Abstract
Background
Maturation of human immunodeficiency virus type 1 (HIV-1) occurs upon activation of HIV-1 protease embedded within GagProPol precursors and cleavage of Gag and GagProPol polyproteins. Although reversible oxidation can regulate mature protease activity as well as retrovirus maturation, it is possible that the effects of oxidation on viral maturation are mediated in whole, or part, through effects on the initial intramolecular cleavage event of GagProPol. In order assess the effect of reversible oxidation on this event, we developed a system to isolate the first step in protease activation involving GagProPol.
Methodology/Principal Findings
To determine if oxidation influences this step, we created a GagProPol plasmid construct (pGPfs-1C) that encoded mutations at all cleavage sites except p2/NC, the initial cleavage site in GagProPol. pGPfs-1C was used in an in vitro translation assay to observe the behavior of this initial step without interference from subsequent processing events. Diamide, a sulfhydral oxidizing agent, inhibited processing at p2/NC by >60% for pGPfs-1C and was readily reversed with the reductant, dithiothreitol. The ability to regulate processing by reversible oxidation was lost when the cysteines of the embedded protease were mutated to alanine. Unlike mature protease, which requires only oxidation of cys95 for inhibition, both cysteines of the embedded protease contributed to this inhibition.
Conclusions/Significance
We developed a system that can be used to study the first step in the cascade of HIV-1 GagProPol processing and show that reversible oxidation of cysteines of HIV-1 protease embedded in GagProPol can block this initial GagProPol autoprocessing. This type of regulation may be broadly applied to the majority of retroviruses.
Introduction
HIV-1 GagProPol autoprocessing is a unique step in the HIV life cycle and involves the dimerization of two GagProPol monomers and subsequent autocleavage of the polyproteins to generate virus structural proteins as well as protease, reverse transcriptase, and integrase [1]. This autoprocessing is usually delayed until GagProPol and Gag polyproteins assemble at the cell membrane and is followed by further cleavage of Gag and other GagProPol polyproteins during and/or just after budding from the plasma membrane [2], [3]. Premature autoprocessing and activation of protease in HIV-1 infected cells or over-expression of GagProPol, impairs viral production and infectivity [4], [5]. Similar results have been described for Rous sarcoma virus (RSV), where a protease-linked dimer results in premature viral processing in the cytoplasm of infected cells in conjunction with decreased virus production [6]. These studies indicate a need for control over the timing of protease activation, particularly when viral expression and therefore protease are at their highest levels.
Previous studies have demonstrated that reversible oxidation can regulate maturation of immature HIV-1 virions [7]. One possible way that this may occur is through modification of the mature protease by oxidation of a cysteine at position 95 (cys95) at the dimer interface; this could then interfere with protease dimerization. In the case of HIV-2, reversible oxidation of methionine 95 (met95) serves to regulate protease activity in the same way [8], [9]. For HTLV-1, two conserved cysteines at positions 90 and 109 serve a similar function [8]. The oxidative modifications of HIV-1 and HIV-2 protease interfere with dimerization [8], [9] and it is predicted that the modifications of HTLV-1 protease may prevent dimerization as well.
A similar process is involved in the regulation of the maturation of Mason-Pfizer monkey virus (M-PMV) capsids [10]. M-PMV, also a retrovirus, is unusual in that the GagPro polyproteins assemble in the cytoplasm but the embedded protease, which contains two methionines and one cysteine at the dimer interface, is still not activated until the time of budding. Experimentally, the M-PMV protease and viral maturation can be activated by the addition of a reducing agent [10]. In fact, essentially all retroviral proteases studied have one or more oxidizable residues at or near the protease dimer interface with the potential to be regulated in this manner [8]. Cellular enzymes, such as thioltransferase and methionine sulfoxide reductase have been shown to reverse these cysteine and methionine oxidative modifications, respectively, leading to protease dimerization and reactivation [8], [11], [12]. It has been suggested that that regulation by reversible oxidation of one or more cysteine or methionine residues at or near the dimer interface is a general property of retroviral proteases [8].
However, it remains unclear whether the regulation of viral maturation by reversible oxidation is effected primarily through oxidation of mature protease or whether the main target is the initial autoprocessing event by the GagProPol polyprotein. The first step in HIV-1 polyprotein processing is an intramolecular cleavage between the p2 protein (also known as spacer peptide 1 or SP1) domain and nucleocapsid (NC) domain or p2/NC [13]–[15]. This step is unique from those carried out by mature protease in that: 1) cleavage of GagProPol in trans by exogenous protease at the p2/NC junction is much more rapid than cleavage at p2/NC by the dimerized precursor polyprotein [13]; 2) the initial processing step is quite resistant to inhibition by active site protease inhibitors when compared to mature protease [14 and our unpublished data]; and 3) even the second cleavage carried out by the embedded protease is 27 fold more sensitive to the potent active site inhibitor, ritonavir, as compared to the initial cleavage at p2/NC by the embedded protease. Due to these differences it was unclear whether oxidation of the embedded protease in GagProPol could reversibly inhibit the initial step in viral maturation.
In this report, we developed a unique assay that enabled us to isolate the initial GagProPol autoprocessing event without interference from subsequent proteolytic events. Through modification of a GagProPol encoding plasmid, we were able to measure the initial cleavage of GagProPol at the P2/NC site. This allowed us to assess conditions that may control this cleavage event. We demonstrate that the initial step in GagProPol processing can be specifically and reversibly regulated through oxidation of the cysteines of the embedded HIV-1 protease, and that this regulation differs in some ways from that of mature HIV-1 protease. The observations reported here indicate reversible oxidation as a potential mechanism to regulate the onset of retroviral polyprotein processing.
