In the field of genomics there has been a move towards the development of novel experimental techniques that enable analysis of all components of a certain kind in a biological system, and this has resulted in the appearance of new methods for analyzing the omes.Obviously, in a given cellular system it is attractive to measure all the mRNAs, all the proteins, a large number of the metabolites, a large fraction of protein-protein or protein-DNA interactions, and so on, but a fundamental problem in functional genomics is integration of the information obtained, i.e., how this information can be integrated and lead to new insights into the functioning of cellular processes. Bioinformatics and advanced computer models are continuously supplying new methods for integration of data, and surely progress in the field of systems biology will eventually result in an ability to describe cellular functions in silico. In the race to obtain large amounts of data for phenotypic characterization of different cellular systems, a relatively simple experimental technique for quantitative determination of metabolic fluxes has escaped the attention of a large part of the biological research community; this technique has been used primarily by researchers in the field of metabolic engineering (1). The technique is based on relatively old principles from biochemistry, namely, feeding of specifically 13C-labeled substrates to the cell for characterization of the metabolism. However, with the development of the necessary mathematical framework for analysis of data obtained from this type of analysis it has become possible to obtain estimates for the fluxes in the different parts of the central carbon metabolism. This information is obviously interesting in connection with improving metabolite production by a given microbial cell, but as demonstrated in a paper in this issue of Journal of Bacteriology (11), it also provides a very powerful tool for functional analysis of different mutant cells. In this short commentary the use of this technique for functional analysis and the advantages and limitations of different techniques for flux quantification are discussed, and some of the underlying methods are reviewed, Finally, some future perspectives are given.
METABOLIC NETWORKS
Cellular metabolism is represented by a large number of metabolic reactions that are involved in the conversion of the carbon source into building blocks needed for macromolecular biosynthesis. Furthermore, there are specific reactions that ensure the constant supply of Gibbs free energy via ATP and redox equivalents (generally in the form of the cofactor NADPH) needed for biosynthesis of macromolecules. This large number of metabolic reactions forms a so-called metabolic network inside the cells, and as a result of reconstruction of the complete metabolic networks in different bacteria (6, 17, 18) and in the yeast Saccharomyces cerevisiae (8), more insight into the function of complete metabolic networks has been obtained. These reconstructed metabolic networks can be used for detailed studies of metabolic functions (4, 16) and the effect of gene deletions (6, 7, 9), and in the context of flux analysis there are two key lessons that can be learned.
The first lesson is that the fraction of open reading frames (ORFs) in a given genome directly involved in cellular metabolism is relatively low. Table 1 lists some statistics on the metabolic networks in four different microorganisms, and it is interesting that a higher percentage of the ORFs encode enzymes involved in metabolism in bacteria with small genomes, like Helicobacter pylori and Haemophilus influenzae (16 to 18%), than in Escherichia coli (15%) and the yeast S. cerevisiae (12%). In E. coli, which has relatively complex regulatory systems, and in eukaryotic cells a larger fraction of the ORFs code for proteins involved in regulation, and the fraction is even larger in higher eukaryotes. However, despite the relatively low fraction of ORFs involved directly in cellular metabolism, many more ORFs do have an impact on cellular metabolism via regulation of gene expression and enzyme activities. Thus, in the MIPS database (http://mips.gsf.de/proj/yeast/index.jsp) there are about twice as many ORFs grouped into carbon and energy metabolism as there are ORFs involved in this part of the metabolism in the reconstructed metabolic network (8), and the majority of the additional ORFs encode proteins involved in regulation.
TABLE 1.
Overview of reactions, metabolites, and ORFs in reconstructed metabolic networksa
| Organism | No. of reactions | No. of metabolites | No. of metabolic ORFs | Total no. of ORFs | % of ORFs involved in metabolism |
|---|---|---|---|---|---|
| H. pylori | 444 | 340 | 268 | 1,638 | 16 |
| H. influenzae | 477 | 343 | 362 | 1,880 | 19 |
| E. coli | 720 | 436 | 695 | 4,485 | 15 |
| S. cerevisiae | 1,175 | 584 | 708 | 5,773 | 12b |
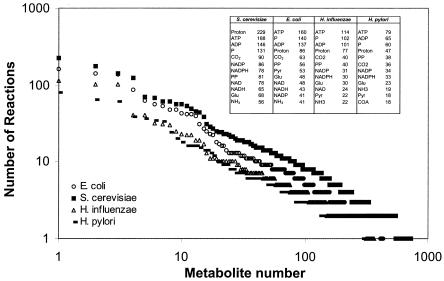
The second lesson is that the reconstructed networks clearly illustrate how the different parts of the cellular metabolism are interconnected, particularly due to usage of common cofactors, like ATP, ADP, NADH, and NADPH. These cofactors are produced in the cellular energy metabolism and are used in a large number of biosynthetic reactions. However, it is not only these cofactors that ensure a tight connection among the different branches of the metabolic network; e.g., in the network for S. cerevisiae there are 86 metabolites (corresponding to 15% of all metabolites in the metabolic network) which are involved in 10 or more reactions. This tight connection of reactions in the cellular metabolism through sharing of metabolites is illustrated in Fig. 1 for the four reconstructed metabolic networks mentioned above. The tight connection of the different parts of the metabolism means that changes in fluxes in one part of the metabolism disseminate to many other parts of the metabolism, resulting in a global response. Thus, measurement of even a few metabolic fluxes may provide valuable information about the function of the complete metabolic network.
