Abstract
Objectives
Morbidity and mortality occurring in patients with multiple myeloma, AL amyloidosis, and light chain deposition disease can result from the pathologic deposition of monoclonal Ig light chains (LCs) in kidneys and other organs. To reduce synthesis of such components, therapy for these disorders typically has involved anti-plasma cell agents; however, this approach is not always effective and can have adverse consequences. We have investigated another means to achieve this objective; namely, RNA interference (RNAi).
Materials and Methods
SP2/O mouse myeloma cells were stably transfected with a construct encoding a λ6 LC (Wil) under control of the CMV promoter, while λ2-producing myeloma cell line RPMI 8226 was purchased from the ATCC. Both were treated with small interfering RNA (siRNA) directed specifically to the V, J, or C portions of the molecules and then analyzed by ELISA, flow cytometry and real time PCR.
Results
Transfected cells were found to constitutively express detectable quantities of mRNA and protein Wil and, after exposure to siRNAs, an ~40% reduction in mRNA and LC production was evidenced at 48 hours. An even greater effect was seen with the 8226 cells.
Conclusion
Our results have shown that RNAi can markedly reduce LC synthesis and provide the basis for testing the therapeutic potential of this strategy using in vivo experimental models of multiple myeloma.
Keywords: RNA interference, small interfering RNA, light chain, Bence Jones protein
Multiple myeloma, AL amyloidosis, and light chain deposition disease are associated with the production by clonal plasma cells of monoclonal light chains (LCs), i.e., Bence Jones proteins (BJPs) [1–6]. These components deposit pathologically as renal tubular casts, amyloid fibrils, or punctate membrane deposits, respectively, in the kidneys or other sites and can eventually lead to organ failure and death.
Therapy of these disorders presently involves use of anti-plasma cell drugs given in conventional amounts or in high doses combined with stem cell transplant and while LC synthesis can be reduced [7, 8], considerable toxicity and even mortality may occur [9]. Due to these complications, another means to suppress LC formation has been investigated experimentally; namely, post-transcriptional gene regulation using an anti-sense oligonucleotide [10]. However, based on recent comparisons of this technique with an alternate approach - RNA interference (RNAi) - the latter may offer a more promising means to achieve this objective [11, 12].
We now report the results of our in vitro studies on the effects of synthetic siRNA molecules that target LC variable (V), joining (J), and constant (C) regions. We found that siRNAs delivered by lipid-based transfection media were non-toxic and also significantly lowered mRNA levels and LC formation in stably transfected human λ6 producing mouse myeloma cells, as well as in human myeloma cells synthesizing λ2 LCs.
Materials and methods
Cell lines
SP2/O mouse myeloma cells (American Type Culture Collection; ATCC, Manassas, VA, USA) were stably transfected by electroporation using a construct encoding the V, J, and C regions of a λ6 LC designated Wil [13] under control of the cytomegalovirus promoter; the modified cells were termed SP2/O-λ6. The human λ2 LC producing myeloma cell line RPMI 8226 [14] was purchased from the ATCC (No. CCL 155), cultured in DMEM F-12 (Lonza, Basel, Switzerland) supplemented with 5% FBS/1% penicillin/streptomycin, and maintained in a humidified incubator at 37°C under 5% CO2. Cell viability and number were determined by trypan blue dye exclusion and hemacytometry, respectively.
Small-interfering RNA (siRNA)
Custom siRNA duplexes corresponding to nucleotide sequences encoding within 3 domains of the V region of λ6 Wil (V1 Wil, V2 Wil, and J1 Wil), 1 of λ2 8226 (V1 8226), and 2 areas common to all λ LCs (C1 λ, C2 λ), as well as 1 unique to the C-terminal portion of 8226 (C3 8226) were purchased from Sigma-Aldrich (St. Louis, MO, USA). SiRNA target homology and specificity were confirmed through a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi); their nucleotide sequences are provided in Table 1. AllStars non-silencing siRNA (Qiagen, Valencia, CA) served as a negative control. For the siRNA studies, cells were washed, collected by centrifugation, and resuspended in 100 µL of phenol red-free DMEM F-12 (Sigma-Aldrich) containing 5% FBS and penicillin/streptomycin. Each of the 24 wells in a microplate received a 100-µL volume that contained 2×105 cells, followed by addition of 1.5 µg of siRNA complexed with 4.5 µL HiPerFect siRNA transfection reagent (Qiagen) in 100 µL phenol red-free medium lacking serum or antibiotics (for control purposes, cells were treated with AllStars siRNA/HiPerFect). The plates were incubated at 37°C for 24, 48, or 72 hours. At each time point, cell viability was assessed using the Cell Titer Blue assay (Promega San Luis Obispo, CA), according to the manufacturer’s instructions.
Table 1.