Results
Development of a GagProPol first-cut assay
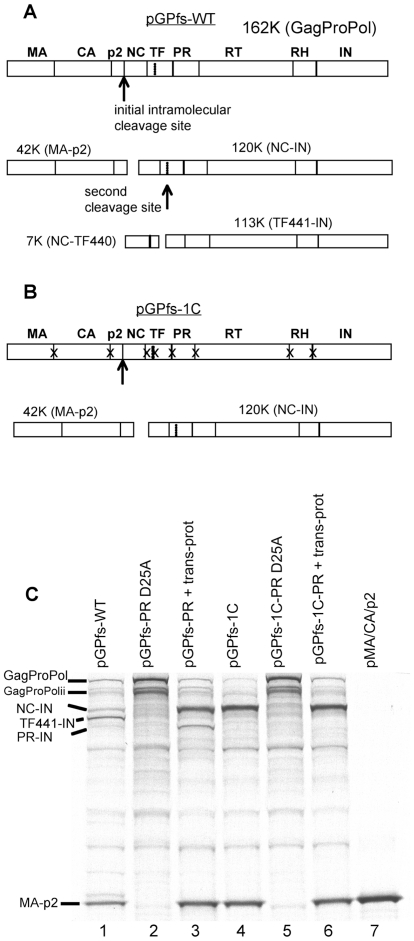
The pGPfs, developed by Pettit et al. [13], contains a frame-shift mutation that causes it to produce full length GagProPol but not p55 Gag protein. The GagProPol produced by this plasmid subsequently undergoes limited autoproteolytic processing. The major products resulting from GagProPol autoprocessing in this system are a result of cleavage at p2/NC and cleavage within the transframe protein (TF) yielding MA-p2, NC-IN, NC-TF440 and TF441-IN (Figure 1A) [13]. We were interested in studying the initial step of polyprotein processing and in determining if this step could be regulated by reversible oxidation. To this end, we produced a modified construct that would only undergo the initial step of polyprotein processing. To ensure processing at only the p2/NC site, we incorporated blocking mutations at all other major processing sites within pGPfs. Amino acid mutations flanking the GagProPol processing sites were chosen based upon previous studies indicating that these substitutions prevented proteolytic processing (Table 1) [16]-[18]. In addition, alanine mutations were placed at the processing sites flanking the N and C termini of protease to prevent processing at these sites by the embedded protease [13], while the protease sequence itself was conserved as wild type (Table 1). While cleavage at these flanking sites was not previously detected [13], these mutations provided additional assurance that the results would not be skewed by a small amount of this product. These modifications also reduced the likelihood that partially processed GagProPol could cleave other polyproteins in trans. The modified pGPfs plasmid had eight processing site mutations and was designated as pGPfs-one cut (pGPfs-1C) (Figure 1B).
Figure 1. Schematic diagram for the GagProPol polyprotein precursor generated from pGPfs, the polyprotein precursor generated from pGPfs-1C and results of GagProPol proteolytic processing of pGPfs (WT) and pGPfs-1C.
A) The GagProPol plasmid construct, pGPfs, encodes a 162 kDa polyprotein. The processing sites for the polyprotein are indicated with vertical lines. The location of the first cut site (p2/NC) is indicated with an arrow. The known secondary cut site (occurring between amino acids 440 and 441 within the transframe protein) is indicated by a dotted line. The resultant products from cleavages at these two sites are indicated. B) The schematic for pGPfs-1C (modified pGPfs plasmid) showing the sites for the blocking mutations designated with an X. C) In vitro transcription/translation with 35S-methionine was carried out for 1 h in the presence of the indicated plasmid construct. Note that plasmids pGPfs-PR D25A and pGPfs-1C-PR D25A encode a protease with a D25A mutation rendering it inactive. pMA/CA/p2, yields the small product (MA-p2) fragment and was used to verify the MA-p2 fragment. Exogenous HIV-1 protease provided in trans (trans-prot, 250 nM) was added to pGPfs-PR D25A or pGPfs-1C-PR D25A constructs following 45 min of translation and was terminated 15 minutes later with 3X LDS sample buffer. Samples were separated by LDS-PAGE and visualized by autoradiography. The location for GagProPol, the internal initiation GagProPol (GagProPol ii) and the products of GagProPol processing (MA-p2, NC-IN, TF441-IN, and PR-IN) occurring through the action of the embedded protease or the exogenous protease are indicated in the figure.
Table 1. Processing site and protease mutations incorporated into pGPfs-1C.
| Processing site | Nucleotide sequence | Amino acid sequence |
| p17/p24 WT | CAA AAT TAC/CCT ATA GTG | Gln Asn Tyr/Pro Ile Val |
| p17/p24 Mut | CAA AAT ATC/CTT ATA GTG | Gln Asn Ile/Leu Ile Val |
| p24/p2 WT | AGA GTT TTG/GCT GAA GCA | Arg Val Leu/Ala Glu Ala |
| p24/p2 Mut | AGA GTT GTG/GCT GAA GCA | Arg Val Val/Ala Glu Ala |
| NC/TF WT | CAG GCT AAC/TTC CTC CGC | Gln Ala Asn/Phe Leu Arg |
| NC/TF Mut | CAG GCT ATC/GAT CTC CGC | Gln Ala Ile/Asp Leu Arg |
| TF440/441 WT | TTG GCC TTC/CTA CAA GGG | Leu Ala Phe/Leu Gln Gly |
| TF440/441 Mut | TTG GCC ATC/CTA CAA GGG | Leu Ala Ile/Leu Gln Gly |
| TF/Protease WT | TTT AAC TTC/CCT CAG GTC | Phe Asn Phe/Pro Gln Val |
| TF/Protease Mut | GCG GCC GCC/CCT CAG GTC | Ala Ala Ala/Pro Gln Val |
| Protease/Pol WT | TTA AAT TTT/CCC ATT AGC | Leu Asn Phe/Pro Ile Ser |
| Protease/Pol Mut | TTA AAT TTT/GCTGCAGCC | Leu Asn Phe/Ala Ala Ala |
| Pol/RNaseH WT | GAA ACC TTC/TAT GTA GAT | Glu Thr Phe/Tyr Val Asp |
| Pol/RNaseH Mut | GAA ACC ATC/TAC GTA GAT | Glu Thr Ile/Tyr Val Asp |
| RNaseH/Integrase WT | AAA GTA CTA/TTT TTA GAT | Lys Val Leu/Phe Leu Asp |
| RNaseH/Integrase Mut | AAA GTA ATA/TTT TTA GAT | Lys Val Ile/Phe Leu Asp |
In vitro translation was carried out using wild type (WT) pGPfs or pGPfs-1C. As expected, in vitro translation of the WT plasmid generated GagProPol, truncated GagProPol from internal initiations (GagProPolii) and expected products of the first and second processing steps (MA-p2, NC-IN, TF441-IN, Figure 1C) [13], [14], [19]. These products (MA-p2, NC-IN, and TF441-IN) were absent when a pGPfs containing a D25A mutation in the protease gene that inactivates the embedded protease (pGPfs-PR D25A) was utilized (Figure 1C, lane 2). A number of weaker non-specific bands were also produced by the WT pGPfs, similar to those seen previously [13], [14], [19]. Most of these weaker bands are unrelated to GagProPol processing as they are generated both when using GagProPol plasmids encoding inactive as well as active protease (Figure 1C, lane 2 and 5). Addition of purified HIV-1 protease (15 min incubation) to pGPfs-PR D25A yielded the major products MA-p2, NC-IN and a band tentatively identified as PR-IN based on studies reported previously (Figure 1C, lane 3) [13].