FIG. 1.
Frequency plot of the number of reactions that each metabolite appears in for four different reconstructed metabolic networks. For each metabolic network the 10 metabolites that appear in the most reactions are listed. PP, pyrophosphate; COA, coenzyme A. The numbers in the box specify the numbers of reactions the 10 most frequently used metabolites participate in for the four different microorganisms.
FLUXES AND PHENOTYPE
As a result of evolution, the function of the central carbon metabolism has been fine-tuned to exactly meet the needs for building blocks and Gibbs free energy in conjunction with cell growth. There is therefore tight regulation of the fluxes through the central carbon metabolism. However, when a given cell experiences a change in its environment, the metabolism has to be adjusted. For example, when the carbon source changes from glucose to acetate, the cells need to down-regulate certain parts of the metabolic network (glycolysis) and activate other parts of the metabolic network (gluconeogenesis). Clearly, a large number of ORFs are involved in this regulation, and modifying the activity of the gene products of these ORFs also influences the fluxes in the metabolic network.
In order to understand the complex regulation of metabolic fluxes, one can specify the flux through a given biochemical reaction as a function of three factors: (i) the activity level of the enzyme catalyzing the reaction; (ii) the properties of the enzyme (i.e., its affinities for the substrates and possible affectors (inhibitors, activators, etc.); and (iii) the concentrations of the metabolites affecting the enzyme activity, including the reactants and products of the enzyme-catalyzed reaction.
The activity level is a function of gene expression, translation, and posttranslational protein modifications. The properties of the enzyme are generally fixed for the biological system under study, but in cases in which heterologous enzymes are inserted in order to redirect carbon fluxes, it is relevant to consider the properties of the heterologous enzyme compared with those of other enzymes interacting with the same metabolite pools. The concentrations of the metabolites are themselves functions of the fluxes in the metabolic network and the properties of the enzymes, and thus there is important feedback regulation imposed on the system.
From the information discussed above it is clear that the metabolic fluxes represent the final outcome of cellular regulation at many different levels, and hence they are an ultimate representation of the cellular phenotype expressed under certain conditions. Analysis of metabolic fluxes is therefore an interesting approach to functional analysis of cells, as illustrated by Hua et al. (11), who analyzed two different knockout mutants of E. coli by quantifying the metabolic fluxes.
FLUXES AND GENOTYPE
As discussed above, metabolic fluxes represent integrative information; i.e., the metabolic fluxes are a function of gene expression, translation, posttranslational protein modifications, and protein-metabolite interactions. In biotechnology it is interesting to obtain integrative information, as one is primarily interested in whether a specific modification results in a higher flux (or a lower flux if this is desirable), whereas understanding the exact molecular mechanisms underlying the change in the flux may be less important. However, for functional analysis of, for example, orphan gene function it is difficult to apply integrative information alone, and in this research area metabolic flux analysis, or fluxome analysis, has to be combined with analysis of other omes (e.g., the transcriptome, the proteome, the interactome, and the metabolome). Thus, it is only through analysis of several omes that it is possible to decode the functions of ORFs involved in overall regulation of cellular metabolism (15). Despite the drawback of representing integrative information, fluxome analysis does represent a method that is attractive for initial screening to determine the functions of orphan genes, as it is a simple method that allows rapid determination of whether deletion of an orphan gene results in modification of the fluxes. Furthermore, fluxome analysis may be used to obtain further insight into the functions of genes with known functions, as illustrated in studies of knockout mutants of S. cerevisiae (5, 10).