Nucleotide sequences of siRNAs
| Oligonucleotide | Length | Sequence |
|---|---|---|
| SP2/O-λ6a | ||
| V1 Wil | 21 | AACAACUAUGUUCACUGGUAC CAUUGUUGAUACAAGUGACCA |
| V2 Wil | 21 | ACUGUGAUCUUUGAGGAUGAC CAUGACACUAGAAACUCCUAC |
| J1 Wil | 21 | AAGUUGACCGUCCUGGGUCAG GAUUCAACUGGCAGGACCCAG |
| RPMI 8226b | ||
| V1 8226 | 21 | GUGACAUUGGUGACUAUCAUU UUCACUGUAACCACUGAUAGU |
| C3 8226 | 21 | CCUGCAGAAUGUUCUUAGUUU UUGGACGUCUUACAAGAAUCA |
| SP2/O-λ6/RPMI 8226c | ||
| C1 λ | 21 | GUCUCCAUAAGUGACUUCUAUU UUCAGAGGUAUUCACUGAAGAU |
| C2 λ | 21 | CCAAACAAAGCAACAACAAUU UUGGUUUGUUUCGUUGUUGUU |
Sequence directed to V- and J-regions of λ6 LC Wil.
Sequences directed to V- and C-regions of λ2 8226.
Sequences directed to C λ regions common to both SP2/O-λ6 and RPMI 8226 LCs.
siRNA optimization and transfection efficiency
Cellular uptake of siRNA was measured with a FACScan flow cytometer (BD Immunocytometry Systems, Franklin Lakes, NJ) using CellQuest Pro software. Optimal transfection conditions for each cell line were determined by titrating the concentration of AllStars siRNA labeled with Alexa 488 and HiperFect. Non-labeled AllStars siRNA was used as a control and Alexa 488-labeled AllStars-treated cells were routinely incorporated to monitor transfection efficiency.
RNA isolation and analysis
Following exposure to siRNA, total cellular RNA was harvested by the RNAeasy Plus RNA isolation kit (Qiagen) and product integrity assessed by agarose gel electrophoresis. RNA concentration was determined by UV spectroscopy.
Sequencing of SP2/O-λ6 LC Wil
Synthesis of first strand cDNA was performed using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). Gene specific primers designed to recognize the leader, V, or C regions of each LC product were purchased from Sigma-Aldrich and included: Wil V1 (forward), 5΄- AATTTTTTACTGACTCAGCCCC-3΄ and Wil C1 (reverse), 5΄-CTGACC CACGACGGTCAACTTG-3. One µg of cDNA was amplified by PCR using Phusion DNA polymerase (New England BioLabs, Ipswich, MA, USA) under the following conditions: DNA denaturation 95°C for 30 seconds, synthesis 55°C for 30 seconds, and elongation 73°C for 30 seconds; reactions were carried out to 40 cycles. PCR products were isolated by agarose gel electrophoresis, visualized by ethidium bromide, and the bands excised. Amplified DNA was purified with the Wizard SV Gel and PCR Clean-Up System (Promega) per manufacturer’s instructions. Product sequence analysis was performed by the University of Tennessee’s Molecular Biology Resource Facility.
Real time RT-PCR
First strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and 500 ng of RNA. Gene specific primers were designed and included: λ6 Wil (forward), 5΄-ACGGTAACCATCTCCTGCA-3΄ and (reverse), 5΄-AGGGTCTGTGGTCATCCTC-3΄ (a 121-bp product); 8226 LC (forward), 5΄-CAA AGCCCCCAAACTCATAA-3΄ and (reverse), 5΄-AACACCACCTCGAAAAGTGC-3΄ (a 180-bp product). Primers to human and mouse GAPDH (Real Time Primers, LLC, Elkins Park, PA, USA) were used to amplify the calibrator gene: human GAPDH (forward), 5΄-GAGTCAACGCGGATTTGGTCGT-3΄ and (reverse), 5΄-TTGATTTTGGAGGGATCTCG-3΄ (a 238-bp product); murine GAPDH (forward), 5΄-CTGGAGAAACCTGCCAAG TA-3΄ and (reverse), 5΄-TGTTGCTGTAGCCGTATTCA-3΄ (a 223-bp product). With each set, amplification efficiency was ~100%. Real-time PCR was performed in 96-well PCR plates with an ICycler PCR unit (Bio-Rad, Hercules, CA, USA) utilizing IQ SYBR Green Supermix that contained 400 nM primer mix and 3 µL cDNA. Fluorescence was detected with an IQ5 Multicolor Real-Time PCR system. Conditions for activation and denaturation were: cycle 1, 95°C for 3 minutes, followed by forty 30-second amplification cycles at 95°C, 63°C, and 72°C. Cells treated with AllStars siRNA acted as controls.
ELISA
SP2/O-λ6 cells obtained 24, 48, and 72 hours post transfection were centrifuged at 400 × g for 5 minutes and the supernatants harvested and diluted 1:10 in PBS containing 1% BSA/0.1% Tween 20. Wells in high-binding affinity ELISA/RIA plates (Fisher Scientific, Pittsburgh, PA, USA) were incubated overnight at 4°C with 100 µL (3.75 µg/mL) of a mouse anti-human λ6 LC mAb [15], washed, and then blocked for 1 hour at 37°C with PBS/1% BSA, followed by addition for 1 hour at 37°C of diluted supernatants. A standard curve was prepared by serial dilution of purified recombinant Vλ6 Wil over a range of 0.3–400 µg/mL. The wells were washed and filled with 100 µL of a rabbit anti-human λ6 LC polyclonal antibody [13] and were pre-blocked in PBS containing 1% BSA/0.5% filtered goat and mouse sera (Sigma-Aldrich)/0.5% MOPC 31c (Sigma-Aldrich)/0.1% Tween 20. Following incubation at 37°C for 1 hour, the plates were washed and bound antibody detected by addition of HRP-conjugated goat anti-rabbit IgG (Jackson Immunoresearch, West Grove, PA, USA); the reaction was developed with ABTS/0.06% H202. Immunoreactivity was quantified by measuring the absorbance at 405 nm with a BioTek Synergy HT Luminescence Reader (BioTek Instruments).