In contrast to the results with WT pGPfs, in vitro translation of pGPfs-1C yielded only the major products of the first processing step and these aligned with the previously identified products obtained from WT pGPfs, namely MA-p2 and NC-IN (Figure 1C, lane 4). In particular, pGPfs-1C did not generate any of the additional processing intermediates that were seen with pGPfs. To confirm that the products from pGPfs-1C processing were indeed due to processing by the embedded protease, we created a D25A mutation in the protease region of pGPfs-1C and designated the plasmid containing inactive protease as pGPfs-1C-PR D25A. In vitro translation of pGPfs-1C-PR D25A produced full length GagProPol (and GagProPolii) but did not produce any cleavage products (Figure 1C, lane 5). As expected, addition of purified HIV-1 protease for 15 minutes (250 nM dimer) to the sample producing D25A GagProPol resulted in almost complete processing only at the p2/NC cut site (Figure 1C, lane 6). To further verify that processing was occurring at the p2/NC cut site, we generated a plasmid encoding MA/CA/p2 and used it as a standard in the in vitro translation assay. The protein product generated from this plasmid (MA-p2) migrated to the same position on the gel as the small product generated from the GagProPol encoded by the first cut plasmid (Figure 1C, lane 7). These studies indicated that in vitro translation of pGPfs-1C produced GagProPol that undergoes a single cleavage event at p2/NC.
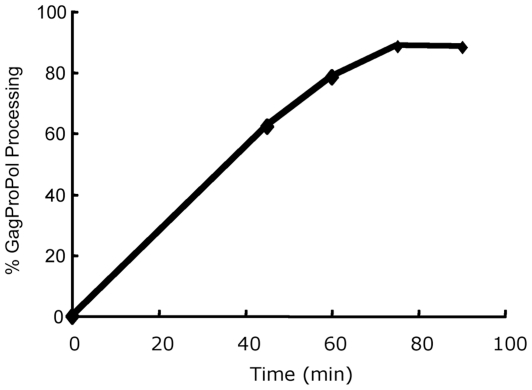
The rate of processing at the initial cleavage site was investigated in a time course experiment. Approximately 60% of the pGPfs-1C GagProPol underwent cleavage at the p2/NC within 45 minutes and 90% underwent cleavage within 75 minutes, at which time it reached a plateau (Figure 2 and Figure S1). After two hours of incubation, minor bands below the large product were evident but these represented less than 5% of the total products produced (data not shown). These likely represent alternate, albeit much less favorable, cleavages occurring through the action of the embedded protease within the large product (NC-IN). These data indicate the embedded HIV-1 protease readily cleaves within pGPfs-1C GagProPol at the p2/NC junction yielding MA-p2 and NC-IN products and that the blocking mutations do not adversely affect processing at the initial site. However, the presence of blocking mutations prevents any significant processing of the polyprotein at the other cleavage sites.
Figure 2. Time course for pGPfs-1C GagProPol processing.
In vitro transcription translation with 35S-methionine was carried out for 45, 60, 75 and 90 minutes. Samples were separated by LDS-PAGE and visualized by autoradiography and the percent processing determined using densitometry. The precursors (GagProPol and GagProPolii) and two products (MA-p2, NC-IN) were scanned and the extent of processing calculated as a percent: [(products)/(precursor plus products) x 100]. The LDS-PAGE for this data is shown Figure S1.
Inhibition of the initial step in GagProPol Processing by oxidation is dependent on the cysteines of HIV-1 protease
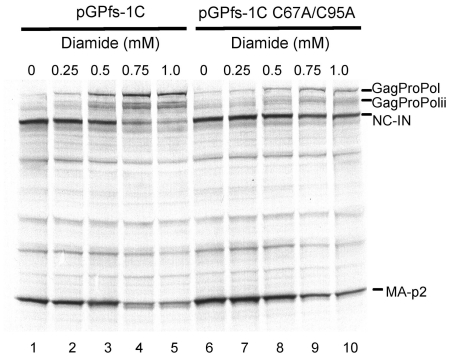
We previously demonstrated that HIV-1 protease could be regulated by reversible oxidation of the conserved cysteines of HIV-1 protease and found that exposure of immature HIV-1 virions to oxidizing agents could inhibit viral maturation [7], [11], [20]. This inhibition required the presence of cys95, but not cys67, since replacing cys95 with alanine led to processing even in the presence of oxidizing agents, while replacing cys67 with alanine had no substantial effect [20]. However, it was unclear if oxidation could similarly affect the action of the embedded protease at the initial step of GagProPol processing. To determine if the initial step in GagProPol processing was sensitive to oxidation, we examined the effect of oxidants on GagProPol processing using pGPfs-1C. Initial studies revealed that hydrogen peroxide (1–10 mM), a nonspecific oxidizing agent, caused partial inhibition of GagProPol processing in a cysteine-dependent manner (data not shown). However, higher concentrations of hydrogen peroxide were detrimental to the translation system, thereby preventing any further assessment on GagProPol processing. Diamide is a thiol-oxidizing agent that can create mixed disulfide bonds between proteins and low molecular weight thiols and therefore we tested the effect of diamide on GagProPol processing [21], [22]. In the absence of diamide, processing with pGPfs-1C proceeded almost to completion as evidenced by the presence of the two GagProPol processing products and the absence of full length GagProPol (Figure 3, lane 1). However, in the presence of diamide, GagProPol processing was inhibited in a dose-dependent fashion at concentrations that only had a minor effect on the in vitro translation system as evidenced by the total product produced in the reactions (Figure 3, lanes 2–5). By contrast, the effect of diamide on the processing of pGPfs-1C C67A, C95A GagProPol was much less pronounced (Figure 3, lanes 6–10). These data indicate that the cysteines of HIV-1 protease are particularly susceptible to diamide oxidation and this oxidation leads to inhibition of the initial cleavage event.
