binding (T domain), elongation (C domain), epimerization (E domain), and release (TE domain) by either cyclization or hydrolysis. The domains are organized in modules (indicated in blue and red), where each module incorporates one dedicated building block into the growing peptide chain. The peptide chain is covalently tethered to the multienzyme via the cofactor phosphopantetheine (ppan), and synthesis proceeds according to the enzymatic template from the N- to the C-terminal end. FA, fatty acid chain.
Beside the A, T, and C domains, which are essential domains of an elongation module, there are also other, optional domains. A common structural motif of nonribosomal peptides is the incorporation of d-amino acids, which is mediated by the epimerization domain (E domain; about 450 amino acids). The E domain catalyzes the racemization of the T-domain-bound amino acid to form an equilibrium between the l and d conformers. However, the C domain incorporates only the d-amino acid into the growing peptide chain (16). In the surfactin synthetase, two epimerization domains in modules 3 and 6 are responsible for the racemization of T-domain-bound l-Leu. The combination of l- and d-amino acids gives the peptide a unique conformation that is important for the specific interaction with its cellular target.
All catalytic domains discussed so far are repeating units of the enzymatic template and contribute to the synthesis of a linear peptide molecule tethered to the multienzyme. In order to reactivate the multienzyme for a next synthesis cycle, the mature peptide must be cleaved once it reaches the end of the assembly line. This reaction usually is accomplished by a thioesterase domain (TE domain; about 280 amino acids) fused to the C-terminal module. The peptide can be released either by hydrolysis as a linear acid or by an intramolecular reaction with an internal nucleophile to give a cyclic peptide. Hydrolytic release is observed, e.g., for vancomycin, whose peptide backbone is constrained by further postsynthetic oxidative cross-linking reactions (10). For surfactin, the peptide backbone is constrained by the intramolecular nucleophilic attack of a β-hydroxyl group of the fatty acid moiety to give a branched-chain lipodepsipeptide (32) (Fig. 1). Many other cyclization strategies give rise to a large and diverse set of cyclic or cyclic branched molecules with distinct biological activities. A TE domain catalyzing a cyclization reaction also is referred to as a peptide cyclase.
CYCLIZATION STRATEGIES: NATURE'S WAY
All known NRPS macrocyclization strategies lead to cyclic or cyclic branched-chain peptides (Fig. 1). In macrolactones, the branch point can be either a hydroxylated amino acid side chain or a hydroxylated fatty acid moiety. For the surfactin peptide cyclase, the ring closure is enzymatically catalyzed between an N-terminal β-hydroxyl fatty acid and the C-terminal peptide end (32). Cyclization was observed only when the (R)-β-hydroxyl fatty acid was used, while the (S)-enantiomer showed only enzymatic hydrolysis, indicating stereoselective recognition (12). The lipodepsipeptide cyclic product is a strong detergent with antiviral, hemolytic, and antibacterial activities (21). A major contribution to the detergent activity is provided by the lipopeptide chain, which is believed to be transferred to the N-terminal residue during the initiation reaction by a fatty acid acyltransferase. In contrast, the syringomycin (Pseudomonas syringae) and fengycin (B. subtilis) lipopeptide cyclases accept serine and tyrosine side chains of the peptide sequence as nucleophiles for cyclization, discriminating the N-terminal β-hydroxyl group of the attached fatty acid moiety (24, 26, 33) (Fig. 1). Moreover, these peptide cyclases display a very high level of regioselectivity by selecting only one specific residue of the substrate from a large source of nucleophiles for cyclization.