For RPMI 8226 cells, 1:100 dilutions of supernatants in PBST were added to wells coated with an 11 µg/mL rabbit anti-human total λ antiserum (DAKO); a mouse anti-human total λ LC mAb (1 µg/mL) served as the secondary reagent. Bound antibody was detected by addition of HRP conjugated goat anti-mouse Ig and absorbance was measured as described above.
Flow cytometry
Histogram overlays of cells exposed to either C1 λ or C2 λ siRNA or sham siRNA molecules were utilized to determine any shift in fluorescence between the experimental and control groups. Regional markers were drawn to exclude untreated cells. RPMI 8226 intracellular LC concentration was determined by flow cytometry [16] where cells treated with experimental or control siRNA were isolated at 24, 48, and 72 hours by centrifugation at 500 × g, washed in PBS/1% BSA, treated with fixative, permeabilized with Cytofix/Cytoperm buffer (BD Biosciences), and blocked for 20 minutes in Cytoperm solution/1% BSA. The cells were resuspended in CytoPerm containing either FITC-labeled rabbit anti-human total λ LC antiserum or, as a control, an unlabeled reagent. After a 1-hour incubation at room temperature, the cells were washed × 3 in PBS and resuspended in 0.5 mL PBS for analysis by flow cytometry (FACScan, BD Biosciences). To measure fluorescence in the control population, gates were set to exclude cells exposed only to AllStars siRNA.
Statistical analysis
ELISA results were normalized and expressed as percent control siRNA treated cells. For each set, data were pooled and analyzed for significance by One-Way ANOVA using Dunnett’s Multiple Comparisons test (GraphPad Prism, GraphPad Software Inc., La Jolla, CA, USA).
Results
Transfection efficiency
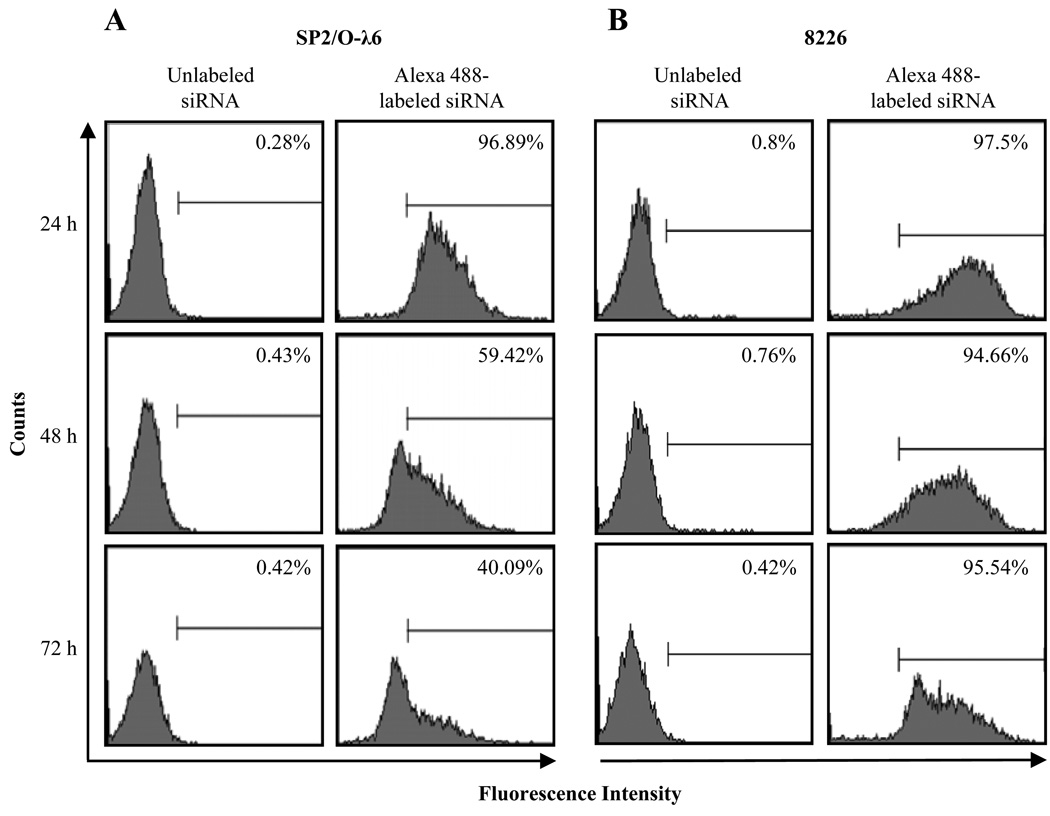
Optimal transfection conditions for both SP2/O-λ6 and 8226 cells were determined by flow cytometry. For each experiment, incorporation of Alexa 488-labeled AllStars siRNA, as well as trypan blue, was used to document transfection efficiency and monitor cytotoxicity. In the case of SP2/O-λ6, ~97% of the cells were transfected at 24 hours; by 48 and 72 hours, this value had decreased to ~59% and ~40%, respectively (Fig. 1). As for 8226, similar results were obtained at 24 hours, but at 72 hours, ~95% of these cells remained labeled. The mean fluorescence intensity of the transfected populations decreased in both lines over the 72-hour period of study, presumably due to dilution by cell division.