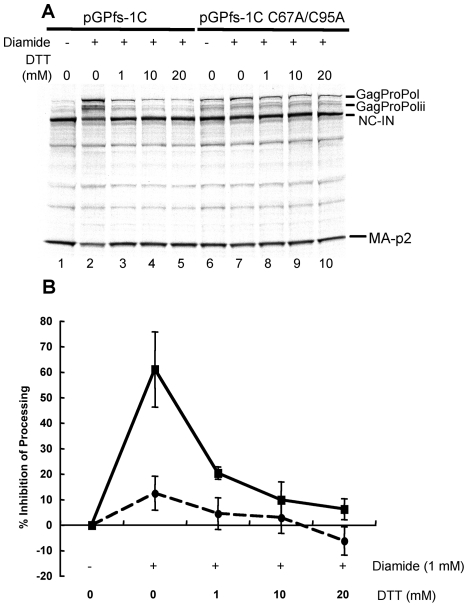
Figure 3. Dose dependent inhibition of pGPfs-1C GagProPol and pGPfs-1C C67A/C95A GagProPol processing by diamide.
In vitro transcription/translation with 35S-methionine was carried out in the presence of increasing concentrations of the sulfhydral oxidant diamide (A) pGPfs-1C (lanes 1–5) or the double cysteine mutant construct pGPfs-1C C67A/C95A (lanes 6–10). Samples were separated by LDS-PAGE and visualized by autoradiography. The precursors (GagProPol and GagProPolii) and products (MA-p2, NC-IN) of GagProPol processing by the embedded protease are indicated.
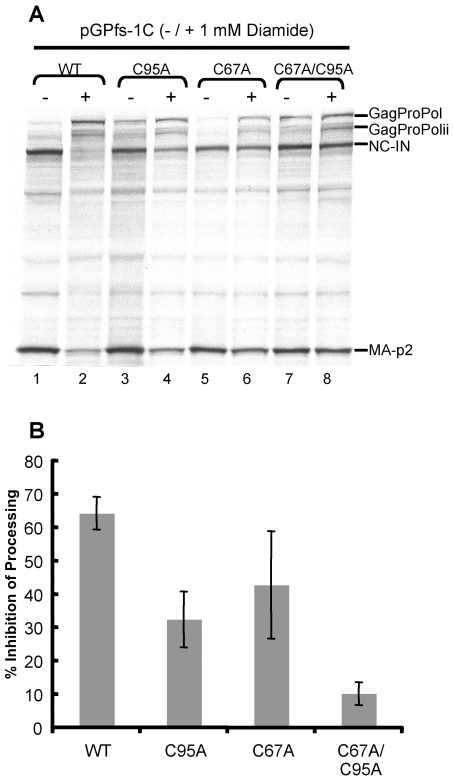
To assess the contribution of each cysteine within the embedded protease to inhibition of processing by diamide, we examined GagProPol processing for the plasmid constructs pGPfs-1C, pGPfs-1C C95A, pGPfs-1C C67A and pGPfs-1C C67A C95A in the presence and absence of 1 mM diamide. In the absence of diamide, all four GagProPols underwent substantial processing, although there were some small differences among them. Of note, the plasmids retaining cys95 (pGPfs-1C and pGPfs-1C C67A) processed nearly to completion (Figure 4A, lanes 1 and 5) while the plasmids lacking cys95 (pGPfs-1C C95A pGPfs-1C C67A C95A) had some residual unprocessed GagProPol remaining (Figure 4A, lanes 3 and 7). This was a consistent finding throughout our studies and indicates that the presence of cys95 is important for maximally efficient cleavage of GagProPol at p2/NC. Diamide (1 mM) substantially inhibited processing of pGPfs-1C GagProPol, but was less effective on each of the single cysteine mutants (Figure 4A, lanes 1–6) and was the least effective at inhibiting processing of the double cysteine mutant GagProPol produced from pGPfs-1C C67A, C95A (Figure 4A, lanes 7–8). Thus, the loss of either cysteine significantly impaired the ability of diamide to block processing. To quantify this effect among the different plasmids and correct for internal differences in translation and processing of the alanine mutants, densitometry was performed on the unprocessed GagProPol bands and product bands from four separate experiments. The extent of processing was first calculated for the untreated controls and then compared to processing following treatment with 1 mM diamide. As shown in Figure 4B, diamide inhibited the processing of GagProPol produced from pGPfs-1C about 65% while processing for C95A, C67A and the double mutant was inhibited by 35%, 42%, and 10%, respectively (Figure 4B). These data demonstrate that optimal inhibition of GagProPol processing by diamide requires the presence of both cys67 and cys95.
Figure 4. Inhibition of GagProPol processing by the oxidant diamide is dependent on both cysteines of the embedded HIV-1 protease.
(A) In vitro transcription/translation with 35S-methionine was carried out for 1 h in the absence (−) or presence (+) of 1 mM diamide. Samples were separated by LDS-PAGE and visualized by autoradiography. The precursors and products of GagProPol processing by the embedded protease are indicated. The pGPfs-1C plasmid construct used is indicated above the wells. (B) Densitometry was carried out on autoradiograms from 4 separate experiments under the conditions as described for Figure 4A and the extent of inhibition of processing calculated. The percent inhibition was calculated as (processing without diamide-processing with diamide)/(processing without diamide) x 100. Percent processing is calculated as in Figure 2. Values represent the average +/− the standard deviation from 4 independent experiments.