In addition to macrolactonization, natural product diversity also is increased by various enzymatic macrolactamization strategies. Basic head-to-tail peptide macrolactamization is observed in the antibiotic tyrocidine from Bacillus brevis and the potent immunosuppressive drug cyclosporin A from Tolypocladium niveum. In cyclosporin A, the final peptide bond is formed by a putative condensation domain (35) instead of a peptide cyclase, emphasizing that nature developed two enzyme species capable of catalyzing product release by cyclization. Besides head-to-tail cyclized lactams, branched-chain lactams also are observed. The peptide cyclase of the antifungal lipopetide mycosubtilin from B. subtilis forms regioselectively an amide bond between an N-terminal β-amino fatty acid and the peptide C-terminal end (Fig. 1). As in surfactin, in mycosubtilin a fatty acid chain is involved in the cyclization process, providing an amine as a nucleophile (8). While the precursors in both cases seem to be β-keto fatty acid residues derived from fatty acid synthases, nature processes these ketones in different ways. They are either reduced to a hydroxyl group, as observed in surfactin, or they are reductively aminated in a process catalyzed by aminotransferases, as observed in mycosubtilin. In both cases, nature uses a common precursor motif, which is subsequently diversified by the application of different synthetic strategies to increase the product outcome. As in fengycin and syringomycin, enzymatic amide bond formation also can occur between an ornithine side chain and the C-terminal peptide end, as observed in the antibiotic bacitracin from Bacillus licheniformis (14). The scope of cyclic lactames and lactones can be further broadened by oligomerization of peptide monomers. This additional strategy enables the B. brevis gramicidin S peptide cyclase to cyclodimerize two linear pentapeptides by catalyzing two subsequent peptide bond formation steps to form the cyclic lactam antibiotic gramicidin S (12) (Fig. 1). Also, cyclotrimerization is observed for the siderophore-forming peptide cyclases of enterobactin and the bacillibactin synthetase from Escherichia coli and B. subtilis, respectively (9, 19). Three units of 2,3-dihydroxybenzoyl-serine are fused together by three subsequent ester bond formation steps between the serine hydroxyl group of one molecule and the C-terminal end of another molecule to give the cyclic trilactone enterobactin. This lactone displays iron-chelating activity, which is closely related to its structure. Three intramolecular catechol ligands provide electron donors required for the coordination of iron, once more emphasizing the close relationship between cyclic structural organization and biological activity.
NRPS peptide cyclases can generate diverse cyclic peptide molecules ranging in size from very small, as in pristinamycin, with 7 residues, to very large, as in syringopeptin, with 22 residues (6, 23). At the large end of the scale is another source of macrocyclic molecules observed in nature, referred to as naturally occurring circular proteins (5). These proteins are of bacterial origin and have a folded three-dimensional structure. In contrast to NRPS, they are produced by the translation of genes. Cyclization occurs posttranslationally only in a head-to-tail fashion to produce a seamless circle of peptide bonds. In contrast to what is known for NRPS, not much is known about the cyclization mechanism of the linear precursors.
The cyclization strategies reported here emphasize that nature has developed a large enzymatic tool set which allows the introduction of diversity into linear peptide sequences by a variety of different cyclization steps. Selection from different nucleophiles, enantiomers, and positions in the peptide sequence makes peptide cyclases unique enzymatic tools with very specific intrinsic stereo- and regioselective recognition elements. Moreover, an understanding of the catalysis of one, two, or three subsequent condensation steps toward cyclization requires further studies of the structural and mechanistic aspects of these enzymes.
CHEMOENZYMATIC CYCLIZATION
The great pharmacological potential of many cyclic peptides emphasizes their role in drug discovery, as they show specific interactions with cellular targets and a high level of resistance to proteolytic enzymes. They are therefore most promising scaffolds for pharmacophores. So far, synthetic chemistry faces several difficulties in the production of cyclic compounds providing sufficiently good yields and regioselectivity. Although cyclization is an entropically favorable process, synthetic macrocyclization is difficult to achieve, since steric repulsion of ring residues as well as the use of protecting groups to ensure proper regiochemistry decreases yields and makes chemical synthetic operations expensive and rather difficult. Since nature developed stereo- and regioselective peptide cyclization enzymes, researchers have aimed to combine chemical linear peptide synthesis with enzymatically catalyzed cyclization. This approach allows easy synthesis of linear peptide sequences by established solid-phase peptide chemistry, followed by selective and efficient enzymatic cyclization without the use of protecting groups and the formation of undesirable by-products.