Figure 1.
Uptake of siRNA by SP2/O-λ6 and 8226 cells. Flow cytometric analyses of (A) SP2/O-λ6 and (B) 8226 cells transfected with Alexa 488-labeled or unlabeled AllStars siRNA 24, 48, or 72 hours post treatment (regional markers were based on cells transfected with non-labeled AllStars siRNA). Numbers represent the percentage of positively transfected cells.
Effect of siRNA treatment on LC mRNA levels
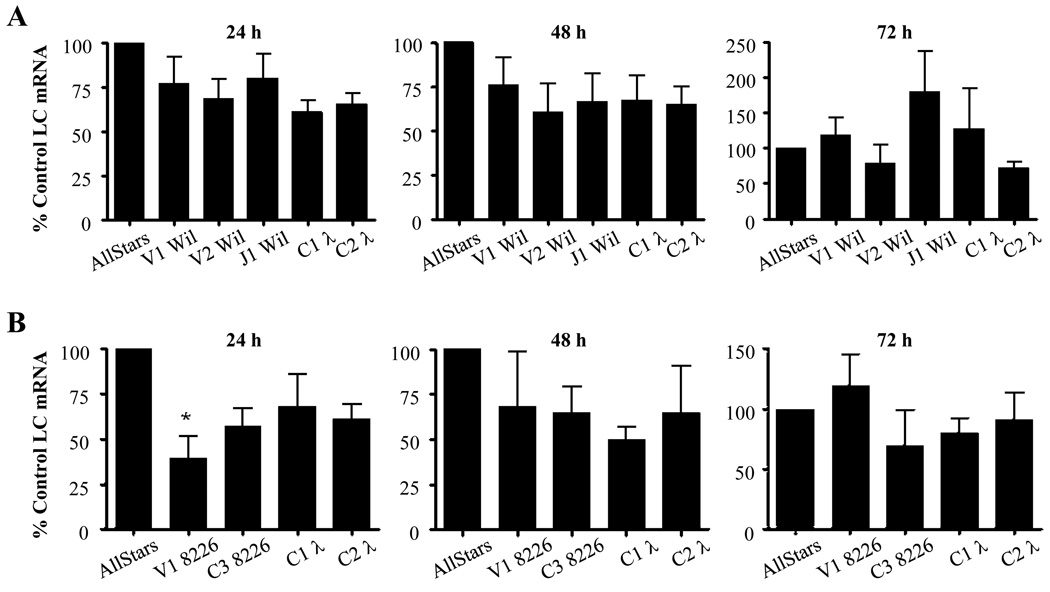
Agarose gel electrophoresis of PCR reaction products from SP2/O-λ6 and 8226 cells revealed bands with the expected size of ~115 and ~180 bp, respectively; no secondary low molecular weight components were observed. At 24 hours, SP2/O-λ6 cells exposed to V1, V2, or J1 Wil had a 23, 32, or 20% reduction in LC mRNA levels and by 48 hours, these values had lowered to 24, 40, or 34%. As for C1 λ and C2 λ siRNAs, a 39 and 35% decrease, respectively, occurred at 24 hours; similar results were seen at 48 hours. 72 hours post-transfection, the findings in all cases were erratic, most likely due to cell doubling and death; however, in general, the levels were equivalent to those of untreated cells, with the exception of V2 Wil and C2 λ which remained 21 and 30% below that found in the controls (Fig. 2).
Figure 2.
Reduction in LC mRNAs in siRNA transfected mouse and human myeloma cells. (A) SP2/O-λ6 cells treated with siRNA specific for the V or J portions of λ6 light chain Wil or to the Cλ region. (B) 8226 cells treated with siRNA directed to either the V- or C-terminus of λ LC 8226. The gene expression levels of LCs Wil and 8226 at 24, 48, and 72 hours were quantified by RT-PCR using murine GAPDH as a calibrator gene (performed in triplicate). Bars indicate the expression of treated versus sham siRNA treated cells as the mean ± SEM of 3 representative experiments.
Exposure of RPMI 8226 cells to V1 or C3 8226 decreased target LC mRNA by 61 and 43%, while a lesser degree of suppression was observed with C1 λ and C2 λ (32 and 39%). Notably, by 48 hours, mRNA levels in cells incubated with C1 λ siRNA were reduced by 51% while those treated with V1 8226, C2 λ, and C3 8226 had values > 32–35%. By 72 hours, results with V1 8226, C1 λ, and C2 λ siRNAs were similar to controls, whereas mRNA levels in C3 8226 treated cells remained low.