Oxidative inhibition of the initial step in GagProPol processing is reversible
Next, we wanted to assess the ability of DTT, a sulfhydral reducing agent, to restore processing after inhibition by diamide treatment. To determine if DTT could reverse the inhibition of GagProPol processing after an initial diamide treatment, the in vitro translation reaction was first carried out for 45 minutes in the absence or presence of 1 mM diamide. Following this, samples were treated with 0, 1, 10, or 20 mM DTT and the reaction was allowed to proceed for 15 more minutes. DTT reversed the diamide-induced inhibition of GagProPol processing at the first cut site (p2/NC) and did so in a dose dependent manner (Figure 5A, lanes 1–5). There was a low level of diamide induced inhibition of processing for the double cysteine mutant pGPfs-C C67A, C95A but this was restored following DTT treatment (Figure 5A, lanes 6–10). Densitometric analyses were done to assess the extent of reversal of diamide inhibited GagProPol processing for pGPfs-1C for three separate experiments. As shown in Figure 5B, diamide inhibited 60% of the processing and much of this (67%) was restored with as little as 1 mM DTT and almost 90% was restored with 20 mM DTT (Figure 5B). Overall, these data indicate that diamide oxidation of the cysteines within the embedded protease leads to inhibition of the initial cleavage step in GagProPol polyprotein processing and this inhibition can be readily reversed with reducing agent.
Figure 5. Dose response restoration of polyprotein processing by DTT following inhibition of pGPfs-1C or pGPfs-1C C67A,C95A processing by diamide.
A) In vitro transcription/translation was carried out with pGPfs-1C (lanes 1–5) or pGPfs-1C C67A,C95A (lanes 6–10) in the absence (−) or presence (+) of 1 mM diamide for 45 min followed by addition 1, 10, or 20 mM DTT for 15 minutes as shown above each lane. Samples were separated by LDS-PAGE and analyzed by autoradiography. The precursors and products of GagProPol processing by the embedded protease are indicated. B) Densitometry was carried out on autoradiograms from 3 separate experiments under the conditions as described for Figure 5A. The percent inhibition of processing was calculated and plotted. for pGPfs-1C (ν) solid line and pGPfs-1C C67A,C95A (λ) dotted line. The following formula was used: Percent inhibition of processing = (processing without diamide-processing with diamide and 0–20 mM DTT)/(processing without diamide) x 100. The values represent the average +/− the standard deviation for 3 independent experiments.
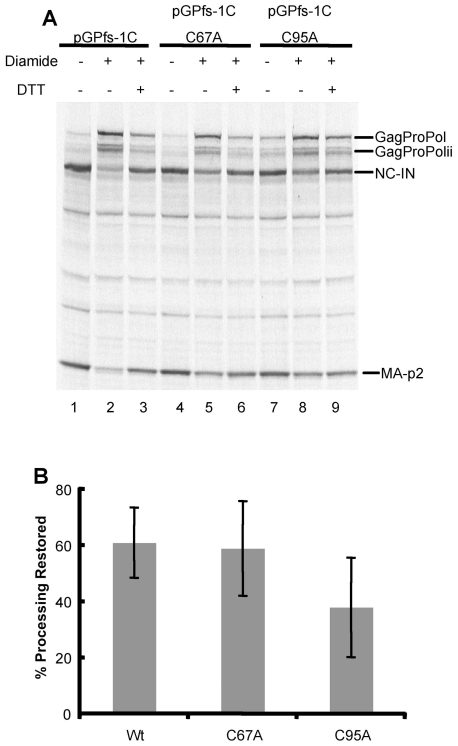
We also investigated the ability of DTT to reverse the diamide-induced inhibition of GagProPol processing for the single cysteine mutants. Once again, diamide blocked the majority of GagProPol processing obtained with pGPfs-1C, but was somewhat less effective at blocking GagProPol processing obtained with single cysteine mutant C67A or C95A pGPfs-1Cs (Figure 6A, lanes 2,5,9). Addition of 1 mM DTT, following inhibition by diamide, restored processing in all cases (Figure 6A, lanes 3,6,and 9). Although processing for pGPfs-1C C67A and pGPfs-1C C95A were similarly inhibited by diamide, restoration of processing with 1 mM DTT was most efficient with pGPfs-1C C67A, and similar to the reversal obtained for pGPfs-1C (Figure 5A). Based on densitometric analysis, DTT restored about 60% of the processing inhibited by diamide (Figure 6B). A similar improvement in GagProPol processing was seen with pGPfs-1C C67A that contains protease with cys95 intact (Figure 6B). However, DTT only restored an average of 40% of processing for the pGPfs-1C C95A GagProPol containing protease with cys67, although this lower level of processing was not statistically significant when compared to the other plasmids (Figure 6B). Together, these data suggest that oxidation of both cysteines can be reversed with DTT. However, cys67 may be more difficult to reverse when compared to cys95 of the embedded protease in GagProPol in the context of the single cysteine mutants.
Figure 6. Restoration of polyprotein processing by DTT following inhibition of pGPfs-1C, pGPfs C95A, or pGPfs C67A processing by diamide.
A) In vitro transcription/translation was carried out for pGPfs-1C, pGPfs C95A, or pGPfs C67A in the absence (−) or presence (+) of 1 mM diamide for 45 min followed by addition of 1 mM DTT. Samples were incubated for an additional 15 min to allow for processing to take place and then separated by LDS-PAGE and analyzed by autoradiography. The precursors and products of GagProPol processing by the embedded protease are indicated. B) Densitometry was carried out on autoradiograms from 3 separate experiments under the conditions as described for Figure 6A. The percent processing restored was calculated as (processing with DTT and diamide-processing with diamide)/(processing without DTT or diamide-processing with diamide) x 100. The values represent the average +/− the standard deviation for 3 independent experiments.