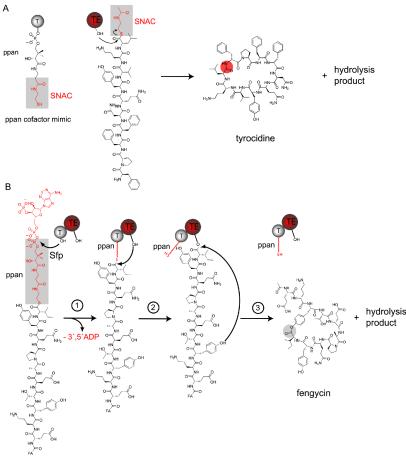
In order to achieve chemoenzymology, translation between the language of chemistry and the language of biology must be established by chemically mimicking the biological pathway as closely as possible. This was first achieved by Trauger et al., who cloned and overexpressed an excised peptide cyclase (28 kDa) from the tyrocidine synthetase (30). In order to prove the activity of this isolated enzyme versus that of the natural enzyme, which is embedded in a 724-kDa multienzyme complex, a short mimicked copy of the natural cofactor phosphopantetheine, N-acetylcysteamine (SNAC), was attached to the C-terminal end of a chemically synthesized linear tyrocidine peptide (Fig. 3A). SNAC represents a link between natural and artificial systems and is compatible with both. Incubation of the mimicked substrate and the excised peptide cyclase revealed activity with an observed cyclization/hydrolysis ratio of 6:1. The turnover of 59 min−1 indicated a very rapid conversion of the linear compound into the cyclic compound, indicating the usefulness of this enzyme as an in vitro biocatalyst (30). Follow-up studies with various peptidyl-SNAC substrates having various lengths, stereochemical properties, and amino acid compositions revealed that the tyrocidine peptide cyclase recognizes only C- and N-terminal residues of the substrate on the basis of identity and stereochemistry, leaving space for making longer and shorter substrates as well as for replacements of residues within the peptide backbone (12). A minimal recognition model was postulated (31). This observed substrate tolerance of the excised tyrocidine peptide cyclase allowed the synthesis of diverse tyrocidine variants in which position 4 (d-Phe) was replaced by 1 of 96 natural and unnatural amino acids. This library of tyrocidine product analogs was subsequently screened for improved or altered bioactivity. In contrast to the natural antibiotic tyrocidine, which does not discriminate between bacterial and eukaryotic cell membranes, the screen revealed that the substitution of d-Phe at position 4 with a positively charged d-amino acid led to a 30-fold increase in the selective recognition of bacterial membranes (13).
FIG. 3.
Chemoenzymatic cyclization strategies. (A) Peptidyl-SNAC. The natural cofactor phosphopantetheine T domain was mimicked for synthetic substrates by attaching the last part of the cofactor, SNAC, to the C-terminal end of the tyrocidine linear peptide chain. This soluble substrate was subsequently incubated with the excised tyrocidine cyclase (TE domain), and cyclic tyrocidine and a hydrolyzed product were observed. (B) Peptidyl-CoA. Synthetically made fengycin CoA is first loaded by the ppan transferase Sfp onto an invariant serine residue of the apo-T domain from an excised fengycin T-TE didomain. In the second step, the substrate is transferred to the peptide cyclase (TE domain) by the nucleophilic attack of an invariant serine residue of the TE domain active site on the peptidyl-ppan thioester. Subsequent cyclization and hydrolysis are catalyzed by the cyclase in the third step of the reaction.
In addition to their use with the tyrocidine peptide cyclase, SNAC substrates also were used to characterize the cyclization of gramicidin S and surfactin (12, 32). In contrast to the tyrocidine peptide cyclase, the surfactin peptide cyclase showed much less substrate tolerance, indicating differences in the binding pockets of these enzymes. While the tyrocidine peptide cyclase was capable of cyclizing shorter and longer SNAC substrates, the surfactin peptide cyclase showed only SNAC hydrolysis for shorter and longer compounds. Moreover, a change in the N-terminal nucleophilic attacking group from an amine to a hydroxyl group in the tyrocidine sequence resulted in no change in the cyclization outcome, while an opposite change from a hydroxyl fatty acid to an amino fatty acid group in the surfactin sequence resulted in only hydrolysis and not cyclization. An important feature of cyclic peptides is the β-sheet content, which is high for molecules with (4n + 2) residues (12, 31). A peptide with a high β-sheet content, such as the dekapeptide tyrocidine (n = 2), facilitates cyclization through substrate preorganization by backbone-to-backbone hydrogen bonds. This intrinsic property of tyrocidine facilitates easy cyclization, which was also reported to occur without catalysis, but at a lower efficiency (3). In contrast, the heptapeptide surfactin displays fewer β sheets and therefore no substantial substrate preorganization. The β-sheet contents in the peptide sequences of surfactin and tyrocidine therefore also may contribute to the observed differences in substrate tolerance.