Effect of siRNAs on LC production
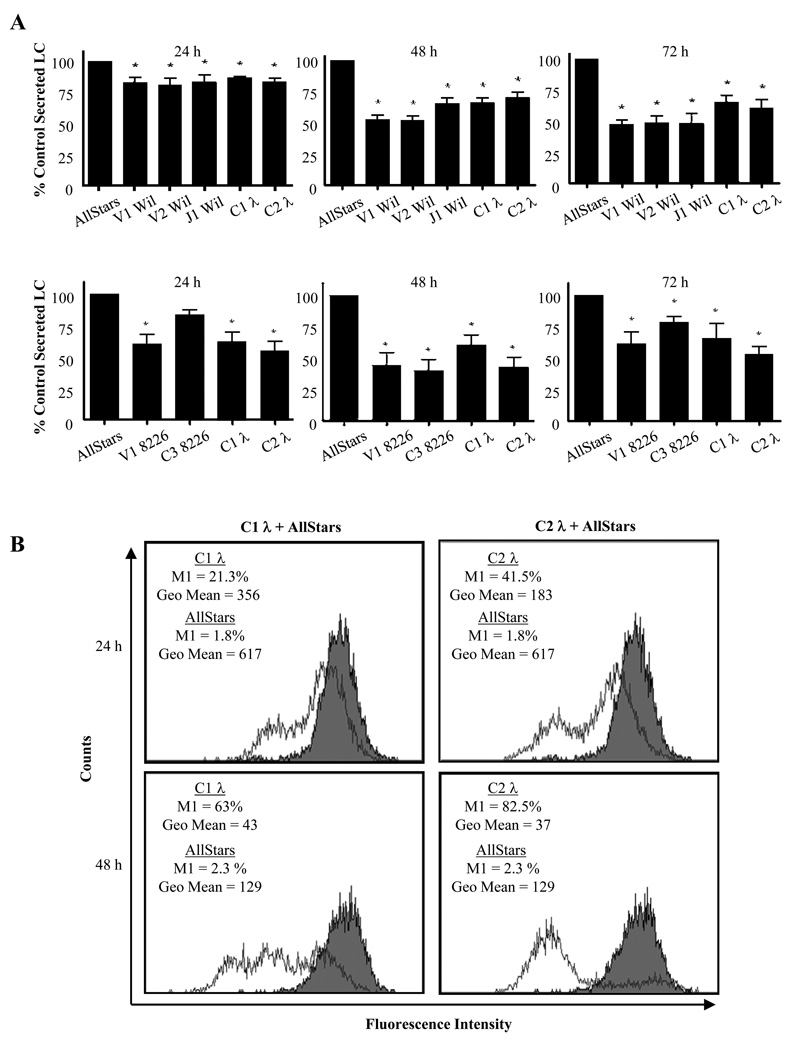
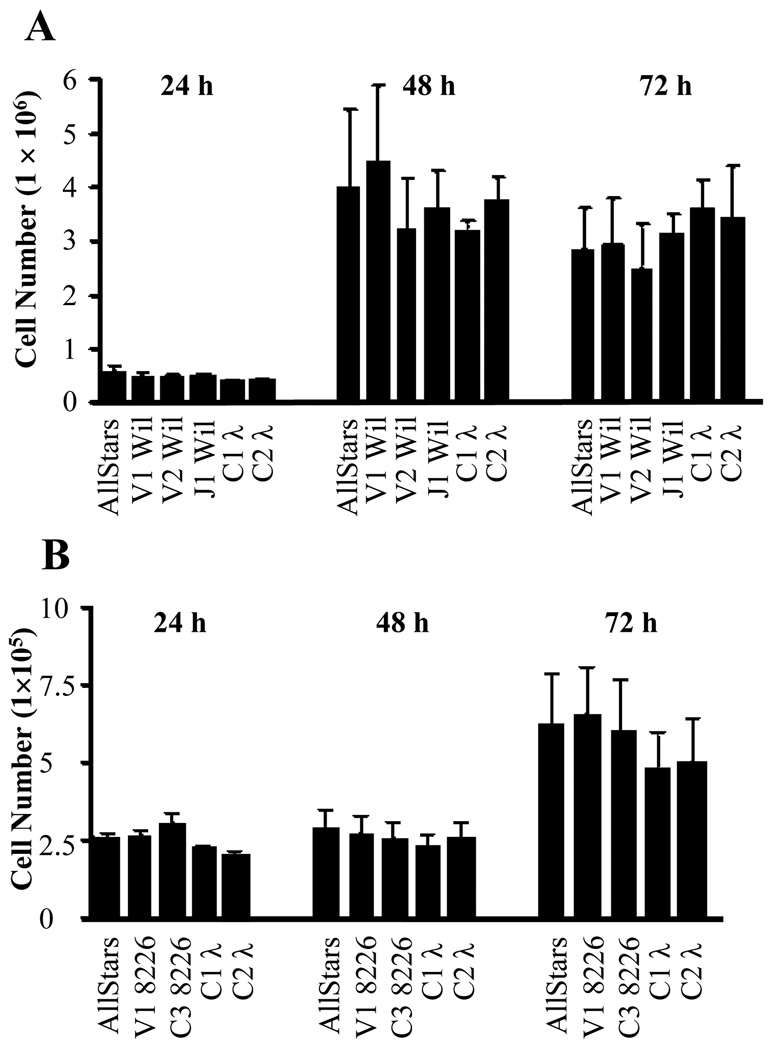
1 × 106 SP2/O-λ6 cells generated ~180 ng of λ6 LC Wil per day. However, when treated with siRNAs targeting Wil V, J, or C regions, LC secretion in culture supernatants at all 3 time points was significantly reduced by 20–50% (p < 0.05) (Fig. 3), whereas exposure to siRNAs specific for LC 8226 had no effect. 72 hours post-transfection, the greatest decrease (~50%) in λ6 LC Wil secretion occurred with the V- and J-region specific siRNAs, as compared to sham-treated cells (C-region siRNA reduced secretion by 40%). Notably, treatment with siRNA had no effect on cell viability or number throughout the 72 hour experimental period, as evidenced in the Cell Titer Blue viability assay (Fig 4).