Discussion
Previous studies demonstrated that reversible oxidation inhibited HIV-1 maturation through the inhibition of protease activity and that the activity of mature protease could be regulated by such oxidation of residues at the dimer interface, thus interfering with dimerization[7], [8], [23]. However, the initial dimerization of HIV protease takes place between two GagProPol monomers that, after dimerization, undergo autoprocessing through the action of the embedded protease of the polyprotein. We hypothesized that regulation of this initial step by reversible oxidation of the dimer interface of GagProPol may account for much or all of the regulation of HIV viral maturation by redox changes. To explore this hypothesis, we used an in vitro GagProPol translation assay system to isolate the dimerization and first step in HIV-1 polyprotein processing. Our modified GagProPol plasmid construct yielded GagProPol that efficiently underwent dimerization and autocleavage at p2/NC without further processing taking place. Using this construct we found that autocleavage at p2/NC within GagProPol was quite sensitive to oxidation by the sulfhydral oxidant, diamide and this was readily reversible with reducing agent. The ability to regulate processing by reversible oxidation was lost when the cysteines of the embedded protease were mutated to alanine and processing proceeded irrespective to the addition of diamide. Although we could not determine the precise mechanism of inhibition using this assay, we suggest that the oxidative modification of cysteines in the embedded protease interferes with GagProPol dimerization, similar to that previously demonstrated for mature protease [8], [9]. Diamide oxidation also blocked the processing for pGPfs-1C C67A/C95A although this was less pronounced than that seen for the pGPfs-1C plasmid encoding a cysteine containing protease. The fact that the low level of inhibition with the double mutant was reversible with DTT (see Figure 5) suggests that other cysteines within the GagProPol also participate to some degree in the inhibition of processing. It has been reported that the nucleocapsid domain contains reactive cysteine residues which, when oxidized, reversibly affect polyprotein processing [24]. Therefore these residues may also play a partial role in the inhibition of GagProPol processing observed with diamide treatment.
There were some key differences between the effects of oxidation on the embedded protease and those observed for mature protease. For mature HIV-1 protease, oxidation of cys95 alone was sufficient to inactivate the enzyme and viral maturation [11], [20]. In contrast, the presence of both cysteines (67 and 95) was required to obtain maximum inhibition/regulation of the initial GagProPol processing step. Also, while oxidation of cys67 of mature protease by glutathionylation increased activity and stabilized the enzyme, oxidation of cys67 of the embedded protease within the in vitro translation system led to partial inhibition of GagProPol processing [11], [20]. This difference in the effect of cys67 oxidation of the embedded protease as compared to the mature protease is likely related to differences in the conformation of protease in GagProPol dimers compared to those for mature dimers. For mature protease, it is more difficult to reverse oxidation of cys67 versus that for cys95 [11]. While oxidation of cys95 in mature WT protease or C67A protease results in complete inactivation of the enzyme [11], [20] and inhibition of protease dimerization [8], [9], oxidation of GagProPol containing only cys95 leads only to partial inhibition of processing. Thus, as suggested previously [13], sequences within GagProPol that lie outside of the protease region may partially compensate for disruption of the protease dimeric interaction. We cannot rule out that the mutations that were made to prevent cleavage at subsequent processing sites had some influence on the differences observed between oxidation of endogenous and versus exogenous protease.
Reversible oxidation of certain amino acids of retroviral proteases has been suggested as one potential mechanism for preventing premature activation of the protease [8], [10]. Studies with immature HIV-1 virions produced by exposure to a protease inhibitor demonstrated that reducing agents could increase viral processing following removal of the inhibitor [7]. Similarly, immature capsids of M-PMV undergo proteolytic processing following the addition of reducing agent [10]. These studies implicated protease oxidation as a mechanism to inhibit protease activation. Our studies here demonstrate clearly that reversible oxidation can regulate the onset of the polyprotein processing cascade for HIV-1.
Evidence for the importance of redox-mediated regulation of protease and/or protease precursors is strongest in the case of M-PMV, in which the GagPro polyproteins assemble in the cytoplasm but do not mature until the time of budding [10]. However, similar regulation of retroviral protease has been shown for several other retroviruses, and essentially all retroviruses studied have at least one cysteine or methionine in the region of the dimer interface, suggesting that this regulation is constrained by viral evolution and therefore important in the retroviral lifecycle [7]–[9]. In the laboratory, HIV-1 does not require the presence of cys67 or cys95 to generate infectious virions from infected cells [7]. Nonetheless, cys67 and cys95 are highly conserved among HIV-1 isolates [25]. Therefore, for HIV-1, it is possible that the regulation of the initial step in processing by reversible oxidation may only be necessary in certain cell types or under certain conditions, such as those involving oxidative stress. Virus infection, including HIV-1 infection, leads to a state of oxidative stress in certain cell types and this is known to enhance HIV-1 replication through the activation of NF-κB [26]–[29]. Oxidative environments, like those encountered in infected cells [26], [30], could lead to reversible cysteine oxidation of the embedded protease and as a result prevent premature protease activation and premature cell death when GagProPol is accumulating to high levels in the cell. Thus, the presence of cysteines within protease may optimize viral production by preventing deleterious effects of the protease on the infected cells. This effect may be particularly important in cells that produce virus for extended periods of time such as monocytes/macrophages. By contrast, the D-type retroviruses, such as M-PMV, may utilize reversible oxidation as the primary means to control protease activity as evidenced by the lack of particle processing in the absence of reducing agent and the activation of such processing with reducing agent [10].
The initial GagProPol processing step provides an additional target for the development of inhibitors of viral replication, a step separate from the processes mediated by the mature protease. While intramolecular enzymatic steps are inherently more difficult to inhibit through a classical mode of competitive inhibition as compared to trans cleavage [14 and references therein], it may be possible to develop inhibitors of GagProPol dimerization that could prevent protease activation and autoprocessing. It is possible the observed diamide inhibition of GagProPol processing in this study results from the inhibition of GagProPol dimerization, although further studies will be necessary to determine this. Recent studies have provided evidence that darunavir and similar compounds have the ability to block protease dimerization within cells [31]. We are currently screening active site inhibitors in this assay to determine what structural properties might favor inhibition of the initial processing step and how these properties relate to their ability to block protease activity and protease dimerization. Moreover, compounds designed as dimerization inhibitors of protease can be screened to assess their potential as inhibitors of the initial processing step. It may be more feasible to inhibit the dimerization of GagProPol than to reverse the dimerization of the mature protease dimer considering the low dissociation constant of mature protease [32]. Such studies can provide an additional target for the pursuit of new therapies, especially in the treatment of multi-drug resistant HIV [33].