HOW TO MEASURE FLUXES
There are no direct methods for analysis of metabolic fluxes. However, based on one key assumption, it is possible to impose a large number of constraints on the fluxes in a given metabolic network. This assumption is as follows: all fluxes into a given intracellular metabolite pool balance all fluxes out of the pool. Basically, this assumption implies that the intracellular concentration of all metabolites is constant at all times, and obviously this is not the case. However, due to the rapid turnover of metabolite pools the intracellular metabolite concentrations can be adjusted rapidly to new levels, and in fact it has been observed that even after drastic changes in the environment the level of intracellular metabolites is adjusted to a new constant value within 1 to 2 min (20).
The key assumption mentioned above means that for a given metabolic network the balances around each metabolite impose a number of constraints on the system. In general, if there are J fluxes and K metabolites, then the degrees of freedom is F = J − K, and through measurement of only F fluxes the remaining fluxes can be calculated. Some fluxes can be measured directly (e.g., the fluxes of substrates into the cells and the fluxes of metabolites that are secreted from the cells), but even though some studies have relied only on measurement of these so-called exchange fluxes (12, 22), it is normally not possible to measure sufficient fluxes to calculate the remaining fluxes with good precision (24). However, if one feeds the cells 13C-labeled glucose (e.g., glucose with enriched 13C in the first position) and subsequently analyzes the 13C enrichment pattern in different intracellular metabolites, one obtains additional experimental data that can be used to obtain solid flux estimates. However, one needs to combine these data with information about the carbon transitions in all biochemical reactions, and the mathematical complexity therefore increases substantially. In recent years solid mathematical frameworks for analysis of this kind of experimental data have been developed (23, 25, 26), and this has resulted in computer algorithms for calculation of the metabolic fluxes from this kind of 13C enrichment data (24). It should, however, be mentioned that currently it is only possible to quantify the fluxes in the central carbon metabolism, but as indicated above, this part of the metabolism is tightly connected to most other parts of the cellular metabolism and it is therefore also the part of the complete metabolic network that is most interesting to study.
Several experimental techniques for analysis of the enrichment patterns in intracellular metabolites have been developed, but all these techniques are currently based on using nuclear magnetic resonance (NMR) (13, 14) or gas chromatography-mass spectrometry (GC-MS) (1). In all methods the enrichment patterns are not measured directly with the intermediates of central carbon metabolism (e.g., pyruvate and oxaloacetate), but rather they are measured with the corresponding amino acids (e.g., alanine and aspartate), as the amino acids are present at much higher levels in the cell both as free amino acids and integrated into proteins. The information content is somewhat different from the information content resulting from an analysis of the enrichment patterns by NMR or GC-MS, but the underlying principle is the same. In order to avoid the relatively complex data analysis required for estimating the metabolic fluxes, a simpler method of estimating flux ratios has been developed based on cofeeding unlabeled and uniformly 13C-labeled [6-13C]glucose (19). The resulting 13C labeling patterns of metabolic intermediates are analyzed by two-dimensional NMR spectroscopy of the amino acids. Since different pathways leading to the same metabolite yield different intact fragments, it is possible to easily calculate flux ratios. In the study of Hua et al. (11) the authors performed a flux ratio analysis, but they also estimated all of the fluxes; this study was the first study in which both methods were used. Both of the methods provide the same results for fluxes at key branch points and therefore basically provide the same kind of information. Determination of the flux ratios is simpler and may therefore seem more attractive, but estimation of all the fluxes provides a better visualization of the results and provides a more complete set of data. However, one should be aware of the fact that not all fluxes may be estimated with the same precision (2).
IDENTIFICATION OF METABOLIC NETWORK TOPOLOGIES
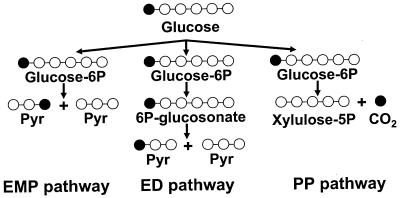
Besides allowing quantification of the metabolic fluxes, the use of 13C-enriched carbon sources is a powerful approach for identifying the metabolic network topology (i.e., which pathways are active under different growth conditions). From analysis of the enrichment patterns in intracellular metabolites one can deduce which pathways are active. This is illustrated in Fig. 2, which shows the enrichment pattern in pyruvate when glucose with 13C enrichment in the first position is metabolized via three different pathways, the Embden-Meyerhof-Parnas pathway, the pentose phosphate pathway, and the Entner-Doudoroff pathway. If glucose is metabolized via the Embden-Meyerhof-Parnas pathway, one-half of the pyruvate molecules are enriched in the third position, whereas one-half of the pyruvate molecules are enriched in the first position if glucose is metabolized via the Entner-Doudoroff pathway. However, if glucose is metabolized via the pentose phosphate pathway, then there is no enrichment of pyruvate as the 13C is lost as carbon dioxide.