Figure 3.
Effect of siRNA on LC synthesis. (A, upper) SP2/O-λ6 and (lower) 8226 cells were cultured 24, 48, or 72 hours with siRNA oligonucleotides, as indicated. At given time points, the centrifuged supernatants were analyzed by ELISA. The data shown represent the ratio of LC concentrations within supernatants of siRNA-treated cells versus controls. For each group, ratios were averaged and analyzed by one-way ANOVA (n = 6; mean ± SEM; *, p < 0.05). (B) Cλ-specific siRNA induced reduction in plasma cell LC concentration. Flow cytometric analysis of 8226 cells transfected with siRNAs C1 λ, C2 λ (open histograms) or AllStars controls (closed histograms). Regional markers that excluded sham treated cells were set for green fluorescence. For each panel, the geometric mean of the gated cell population and the percent of cells falling within the marker region are indicated.
Figure 4.
Effect of siRNA on cell viability. After exposure to V-, J-, and C- region specific siRNAs or AllStars control siRNA, transfected SP2/O-λ6 (A) and 8226 cells (B) were assessed for viability using the CellTiter Blue assay. Data were pooled from 5 and 6 experimental replicates, respectively, and analyzed by one-way ANOVA (mean ± SEM; *, p < 0.05).
In the case of RPMI 8226, 1 × 106 cells produced ~13.4 µg of LC per day. 24 hours post-transfection, exposure to V1 8226, C1, and C2 siRNA constructs significantly lowered LC production by 40, 39, and 61% (p < 0.05), while treatment with AllStars non-silencing control siRNA had no effect (Fig. 3). At 48 hours, λ2 protein secretion for all treatment groups was significantly reduced by ~40–65% (p < 0.05) and this phenomenon was still apparent at 72 hours. In contrast, exposure of 8226 cells to siRNAs that were specific for LC Wil did not repress LC production at any time point.
Reduction in cytoplasmic LC production by 8226 cells exposed to siRNA also was documented by flow cytometry (Fig. 3), where a gate region was drawn that excluded control siRNA-treated cells (cells within this area were considered negative). The geometric mean of each population was used to estimate relative fluorescence intensity between cells exposed to experimental versus control siRNAs. At 24 and 48 hours, 21.3 and 63% of C1 λ treated cells were negative, as compared with controls (1.8 and 2.3%); further, exposure to C1 λ siRNA elicited a 58 and 33% reduction in fluorescence intensity at 24 and 48 hours. C2 λ siRNA caused an even greater decrease in cellular LC concentration, with a 42 and 70% loss of fluorescence at 24 and 48 hours, respectively. As with the SP2/O-λ6 cells, the siRNAs did not affect cell viability or number (Fig 4).
Discussion
A major factor contributing to the morbidity and mortality of patients with clonal plasma cell dyscrasias is the overproduction of Ig free LCs and their subsequent aggregation and deposition in tissues and organs. Current standard of care involves use of conventional or dose-intensive anti-plasma cell chemotherapy to reduce the synthesis of toxic LCs. A recent retrospective survey of individuals diagnosed with multiple myeloma and LC cast nephropathy indicated that a 50% reduction in circulating free LC levels correlated with an improved clinical outcome in patients who responded to such treatments as compared to those who did not [17]. Use of similar therapies in AL amyloidosis or light chain deposition disease also has resulted in clinical improvement [18].
Another approach to reduce protein synthesis is the use of anti-sense oligonucleotides, which has been shown experimentally to effect a modest reduction in LC secretion by human myeloma cells and xenografts [10], as well as amyloidogenic transthyretin by hepatocytes [19]. This technology, though, has been superseded by RNAi due to the latter’s greater efficiency [11] and specificity in causing post-transcriptional “silencing” of protein expression [20,21].