To expand the set of cyclization catalysts, peptide cyclases from other NRPS systems, such as mycosubtilin and fengycin from B. subtilis and syringomycin from P. syringae, recently were cloned and overexpressed. Contrary to the observations for the surfactin and tyrocidine cyclases, no activity was observed for the mycosubtilin, fengycin, and syringomycin cyclases with synthetically made peptidyl-SNAC substrates, indicating a limitation in the chemoenzymatic potential of the latter cyclases. The inability to recognize or bind SNAC substrates in the active site of the excised peptide cyclase could be affected by the manner in which the short mimicked copy of SNAC is presented to the enzyme. To overcome this limitation, the cyclase domain (TE domain) was excised with the preceding cofactor-binding T domain as a T-TE didomain. Recombinant apo-T-TE cyclases then could be loaded in vitro with chemically synthesized peptidyl-CoA by using the ppan transferase Sfp. The resulting peptidyl-ppan-T-TE holocyclase carried the covalent cofactor-bound substrate in a way that mimicked the natural substrate presentation in the NRPS assembly line (28) (Fig. 3B). Incubation of fengycin T-TE cyclase with fengycin CoA and Sfp revealed cyclization and hydrolysis activities, which were not observed with SNAC substrates alone. These results indicate that the peptide needs to be directed into the peptide cyclase active site by the cofactor ppan to ensure correct enzyme recognition; in contrast, soluble substrates are not properly directed into the active site by diffusion. Cyclases which do not show activity with SNAC substrates seem to require covalent binding of the peptide substrate to the ppan T-domain in order to catalyze cyclic product formation.
STRUCTURAL AND MECHANISTIC ASPECTS OF NRPS PEPTIDE CYCLASES
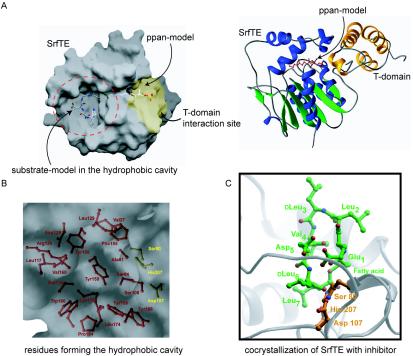
The three-dimensional organization of enzyme residues encodes all information required to understand the principles and general features of macrocyclization. Regiospecific selection of only one nucleophile for cyclization and the exclusion of water to prevent undesired hydrolysis are features which are embedded in the structural fold. Moreover, the question of how thioesterases from different NRPS systems catalyze termination in one case by cyclization and in another case by hydrolysis needs to be elucidated. Crystallographic data for the excised surfactin peptide cyclase (TE domain) showed that this enzyme is a member of the α,β-hydrolase family (2) (Fig. 4A). Since this was the first crystal structure determined for an NRPS peptide cyclase, it served as a prototype for detailed mechanistic investigations (32). The structural similarity to serine hydrolases suggested that an active-site catalytic triad is responsible for the macrocyclization activity. This notion is in agreement with a recent structural model of the surfactin cyclase which suggested that the ppan T-domain-bound peptidyl chain is directed through a cleft into the active site of the peptide cyclase and transferred to an invariant serine residue (Ser80), which is activated by histidine (His207) and aspartate (Asp107) (2) (Fig. 4B). The identity of this catalytic triad was confirmed by mutational analysis, which showed that all three residues were essential for enzyme activity (32). The hydrophobic surfactin peptidyl chain of the acyl-O-enzyme intermediate is accommodated in a predominantly hydrophobic binding pocket with two cationic residues predicted to direct cyclization through specific interactions with the substrate (Fig. 4B). Mutation of these residues to alanine resulted in a dramatic decrease in overall activity, indicating their relevance for peptide recognition.