FIG. 2.
Illustration of how measurement of the 13C enrichment patterns can be used to identify active pathways. EMP, Embden-Meyerhof-Parnas; ED, Entner-Doudoroff; PP, pentose phosphate.
The principles illustrated above can be taken much further, and it may even be possible to locate specific biochemical reactions in different compartments in eukaryotic cells (5, 21). Hua et al. (11) identified the topology of the metabolic network in wild-type E. coli and in mutants with disruptions of phosphoglucomutase and glucose-6-phosphate dehydrogenase and subsequently quantified the metabolic fluxes in the networks identified. Of particular interest, they identified some Entner-Doudoroff pathway activity in a phosphoglucose isomerase knockout strain and also activity in the glyoxylate shunt.
A NEW TOOL IN FUNCTIONAL GENOMICS?
As mentioned above, so far metabolic flux analysis has primarily been used for quantification of fluxes in connection with metabolic engineering of microbial overproducing strains, but as discussed here, it is obvious that this technique offers some interesting possibilities for performing functional analyses of different mutants in the field of functional genomics, as illustrated by Hua et al. (11) and other workers (5). In order for the technique to gain wider application in functional genomics, however, it is necessary to develop the technique further, and among other things this may involve ease of experimentation, direct analysis of metabolites, and high-throughput analysis.
Ease of experimentation.
The experimental technique that has been developed is relatively easy to perform, and in particular, the introduction of simple GC-MS methods has allowed analyses to be performed in many laboratories. However, interpretation of the experimental data is relatively complicated, and in particular, identification of the metabolic network requires substantial insight into cellular metabolism. This problem may be solved in the future with better computer algorithms for rapid testing of different metabolic networks and at the same time quantification of the metabolic fluxes.
Direct analysis of metabolites.
The technique of measuring the enrichment pattern in amino acids rather than in intracellular metabolites facilitates the analysis substantially, but it would be interesting to use novel methods for direct analysis of the enrichment patterns in intracellular metabolites, like pyruvate and oxaloacetate. This would enable analysis of the fluxes during rapid transients, something that is not possible with the current techniques (due to the slow dynamics in turnover of the protein pool). However, introduction of new analytical techniques will also require more advanced models for data interpretation as issues related to turnover of amino acids will become relevant.
High-throughput analysis.
In principle, there is nothing that prevents the use of the current techniques for high-throughput analysis; these techniques include, e.g., using microtiter plates for growth of different cells and subsequent analysis of a large number of mutants. Such techniques should enable screening of a large number of mutants and thus enable the development of large databases that can be used for more detailed functional analysis.
There have been recent developments in all three areas described above, and it is therefore predicted that metabolic flux analysis will be used much more widely for functional analysis in the future.
Acknowledgments
I acknowledge Jochen Förster (Fluxome Sciences A/S), Thomas Grotkjær (DTU), and Mats Åkesson (DTU) for fruitful comments. Iman Famili (University of California at San Diego) is acknowledged for putting Fig. 1 together. I also acknowledge a good and friendly collaboration with Bernhard Palsson (University of California at San Diego) in modeling of cellular metabolism.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Christensen, B., and J. Nielsen. 1999. Isotopomer analysis using GC-MS. Metab. Eng. 1:282-290. [DOI] [PubMed] [Google Scholar]
- 2.Christensen, B., A. K. Gombert, and J. Nielsen. 2002. Analysis of flux estimates based on 13C-labeling experiments. Eur. J. Biochem. 269:2795-2800. [DOI] [PubMed] [Google Scholar]
- 3.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 4.Covert, M. W., C. H. Schilling, I. Famili, J. S. Edwards, I. I. Goryanin, E. Selkov, and B. O. Palsson. 2001. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26:179-186. [DOI] [PubMed] [Google Scholar]