In this regard, we have demonstrated that exposure of 2 different human LC synthesizing cell lines to siRNAs directed to the V, J, or C portions of the molecules resulted in reduction of mRNA and protein production. Because our siRNAs were in the form of small synthetic oligonucleotides, we used, in lieu of a viral delivery system, a lipid based transfection media, namely HiPerFect, which proved to be highly efficient and well tolerated, based on cell counts and viability. Indeed, other similar systems have produced little or no toxicity in animal models [10, 22–24]. Additionally; we investigated cellular uptake of the siRNAs by flow cytometry and found that both SP2/O-λ6 and 8226 cells had equally high incorporation of the labeled reagents. For SP2/O-λ6, > 40% of the cells remained positive over the course of the 72-hour period whereas, for 8226, the fluorescence intensity was greater (> 90% positive). We attribute these differences to varying cell growth rates for each line. The 8226 cell numbers remained relatively stable over the initial 48 hour period, approximating the 2 × 105 cells that were originally plated. However, by 72 hours these values had significantly increased, equating a doubling event. Further, a sub-population of negative 8226 cells had begun to appear, as evidenced by a distinct “shoulder” in the flow cytometry histogram. This contrasted with the SP2/O-λ6 cells that had already doubled at 24 hours and, by 48 hours, had increased ~10-fold. At 72 hours, the number receded, most likely due to overcrowding. We hypothesize that, for both lines, negative cells exhibited reduced fluorescence due to dilution of the siRNAs oligonucleotides that occurred during cell doublings. As the SP2/O-λ6 cells underwent a more rapid turnover, the negative population was apparent at an earlier time point. Nonetheless, in both cases, there was no significant decrease in cell count after treatment with LC specific siRNAs.
Reduction in target mRNA levels was most apparent in the 8226 cells transfected with V1 8226 siRNA where a significant (p < 0.05) drop in LC mRNA occurred, as compared with sham treated controls. Exposure to the other siRNA oligonucleotides was somewhat less effective, with only a 30–45% reduction in target LC mRNA. This trend persisted throughout 48 hours with an apparent recovery of mRNA by 72 hours. Although not as dramatic, the SP2/O-λ6 cells had reduced LC mRNAs by 24 hours after exposure to siRNAs specific for either the V or C region of the λ6 LC which persisted through 48 hours.
With regard to LC levels, exposure of either SP2/O-λ6 or 8226 cells to the siRNAs significantly lowered target protein expression. Treatment of SP2/O-λ6 cells with any of the siRNA molecules induced, by 24 hours, a moderate, but still significant (p < 0.05), reduction in LC production that was even greater at 48 and 72 hours. Likewise, exposure of the 8226 cells to the siRNAs significantly (p < 0.05) reduced the concentration of secreted protein in all groups, except the C3 treated cells where there was a less pronounced effect. We attribute this to the presence in the C3 siRNA of a single nucleotide mismatch between the 2 RNA sequences, apparently due to a silent mutation in the germline λ2 gene as compared to the actual LC product of the 8226 line. This result emphasizes the need for perfectly matched siRNA molecules [21].
Immunostaining of fixed and permeabilized RPMI 8226 cells exposed to C1, C2, or AllStars siRNA revealed a substantial population that exhibited reduced intracellular LC. This response did not occur in untreated cells or those transfected with AllStars control siRNA. We posit that this reduction was due to the silencing effect of the siRNA molecules on mRNA synthesis and, as a consequence, the cellular LC pool was depleted by secretion of previously synthesized protein. This combination, i.e., blockage of LC gene translation and loss of cytoplasmic protein by secretion, resulted in cells devoid of LCs. These results indicate that measurement by flow cytometry of cellular protein levels alone may provide a more accurate depiction of the dynamic process of LC depletion that results from siRNA-mediated silencing.
Our data have demonstrated that siRNA molecules directed towards V, J, or C regions of LCs can lead to a reduction in target mRNA, as well as cellular and secreted protein, in human LC producing cells. These results, albeit transient, were highly specific for the target mRNA and proteins and demonstrated that treatment with these molecules, as well as the transfection reagents themselves, was non-cytotoxic. Given these encouraging findings, our future efforts will be directed towards determining the in vivo efficacy of RNAi in inhibiting LC production in mice bearing SP2/O-λ6 or 8226 plasmacytomas. The ability to eliminate or reduce synthesis of nephrotoxic or amyloidogenic LCs would provide an invaluable therapeutic adjuvant in management of clonal plasma cell dyscrasias.
Acknowledgements
The author would like to acknowledge Luis Acero at the MD Anderson Cancer Research Institute for assistance with real-time PCR and experimental design. This research was supported in part by UHPSS Research grant CA10056 from the National Cancer Institute and the Physicians Medical Education Research Foundation. AS is an American Cancer Society Clinical Research Professor.