FIG. 4.
Structural data for surfactin cyclase SrfTE. (A) Surface (right) and ribbon (left) representations of surfactin cyclase (α,β-hydrolase fold). The peptide substrate and the ppan arm are modeled into a hydrophobic binding pocket and into a substrate channel, respectively. Modeling with the T domain (yellow) revealed a putative T-TE didomain docking site. (B) Magnification of the binding pocket shows the catalytic triad (Ser80, His207, and Asp107) and several hydrophobic residues, with the exception of Lys111 and Arg120. (C) Cocrystallization study of SrfTE with a boronic acid inhibitor substrate revealed specific binding pockets for d-Leu6 and Leu7. Based on modeling experiments, the rest of the peptide sequence seems to loop out of the enzyme.
Further studies of enzymatic substrate recognition elements for the surfactin peptide cyclase were carried out by using a detailed substrate scan and cocrystallization analysis for enzyme-bound inhibitor-substrate. The crystallization studies revealed well-defined binding pockets for the two C-terminal leucine residues in the enzyme, while the rest of the peptide sequence seems to be less well coordinated (32) (Fig. 4C). In the deacylation step of the reaction, the β-hydroxyl group of the fatty acid moiety is activated by the same histidine and aspartate to facilitate an intramolecular nucleophilic attack on the acyl-enzyme ester bond to release the final lactone product. Many thioesterase domains from other NRPS systems, e.g., vancomycin, as well as structurally related lipases only hydrolyze and do not cyclize their products. A sequence alignment between the surfactin peptide cyclase and other members of the α,β-hydrolase enzyme family that catalyze only hydrolysis revealed the conservation among lipases of a glycine residue involved in the formation of the oxyanion hole. In the surfactin peptide cyclase, a proline residue is located at this position. A mutation of proline to glycine resulted in a 12-fold change in the product ratio in favor of hydrolysis, indicating that the change from a rigid proline to a flexible glycine increases the conformational freedom in this region of the active site and creates more access for water to capture the acyl-enzyme intermediate (32). This residue seems to be a switch between hydrolysis and cyclization among α,β-hydrolases.
Detailed investigations of the surfactin peptide cyclase provided insights into mechanistic and architectural features of an enzyme which produces a branched cyclic lipodepsipeptide. Less is known about the mechanism and structure of other cyclases, particularly oligomerizing cyclases. A detailed mass spectrometric analysis was carried out for the last module of the enterobactin assembly line (EntF), containing a C-terminal peptide cyclase which catalyzes the cyclotrimerization of three 2,3-dihydroxybenzoyl-serine (DHB-Ser) units to give the cyclic trilactone enterobactin (27). In order to localize acyl-enzyme intermediates, an active-site histidine-to-alanine mutant enzyme with a very low substrate turnover was used. With this approach, it was possible to provide evidence for a covalent acyl-O-TE domain intermediate and demonstrate that the peptide cyclase is involved in two reactions: acyl-chain growth and cyclization. In the first steps of acyl-chain growth, DHB-Ser is transferred to the active-site serine of the peptide cyclase by the nucleophilic attack of active-site serine on the acyl-thioester of the upstream holo-T domain. The second step requires catalytic generation of a DHB-Ser alkoxide, which in turn allows nucleophilic attack on another DHB-Ser thioester-bound T domain to form a dimeric ester. The elongation step is repeated a third time before the final cyclic trilactone is released by the intramolecular nucleophilic attack of serine on the acyl-O-TE domain ester bond. This mechanistic analysis suggests that the enterobactin cyclase serves as a “waiting room” while the phosphopantetheinyl T domain is reacylated, a process which requires a stable ester bond and the exclusion of any water from the active site. Crystallographic data for this peptide cyclase and others will provide more insight into the overall mechanisms and allow for a comparison of their features.