- 5.dos Santos, M. M., A. K. Gombert, B. Christensen, L. Olsson, and J. Nielsen. 2003. Identification of in vivo enzyme activities in the co-metabolism of glucose and acetate by Saccharomyces cerevisiae using 13C-labeled substrates. Eukaryot. Cell 2:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, J. S., and B. O. Palsson. 2000. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics and capabilities. Proc. Natl. Acad. Sci. USA 97:5528-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Famili, I., J. Förster, J. Nielsen, and B. Ø. Palsson 2003. Saccharomyces cerevisiae phenotypes can be predicted using constraint based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. USA, 100:13134-13139. [DOI] [PMC free article] [PubMed]
- 8.Förster, J., I. Famili, P. Fu, B. Ø. Palsson, and J. Nielsen. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Förster, J., I. Famili, B. O. Palsson, and J. Nielsen. 2003. Large-scale evaluation of in silico gene deletions in Saccharomyces cerevisiae. Omics J. Integr. Biol. 7:193-202. [DOI] [PubMed] [Google Scholar]
- 10.Gombert, A. K., M. M. dos Santos, B. Christensen, and J. Nielsen. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae at different conditions of glucose repression. J. Bacteriol. 183:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua, Q., C. Yang, T. Baba, H. Mori, and K. Shimizu. 2003. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 185:7053-7067. [DOI] [PMC free article] [PubMed]
- 12.Jørgensen, H., J. Nielsen, J. Villadsen, and H. Mølgaard. 1995. Metabolic flux distributions in Penicillium chrysogenum during fed-batch cultivations. Biotechnol. Bioeng. 46:117-131. [DOI] [PubMed] [Google Scholar]
- 13.Maaheimo, H., J. Fiaux, Z. P. Cakar, J. E. Bailey, U. Sauer, and T. Szyperski. 2001. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional 13C labeling of common amino acids. Eur. J. Biochem. 268:2464-2479. [DOI] [PubMed] [Google Scholar]
- 14.Marx, A., A. A. de Graaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen, J., and L. Olsson. 2002. An expanded role for microbial physiology in metabolic engineering and functional genomics: moving towards systems biology. FEMS Yeast Res. 2:175-181. [DOI] [PubMed] [Google Scholar]
- 16.Reed, J. L., and B. O. Palsson. 2003. Thirteen years of building constraints-based in silico models of Escherichia coli. J. Bacteriol. 185:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling, C. H., M. W. Covert, I. Famili, G. M. Church, J. S. Edwards, and B. O. Palsson. 2002. Genome-scale metabolic models of less-characterized organisms a case study for Helicobacter pylori, J. Bacteriol. 184:4582-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schilling, C. H., and B. O. Palsson. 2000. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J. Theor. Biol. 203:249-283. [DOI] [PubMed] [Google Scholar]
- 19.Szyperski, T., R. W. Glaser, M. Hochuli, J. Fiaux, U. Sauer, J. E. Bailey, and K. Wüthrich. 1999. Bioreaction network topology and metabolic flux ratio analysis by biosynthetic fractional 13C labeling and two-dimensional NMR spectroscopy. Metab. Eng. 1:189-197. [DOI] [PubMed] [Google Scholar]
- 20.Theobald, U., W. Mailinger, M. Baltes, M. Rizzi, and M. Reuss. 1997. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae. I. Experimental observations. Biotechnol. Bioeng. 55:305-316. [DOI] [PubMed] [Google Scholar]
- 21.Thykaer, J., B. Christensen, and J. Nielsen. 2002. Metabolic network analysis of an adipoyl-7-ADCA producing strain of Penicillium chrysogenum: elucidation of adipate degradation. Metab. Eng. 4:151-158. [DOI] [PubMed] [Google Scholar]
- 22.Vallino, J. J., and G. Stephanopoulos. 1993. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng, 41:633-646. [DOI] [PubMed] [Google Scholar]
- 23.Van Winden, W. A., W. M. Van Gulik, D. Schipper, P. J. Verheijen, P. Krabben, J. L. Vinke, and J. J. Heijnen. 2003. Metabolic flux and metabolic network analysis of Penicillium chrysogenum using 2D [13C, 1H] COSY NMR measurements and cumulative bondomer simulation. Biotechnol. Bioeng. 83:75-92. [DOI] [PubMed] [Google Scholar]
- 24.Wiechert, W. 2001. 13C metabolic flux analysis. Metab. Eng. 3:195-206. [DOI] [PubMed] [Google Scholar]
- 25.Wiechert, W., and A. A. de Graaf. 1997. Bidirectional reaction steps in metabolic networks. I. Modeling and simulation of carbon isotope labeling experiments. Biotechnol. Bioeng. 55:101-117. [DOI] [PubMed] [Google Scholar]
- 26.Wiechert, W., C. Siefke, A. A. de Graaf, and A. Marx. 1997. Bidirectional reaction steps in metabolic networks. II. Flux estimation and statistical analysis. Biotechnol. Bioeng. 55:118-135. [DOI] [PubMed] [Google Scholar]