Materials and Methods
Plasmid construction and mutagenesis
The plasmids pGPfs and pGPfs-PR D25Awere obtained from Dr. Ronald Swanstrom (UNC-Chapel Hill, NC). pGPfs contains a T7 promoter and has a forced frame-shift mutation leading to the production of full length GagProPol in an in vitro translation system [13]. pGPfs-PR D25A contains a D25A mutation in the protease coding region to inactivate protease activity and prevent proteolytic processing of the GagProPol precursor generated during in vitro translation. Further mutations were introduced into pGPfs by site directed mutagenesis using a Stratagene QuikChange Multi Site-Directed Mutagenesis Kit. The primers used to introduce processing site and protease mutations are shown in Table 1. A DNA fragment encoding the matrix-capsid-p2 polyprotein (MA-p2) was produced using PCR to add NdeI and BamHI restriction sites [34]. This DNA fragment was then ligated into expression vector pET11 creating plasmid pMA/CA/p2 and its product was used as a control to verify the identity of the small product from the first processing step. Plasmids were also produced with single and double cysteine-to-alanine mutations and/or a D25A mutation introduced in the protease-coding region of pGPfs-1C. The primers used for these constructs are shown in Table 1.
In-vitro translation reactions for studying GagProPol proteolytic processing
Transcription and translation reactions were performed similar to those described previously using a TnT T7 Coupled Reticulocyte Lysate System (Promega) except when noted below [13]. The reactions (25 µL) were carried out with Easy-Tag L-35S-methionine (>1000Ci/mM, Perkin Elmer) rather than 35S-cysteine. This was done since there are a similar number of methionines in the two products from the first cut and therefore they would yield bands with similar intensity in the autoradiograms. In addition, mutating the cysteines of the encoded protease would not alter the intensity of the product bands in experiments using these constructs. A master mix was prepared using the TnT rabbit reticulocyte lysate (RRL), reaction buffer, T7 polymerase, and amino acid mixture (minus methionine) provided with the system at ratios suggested by the technical bulletin. Additionally, RNasin Ribonuclease Inhibitor (Promega) was added to the master mix at 1 µL per 25 µL reaction mixture. Nuclease-free DEPC water was used to bring the final volume of each reaction to 25 µL. The master mix was incubated on a heat block for 15 minute at 30°C. The DNA plasmid constructs were added to the master mix at 0.5 µg/reaction or at 0.125 µg/reaction for pMA/CA/p2. When multiple plasmids were tested in the same experiment, the master mix was distributed among the treatment wells before the addition of DNA. Purified HIV-1 protease was prepared as described previously [35] and when indicated added at a final concentration of 250 nM active protease dimer. After incubation at 30°C, an equal volume of 3x lithium dodecyl sulfate-polyacrylamide gel electrophoresis (LDS-PAGE) loading buffer containing 150 mM dithiothreitol (DTT) (Invitrogen) was added to each reaction tube and the samples were heated at 70°C for 15 minutes. The samples were cooled at room temperature for 30 minutes and then loaded onto a 1 mm 10-well 4–12% Bis-Tris NuPAGE gel (Invitrogen). The gel was run at 100 volts for the first 10 minutes and then run at 200 volts for 70 minutes. The gel was washed in the following sequence; distilled water for 10 minutes, 10% acetic acid for 30 minutes, 8–10 washes with distilled water, and finally 10% glycerol for 15 minutes. Subsequently, the gel was placed under a conventional dryer/vacuum for 90 minutes at 54°C and exposed to film for autoradiography using Kodak Scientific Imaging film (Kodak). For quantification, GagProPol related bands on the film were scanned and densitometry performed using Un-Scan-It (Silk Scientific Corporation). Ratios of product formation to total GagProPol protein production (substrates and products) for each sample were determined, thus correcting for any internal differences in the overall extent of translated GagProPol protein that was obtained for each sample.
Supporting Information
In vitro transcription/translation with 35S-methionine was carried out for 45, 60, 75 and 90 minutes without (lanes 1,3,5,and 7) or with (lanes 2,4,6, and 8) 1 mM DTT. Samples were separated by LDS-PAGE and visualized by autoradiography and the percent processing determined using densitometry. The precursors (GagProPol and GagProPolii) and two products (MA-p2, NC-IN) are indicated.
(2.18 MB TIF)
Acknowledgments
We thank Steve Pettit for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors have no support or funding to report.
References
- 1.Coffin JM, Hughes SH, Vatmus HE, editors. New York: Cold Spring Harbor; 1997. Retroviruses.843 [Google Scholar]
- 2.Peng C, Ho BK, Chang TW, Chang NT. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. Journal of virology. 1989;63(6):2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(15):5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karacostas V, Wolffe EJ, Nagashima K, Gonda MA, Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193(2):661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 5.Krausslich H-G. Human Immunodeficiency Virus Proteinase Dimer as Component of the Viral Polyprotein Prevents Particle Assembly and Viral Infectivity. Proceddings of the National Acadamy of Sciences. 1991;88:3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burstein H, Bizub D, Skalka AM. Assembly and processing of avian retroviral gag polyproteins containing linked protease dimers. Journal of virology. 1991;65(11):6165–6172. doi: 10.1128/jvi.65.11.6165-6172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis DA, Yusa K, Gillim LA, Newcomb FM, Mitsuya H, et al. Conserved cysteines of the human immunodeficiency virus type 1 protease are involved in regulation of polyprotein processing and viral maturation of immature virions. Journal of virology. 1999;73(2):1156–1164. doi: 10.1128/jvi.73.2.1156-1164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis DA, Brown CA, Newcomb FM, Boja ES, Fales HM, et al. Reversible oxidative modification as a mechanism for regulating retroviral protease dimerization and activation. Journal of virology. 2003;77(5):3319–3325. doi: 10.1128/JVI.77.5.3319-3325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis DA, Tebbs IR, Daniels SI, Stahl SJ, Kaufman JD, et al. Analysis and characterization of dimerization inhibition of a multi-drug-resistant human immunodeficiency virus type 1 protease using a novel size-exclusion chromatographic approach. The Biochemical journal. 2009;419(2):497–506. doi: 10.1042/BJ20082068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker SD, Hunter E. Activation of the Mason-Pfizer monkey virus protease within immature capsids in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14631–14636. doi: 10.1073/pnas.251460998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis DA, Newcomb FM, Starke DW, Ott DE, Mieyal JM, et al. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. Journal of Biological Chemistry. 1997;272(41):25935–25940. doi: 10.1074/jbc.272.41.25935. [DOI] [PubMed] [Google Scholar]