CONCLUDING REMARKS
Research efforts of the past few years in the growing field of enzymatic peptide cyclization in nonribosomal peptide synthesis, summarized in this article, have revealed substantial insights into the architectural, mechanistic, and functional organization of NRPS peptide cyclases. Based on the diversity of natural cyclization strategies, chemoenzymatic approaches were developed to allow cross talk between biology and chemistry to reprogram natural peptide sequences by chemical peptide synthesis and subsequent enzymatic cyclization. This method can serve as a new source of small cyclic peptide molecules with altered or improved pharmacological activities. Since these enzyme catalysts are valuable tools for the synthesis of cyclic molecules, future research efforts also will concentrate on in vitro protein evolution to generate custom-made catalysts for cyclization of a given peptide sequence.
Acknowledgments
This work was supported by Studienstiftung des Deutschen Volkes (SAS) and Deutsche Forschungsgemeinschaft (MAM).
REFERENCES
- 1.Belshaw, P. J., C. T. Walsh, and T. Stachelhaus. 1999. Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284:486-489. [DOI] [PubMed] [Google Scholar]
- 2.Bruner, S. D., T. Weber, R. M. Kohli, D. Schwarzer, M. A. Marahiel, C. T. Walsh, and M. T. Stubbs. 2002. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure (Cambridge) 10:301-310. [DOI] [PubMed] [Google Scholar]
- 3.Bu, X., X. Wu, G. Xie, and Z. Guo. 2002. Synthesis of tyrocidine A and its analogues by spontaneous cyclization in aqueous solution. Org. Lett. 4:2893-2895. [DOI] [PubMed] [Google Scholar]
- 4.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16:4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craik, D. J., N. L. Daly, I. Saska, M. Trabi, and K. J. Rosengren. 2003. Structures of naturally occurring circular proteins from bacteria. J. Bacteriol. 185:4011-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Crecy-Lagard, V., W. Saurin, D. Thibaut, P. Gil, L. Naudin, J. Crouzet, and V. Blanc. 1997. Streptogramin B biosynthesis in Streptomyces pristinaespiralis and Streptomyces virginiae: molecular characterization of the last structural peptide synthetase gene. Antimicrob. Agents Chemother. 41:1904-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieckmann, R., Y. O. Lee, H. van Liempt, H. von Dohren, and H. Kleinkauf. 1995. Expression of an active adenylate-forming domain of peptide synthetases corresponding to acyl-CoA-synthetases. FEBS Lett. 357:212-216. [DOI] [PubMed] [Google Scholar]
- 8.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehring, A. M., I. Mori, and C. T. Walsh. 1998. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37:2648-2659. [DOI] [PubMed] [Google Scholar]
- 10.Hubbard, B. K., and C. T. Walsh. 2003. Vancomycin assembly: nature's way. Angew. Chem. Int. Ed. Engl. 42:730-765. [DOI] [PubMed] [Google Scholar]
- 11.Keating, T. A., C. G. Marshall, C. T. Walsh, and A. E. Keating. 2002. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat. Struct. Biol. 9:522-526. [DOI] [PubMed] [Google Scholar]
- 12.Kohli, R. M., J. W. Trauger, D. Schwarzer, M. A. Marahiel, and C. T. Walsh. 2001. Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry 40:7099-7108. [DOI] [PubMed] [Google Scholar]
- 13.Kohli, R. M., C. T. Walsh, and M. D. Burkart. 2002. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature 418:658-661. [DOI] [PubMed] [Google Scholar]
- 14.Konz, D., A. Klens, K. Schorgendorfer, and M. A. Marahiel. 1997. The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem. Biol. 4:927-937. [DOI] [PubMed] [Google Scholar]
- 15.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 16.Linne, U., and M. A. Marahiel. 2000. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry 39:10439-10447. [DOI] [PubMed] [Google Scholar]