Abbreviations
- BJP
Bence Jones protein
- RNAi
RNA interference
- siRNA
small interfering RNA
- LC
Ig light chain
- V
variable region
- J
joining region
- C
constant region
- mAb
monoclonal antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Basic-Jukic N, Kes P, Labar B. Myeloma kidney: pathogenesis and treatment. Acta Med Croatica. 2001;55:169–175. [PubMed] [Google Scholar]
- 2.Kyle RA, Gertz MA, Witzig TE, et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin Proc. 2003;78:21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 3.Pozzi C, Locatelli F. Kidney and liver involvement in monoclonal light chain disorders. Semin Nephrol. 2002;22:319–330. [PubMed] [Google Scholar]
- 4.Tanenbaum ND, Howell DN, Middleton JP, Spurney RF. Lambda light chain deposition disease in a renal allograft. Transplant Proc. 2005;37:4289–4292. doi: 10.1016/j.transproceed.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 5.Gertz MA, Lacy MQ, Dispenzieri A, Hayman SR. Amyloidosis. Best Pract Res Clin Haematol. 2005;18:709–727. doi: 10.1016/j.beha.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Rajkumar SV, Dispenzieri A, Kyle RA. Monoclonal gammopathy of undetermined significance, Waldenstrom macroglobulinemia, AL amyloidosis, and related plasma cell disorders: diagnosis and treatment. Mayo Clin Proc. 2006;81:693–703. doi: 10.4065/81.5.693. [DOI] [PubMed] [Google Scholar]
- 7.Durie BG, Kyle RA, Belch A, et al. Myeloma management guidelines: a consensus report from the Scientific Advisors of the International Myeloma Foundation. Hematol J. 2003;4:379–398. [PubMed] [Google Scholar]
- 8.Rajkumar SV, Kyle RA. Conventional therapy and approach to management. Best Pract Res Clin Haematol. 2005;18:585–601. doi: 10.1016/j.beha.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Fenk R, Schneider P, Kropff M, et al. High-dose idarubicin, cyclophosphamide and melphalan as conditioning for autologous stem cell transplantation increases treatment-related mortality in patients with multiple myeloma: results of a randomised study. Br J Haematol. 2005;130:588–594. doi: 10.1111/j.1365-2141.2005.05641.x. [DOI] [PubMed] [Google Scholar]
- 10.Ohno S, Yoshimoto M, Honda S, et al. The antisense approach in amyloid light chain amyloidosis: identification of monoclonal Ig and inhibition of its production by antisense oligonucleotides in in vitro and in vivo models. J Immunol. 2002;169:4039–4045. doi: 10.4049/jimmunol.169.7.4039. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand J-R, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commum. 2002;296:1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- 12.Bilanges B, Stokoe D. Direct comparison of the specificity of gene silencing using antisense oligonucleotides and RNAi. Biochem J. 2005;388:573–583. doi: 10.1042/BJ20041956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wall JS, Gupta V, Wilkerson M, et al. Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four Vλ6 proteins. J Mol Recognit. 2004;17:323–331. doi: 10.1002/jmr.681. [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka Y, Moore GE, Yagi Y, Pressman D. Production of free light chains of immunoglobulin by a hematopoietic cell line derived from a patient with multiple myeloma. Proc Soc Exp Biol Med. 1967;125:1246–1250. doi: 10.3181/00379727-125-32327. [DOI] [PubMed] [Google Scholar]
- 15.Abe M, Goto T, Kennel SJ, et al. Production and immunodiagnostic applications of antihuman light chain monoclonal antibodies. Am J Clin Pathol. 1993;100:67–74. doi: 10.1093/ajcp/100.1.67. [DOI] [PubMed] [Google Scholar]
- 16.Lee GM, Kim JH, Lee HG. Flow cytometric analysis of antibody producing cells using double immunofluorescent staining. Biotechnol Tech. 1996;10:615–620. [Google Scholar]
- 17.Leung N, Gertz MA, Zeldenrust SR, et al. Improvement of cast nephropathy with plasma exchange depends on the diagnosis and on reduction of serum free light chains. Kidney Int. 2008;73:1282–1288. doi: 10.1038/ki.2008.108. [DOI] [PubMed] [Google Scholar]
- 18.Lachmann HJ, Gallimore R, Gillmore JD, et al. Outcome in systemic AL amyloidosis in relation to changes in concentration of circulating free immunoglobulin light chains following chemotherapy. Br J Haematol. 2003;122:78–84. doi: 10.1046/j.1365-2141.2003.04433.x. [DOI] [PubMed] [Google Scholar]
- 19.Benson MD, Kluve-Beckerman B, Zeldenrust SR, et al. Targeted suppression of an amyloidogenic transthyretin with antisense oligonucleotides. Muscle Nerve. 2006;33:609–618. doi: 10.1002/mus.20503. [DOI] [PubMed] [Google Scholar]
- 20.Feng X, Zhao P, He Y, Zuo Z. Allele-specific silencing of Alzheimer's disease genes: The amyloid precursor protein genes with Swedish or London mutations. Gene. 2006;371:68–74. doi: 10.1016/j.gene.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Kurosawa T, Igarashi S, Nishizawa M, Onodera O. Selective silencing of a mutant transthyretin allele by small interfering RNAs. Biochem Biophys Res Commun. 2005;337:1012–1018. doi: 10.1016/j.bbrc.2005.09.142. [DOI] [PubMed] [Google Scholar]
- 22.Sioud M. siRNA Delivery In Vivo. Methods Mol Biol. 2005;309:237–249. doi: 10.1385/1-59259-935-4:237. [DOI] [PubMed] [Google Scholar]
- 23.Ohara PT, Vit J-P, Bhargava A, Jasmin L. Evidence for a role of connexin 43 in trigeminal pain using RNA interference in vivo. J Neurophysiol. 2008;100:3064–3073. doi: 10.1152/jn.90722.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank-Kamenetsky M, Grefhorst A, Anderson NN, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad.Sci U.S.A. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]