Abstract
The advent of mammalian gene engineering and genetically modified mouse models has led to renewed interest in developing resources for referencing and quantitative analysis of mouse brain anatomy. In this study, we used diffusion tensor imaging (DTI) for quantitative characterization of anatomical phenotypes in the developing mouse brain. As an anatomical reference for neuroscience research using mouse models, this paper presents DTI based atlases of ex vivo C57BL/6 mouse brains at several developmental stages. The atlas complements existing histology and MRI-based atlases by providing users access to three-dimensional, high-resolution images of the developing mouse brain, with distinct tissue contrasts and segmentations of major gray matter and white matter structures. The usefulness of the atlas and database was demonstrated by quantitative measurements of the development of major gray matter and white matter structures. Population average images of the mouse brain at several postnatal stages were created using large deformation diffeomorphic metric mapping and their anatomical variations were quantitatively characterized. The atlas and database enhance our ability to examine the neuroanatomy in normal or genetically engineered mouse strains and mouse models of neurological diseases.
Introduction
The mammalian brain is one of the most complicated structures within living organisms. The three-dimensional organization (comprising the spatial dimensions), which consists of more than 100 billion neurons in the adult brain, combined with the developmental processes (comprising a temporal dimension), constitute a vast four-dimensional (4D) entity. Although there is much literature that describes the anatomy of the rodent brain and its development, for example, the development of the cortex (Bayer and Altman, 1991; Sidman and Rakic, 1973), thalamus (Altman and Bayer, 1988), and cerebellum (Altman and Bayer, 1985), the spatiotemporal anatomy of the developing rodent brain remains poorly documented. The need for such information has become even more urgent because of increasingly sophisticated gene and cell engineering technologies. For example, mapping gene and protein expression, detecting anatomical changes caused by gene mutations, and delivery and extraction of cells in the brain all require precise 4D anatomical information of the rodent brain. Existing atlases contain only limited coverage of developmental stages. Most of these atlases are histology-based and two-dimensional, and therefore, have limited slice locations and orientations. As a result, several key aspects of the spatiotemporal development of the mouse brain have not been well documented. Questions about the volume and coordinates of specific brain structures in the developing C57BL/6 mice, and the range of their anatomical variation can only be answered with a three-dimensional atlas of the developing mouse brain.
With modern magnetic resonance imaging (MRI), we can now generate three-dimensional images of the mouse brain with high spatial resolution (Benveniste et al., 2000; Jacobs et al., 1999; Johnson et al., 2002; Kovacevic et al., 2005; Natt et al., 2002; Petiet et al., 2008). This opens up new possibilities for establishing 4D atlases of the developing mouse brain. The advantages of the MRI-based atlases are clear. The image is inherently three-dimensional (3D) without distortions due to embedding and sectioning, and the entire mouse brain can be imaged with a one-to-one correspondence between image pixels and tissue regions. The data are collected and stored in digitized formats, which makes operations such as rotation, segmentation, quantification, and standardization straightforward. The accuracy (less distortion) and the time efficiency (the entire 3D image reconstruction requires only hours) also allow investigations of anatomical variation among individuals and the creation of a probabilistic atlas. MRI-based atlases, however, have a limited resolution (up to 20 ~ 30 μm) and a limited range of image contrasts, meaning many brain structures, such as various nuclei in the thalamus, are difficult to delineate. Nonetheless, MRI-based atlases carry valuable information that is complementary to histology-based atlases and could be an important tool for future brain research.
Several MRI-based atlases have been established (Aggarwal et al., 2009; Badea et al., 2007; Dorr et al., 2008; Jacobs et al., 1999; Ma et al., 2005; MacKenzie-Graham et al., 2004; Petiet et al., 2008). Most recently, Chan et al. introduced a combined MRI-CT 3D atlas of the adult mouse brain (Chan et al., 2007), demonstrating exciting opportunities for three-dimensional navigation of brain anatomy. Our group recently also published a stereotaxic mouse brain atlas that combines CT and diffusion tensor imaging (DTI) data (Aggarwal et al., 2009). Promising applications for MRI-based atlases, such as registration of histology-based information (e.g., gene expression maps), have also been demonstrated (MacKenzie-Graham et al., 2004).
The atlas described here is an extension of these previous studies with several important advancements. It was created from a database of 3D high resolution T2-weighted and diffusion tensor images of the developing C57BL/6 mouse brains, beginning at embryonic day 12 (E12). The inclusion of DTI data, of which the contrasts mainly reflect tissue microstructural organization (Basser and Pierpaoli, 1996; Le Bihan, 2003; Mori and Zhang, 2006), are key for delineation of various white matter structures and certain gray matter structures in the developing mouse brains (Mori et al., 2001; Zhang et al., 2003). Based on this database, major gray and white matter structures were identified, labeled, and segmented in MR images based on existing histology-based atlases. Quantitative characterization of normal brain development, including the sizes of the whole brain, neocortex, cerebellum, hippocampus, and several white matter tracts, was performed. Finally, we characterized anatomical variability at several developmental stages. The atlas and database will be made publicly available at http://lbam.med.jhmi.edu to facilitate research using this 4D mouse brain MRI database.