- 17.Lipmann, F., W. Gevers, H. Kleinkauf, and R. Roskoski, Jr. 1971. Polypeptide synthesis on protein templates: the enzymatic synthesis of gramicidin S and tyrocidine. Adv. Enzymol. Relat. Areas Mol. Biol. 35:1-34. [DOI] [PubMed] [Google Scholar]
- 18.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in non-ribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 19.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 20.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochem 3:490-504. [DOI] [PubMed] [Google Scholar]
- 21.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 22.Reuter, K., M. R. Mofid, M. A. Marahiel, and R. Ficner. 1999. Crystal structure of the surfactin synthetase-activating enzyme Sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J. 18:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholz-Schroeder, B. K., J. D. Soule, and D. C. Gross. 2003. The sypA, sypS, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant-Microbe Interact. 16:271-280. [DOI] [PubMed] [Google Scholar]
- 24.Scholz-Schroeder, B. K., J. D. Soule, S. E. Lu, I. Grgurina, and D. C. Gross. 2001. A physical map of the syringomycin and syringopeptin gene clusters localized to an approximately 145-kb DNA region of Pseudomonas syringae pv. syringae strain B301D. Mol. Plant-Microbe Interact. 14:1426-1435. [DOI] [PubMed] [Google Scholar]
- 25.Schwarzer, D., R. Finking, and M. A. Marahiel. 2003. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20:275-287. [DOI] [PubMed] [Google Scholar]
- 26.Segre, A., R. C. Bachmann, A. Ballio, F. Bossa, I. Grgurina, N. S. Iacobellis, G. Marino, P. Pucci, M. Simmaco, and J. Y. Takemoto. 1989. The structure of syringomycins A1, E and G. FEBS Lett. 255:27-31. [DOI] [PubMed] [Google Scholar]
- 27.Shaw-Reid, C. A., N. L. Kelleher, H. C. Losey, A. M. Gehring, C. Berg, and C. T. Walsh. 1999. Assembly line enzymology by multimodular nonribosomal peptide synthetases: the thioesterase domain of E. coli EntF catalyzes both elongation and cyclolactonization. Chem. Biol. 6:385-400. [DOI] [PubMed] [Google Scholar]
- 28.Sieber, S. A., C. T. Walsh, and M. A. Marahiel. 2003. Loading peptidyl-coenzyme A onto peptidyl carrier proteins: a novel approach in characterizing macrocyclization by thioesterase domains. J. Am. Chem. Soc. 125:10862-10866. [DOI] [PubMed] [Google Scholar]
- 29.Stachelhaus, T., A. Hüser, and M. A. Marahiel. 1996. Biochemical characterization of peptidyl carrier protein (PCP), the thiolation domain of multifunctional peptide synthetases. Chem. Biol. 3:913-921. [DOI] [PubMed] [Google Scholar]
- 30.Trauger, J., R. Kohli, H. Mootz, M. Marahiel, and C. Walsh. 2000. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature 407:215-218. [DOI] [PubMed] [Google Scholar]
- 31.Trauger, J. W., R. M. Kohli, and C. T. Walsh. 2001. Cyclization of backbone-substituted peptides catalyzed by the thioesterase domain from the tyrocidine nonribosomal peptide synthetase. Biochemistry 40:7092-7098. [DOI] [PubMed] [Google Scholar]
- 32.Tseng, C. C., S. D. Bruner, R. M. Kohli, M. A. Marahiel, C. T. Walsh, and S. A. Sieber. 2002. Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase. Biochemistry 41:13350-13359. [DOI] [PubMed] [Google Scholar]
- 33.Vanittanakom, N., W. Loeffler, U. Koch, and G. Jung. 1986. Fengycin—a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. (Tokyo) 39:888-901. [DOI] [PubMed] [Google Scholar]
- 34.Walsh, C. T. 2003. Antibiotics: actions, origins, and resistance, p. 195-220. ASM Press, Washington, D.C.
- 35.Weber, G., and E. Leitner. 1994. Disruption of the cyclosporin synthetase gene of Tolypocladium niveum. Curr. Genet. 26:461-467. [DOI] [PubMed] [Google Scholar]
- 36.Weber, T., R. Baumgartner, C. Renner, M. A. Marahiel, and T. A. Holak. 2000. Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases. Struct. Fold Des. 8:407-418. [DOI] [PubMed] [Google Scholar]