- 12.Davis DA, Newcomb FM, Moskovitz J, Fales HM, Levine RL, et al. Reversible Oxidation of HIV-2 Protease. In: Sies H, Packer L, editors. Methods in Enzymology. New York: Academic Press; 2002. pp. 249–259. [DOI] [PubMed] [Google Scholar]
- 13.Pettit SC, Gulnik S, Everitt L, Kaplan AH. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. Journal of virology. 2003;77(1):366–374. doi: 10.1128/JVI.77.1.366-374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. Journal of virology. 2004;78(16):8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, et al. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. Journal of virology. 1998;72(4):2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettit CS, Simsic J, Loeb DD, Everitt L, Hutchison CA, III, et al. Analysis of Retroviral protease Cleavage Sites Reveals Two Types of Cleavage Sites and the Structural requirements of the P1 Amino Acid. The Journal of biological chemistry. 1991;266(22):14539–14547. [PubMed] [Google Scholar]
- 17.Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. Journal of virology. 1994;68(12):8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards AD, Phylip LH, Farmerie WG, Scarborough PE, Alvarez A, et al. Sensitive, soluble chromogenic substrates for HIV-1 proteinase. The Journal of biological chemistry. 1990;265(14):7733–7736. [PubMed] [Google Scholar]
- 19.Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. Journal of virology. 2005;79(16):10601–10607. doi: 10.1128/JVI.79.16.10601-10607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis DA, Dorsey K, Wingfield PT, Stahl SJ, Kaufman J, et al. Regulation of HIV-1 protease activity through cysteine modification. Biochemistry. 1996;35(7):2482–2488. doi: 10.1021/bi951525k. [DOI] [PubMed] [Google Scholar]
- 21.Kosower EM, Kosower NS. Lest I forget thee, glutathione. Nature. 1969;224(5215):117–120. doi: 10.1038/224117a0. [DOI] [PubMed] [Google Scholar]
- 22.Schuppe I, Moldeus P, Cotgreave IA. Protein-specific S-thiolation in human endothelial cells during oxidative stress. Biochemical pharmacology. 1992;44(9):1757–1764. doi: 10.1016/0006-2952(92)90069-u. [DOI] [PubMed] [Google Scholar]
- 23.Davis DA, Newcomb FM, Moskovitz J, Wingfield PT, Stahl SJ, et al. HIV-2 protease is inactivated after oxidation at the dimer interface and activity can be partly restored with methionine sulphoxide reductase. The Biochemical journal. 2000;346:305–311. [PMC free article] [PubMed] [Google Scholar]
- 24.Turpin JA, Terpening SJ, Schaeffer CA, Yu G, Glover CJ, et al. Inhibitors of human immunodeficiency virus type 1 zinc fingers prevent normal processing of gag precursors and result in the release of noninfectious virus particles. Journal of virology. 1996;70(9):6180–6189. doi: 10.1128/jvi.70.9.6180-6189.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuiken CL, Foley B, Hahn B, Korber B, McCutchan F, et al., editors. Los Alamos National Laboratory. Los Alamos, NM: Theoretical Biology and Biophysics Group; 2004. HIV Sequence Compendium 2004. [Google Scholar]
- 26.Ciriolo MR, Palamara AT, Incerpi S, Lafavia E, Bue MC, et al. Loss of GSH, oxidative stress, and decrease of intracellular pH as sequential steps in viral infection. The Journal of biological chemistry. 1997;272(5):2700–2708. doi: 10.1074/jbc.272.5.2700. [DOI] [PubMed] [Google Scholar]
- 27.Kurata S. Sensitization of the HIV-1-LTR upon long term low dose oxidative stress. The Journal of biological chemistry. 1996;271(36):21798–21802. doi: 10.1074/jbc.271.36.21798. [DOI] [PubMed] [Google Scholar]
- 28.Sappey C, Boelaert JR, Legrand-Poels S, Forceille C, Favier A, et al. Iron chelation decreases NF-kappa B and HIV type 1 activation due to oxidative stress. AIDS Res Hum Retroviruses. 1995;11(9):1049–1061. doi: 10.1089/aid.1995.11.1049. [DOI] [PubMed] [Google Scholar]
- 29.Westendorp MO, Shatrov VA, Schulze-Osthoff K, Frank R, Kraft M, et al. HIV-1 Tat potentiates TNF-induced NF-kappa B activation and cytotoxicity by altering the cellular redox state. The EMBO journal. 1995;14(3):546–554. doi: 10.1002/j.1460-2075.1995.tb07030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aukrust P, Svardal AM, Muller F, Lunden B, Berge RK, et al. Increased levels of oxidized glutathione in CD4+ lymphocytes associated with disturbed intracellular redox balance in human immunodeficiency virus type 1 infection. Blood. 1995;86(1):258–267. [PubMed] [Google Scholar]
- 31.Koh Y, Matsumi S, Das D, Amano M, Davis DA, et al. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. The Journal of biological chemistry. 2007;282(39):28709–28720. doi: 10.1074/jbc.M703938200. [DOI] [PubMed] [Google Scholar]
- 32.Darke PL, Jordan SP, Hall DL, Zugay JA, Shafer JA, et al. Dissociation and association of the HIV-1 protease dimer subunits: equilibria and rates. Biochemistry. 1994;33(1):98–105. doi: 10.1021/bi00167a013. [DOI] [PubMed] [Google Scholar]
- 33.Mascolini M, Richman D, Larder B, Mellors J, Boucher CA. Clinical implications of resistance to antiretrovirals: new resistance technologies and interpretations. Antiviral therapy. 2008;13(2):319–334. [PubMed] [Google Scholar]
- 34.Scharf SJ, Horn GT, Erlich HA. Science. 4768. Vol. 233. New York, NY: 1986. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. pp. 1076–1078. [DOI] [PubMed] [Google Scholar]
- 35.Davis DA, Brown CA, Singer KE, Wang V, Kaufman J, et al. Inhibition of HIV-1 replication by a peptide dimerization inhibitor of HIV-1 protease. Antiviral Res. 2006;72(2):89–99. doi: 10.1016/j.antiviral.2006.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro transcription/translation with 35S-methionine was carried out for 45, 60, 75 and 90 minutes without (lanes 1,3,5,and 7) or with (lanes 2,4,6, and 8) 1 mM DTT. Samples were separated by LDS-PAGE and visualized by autoradiography and the percent processing determined using densitometry. The precursors (GagProPol and GagProPolii) and two products (MA-p2, NC-IN) are indicated.
(2.18 MB TIF)