Methods and Materials
Experimental animals
All experiments and procedures were approved by the Animal Research Committee of the Johns Hopkins University School of Medicine and the University of Maryland School Of Medicine. C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME, USA) at various developmental stages (Table 1) were perfusion-fixed with 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer saline (PBS) either by immersion fixation for embryonic and neonatal samples, or by trans-cardiac perfusion fixation for postnatal and adult (P60 and older) mice. Mouse heads were removed and immersed in 4% PFA for more than 48 hours. To preserve the shape of the brains during imaging, the brains were left inside the skull. Before imaging, the samples were placed in PB with 0.1 mM gadopentetate dimeglumine (Magnevist, Berlex Imaging, Wayne, NJ, USA) for more than 48 hours. The addition of gadolinium enhanced MR signals and reduced imaging time (Johnson et al., 2002). We then transferred the samples into custom-built, MR-compatible tubes. The tubes were filled with fomblin (Fomblin Profludropolyether, Ausimont, Thorofare, New Jersey, USA), a non-hydrogen containing liquid for susceptibility matching and to prevent dehydration.
Table 1.
Number of animals and imaging resolution for data included in the developing mouse brain atlas.
| Age | Number of animals | Imaging Resolution (mm3) |
|---|---|---|
| E12 | 2 | 0.080 × 0.080 × 0.080 |
| E13 | 2 | 0.080 × 0.080 × 0.080 |
| E14 | 2 | 0.080 × 0.080 × 0.080 |
| E15–E16 | 5 | 0.085 × 0.085 × 0.085 |
| E17–E18 | 7 | 0.090 × 0.090 × 0.090 |
| P0 | 4 | 0.090 × 0.090 × 0.090 |
| P1–P2 | 5 | 0.090 × 0.090 × 0.090 |
| P4–P5 | 4 | 0.10 × 0.10 × 0.10 |
| P6–P7 | 7 | 0.10 × 0.10 × 0.10 |
| P10 | 6 | 0.10 × 0.10 × 0.10 |
| P14–P15 | 6 | 0.12 × 0.12 ×0.12 |
| P21 | 8 | 0.125 × 0.125 × 0.125 |
| P28–P30 | 8 | 0.125 × 0.125 × 0.125 |
| P42 | 8 | 0.125 × 0.125 × 0.125 |
| P60 | 10 | 0.125 × 0.125 × 0.125 |
| P80 and older | 10 | 0.125 × 0.125 × 0.125 |
Data Acquisition
Ex vivo imaging was performed on an 11.7 Tesla NMR spectrometer (Bruker Biospin, Billerica, MA, USA). Bird-cage RF coils with diameters of 10 ~ 20 mm were used for transmission and reception. A 3D diffusion-weighted multiple spin echo sequence was used to acquire images with four spin echoes (six spin echoes for embryonic and neonatal brains), a repetition time (TR) of 0.7s, an echo time (TE) of 35 ms, and two signal averages. The field of view ranged from 9 mm × 6 mm × 6 mm for the smallest sample (E12) to 20 mm × 14 mm × 10 mm for the adult samples. The native imaging resolution ranged from 0.08 mm × 0.08 mm × 0.08 mm for embryonic samples to 0.125 mm × 0.125 mm × 0.125 mm for adult samples. Images reconstructed from each echo were added together to form one image to enhance the signal-to-noise-ratio (SNR). For each sample, eight diffusion-weighted images were acquired with a diffusion gradient duration (δ) of 6 ms and a separation between the pair of diffusion gradients (Δ) of 14 ms for postnatal and adult brains and δ = 5 ms / Δ = 11 ms for embryonic and neonatal brains (P7 and younger). Two images were acquired with a minimum b value (50 s/mm2) and six images with a maximum b value (1,000~1,200 s/mm2 for postnatal and adult samples and 700 ~ 800 s/mm2 for embryonic and neonatal samples). Diffusion sensitizing gradients were applied along six different orientations: [0.707, 0.707, 0], [0.707, 0, 0.707], [0, 0.707, 0.707], [−0.707, 0.707, 0], [0.707, 0, −0.707], and [0, −0.707, 0.707]. The total imaging time was approximately 24 hours. Co-registered 3D T2-weighted images were acquired with the same field of view and resolution using a fast spin echo sequence, with a TE/TR of 50/900ms, a flip angle of 40 degrees, four signal averages, an echo train length of four (the first echo was located at the center of the k-space), and an imaging time of less than two hours.
Image processing
The ex vivo 3D T2-weighted and diffusion-weighted images were reconstructed on an off-line PC workstation using IDL (ITT Visual Information Solutions, Boulder, CO, USA). Diffusion tensor fitting was performed using DtiStudio (Jiang et al., 2006). The six elements of diffusion tensor were determined by log-linear fitting. The tensor was diagonalized to obtain three eigenvalues (λ1–3) and corresponding eigenvectors (ν1–3). Fractional anisotropy (FA) (Basser and Pierpaoli, 1996) was calculated.
Direction-encoded color map (DEC) images were generated by combining the images of the primary eigenvector (V1) and FA into RGB images (V1·FA). In DEC images, the ratio among the G(reen), R(ed), and B(lue) components of each pixel was defined by the ratio of the absolute values of the x, y, and z components of the primary eigenvector, and the intensity was proportional to the FA. In our atlas, red was assigned to the medial-lateral axis, green to the rostral-caudal axis, and blue to the dorsal-ventral axis. Average diffusion-weighted (aDW) images are the average of all diffusion-weighted images from each specimen. Signals from skull tissues in these images were manually removed and the total brain volume of each specimen was measured. At each developmental stage, a representative brain with the median brain volume was selected as the template and manually re-oriented to the standard orientations as defined in two histology-based atlases ((Foster, 1998) for embryonic brains and (Paxinos and Franklin, 2003) for postnatal and adult brains). In the re-oriented images, the left-right and rostral-caudal axes of the brain were along the x and y axes, respectively. For postnatal and adult brains, the dorsal, flat part of the cortex was perpendicular to the z axis. For embryonic brains, the line that connected the bases of the hypothalamus and brainstem in the mid-sagittal plane was perpendicular to the z axis. The aDW images from other specimens at the same developmental stage were rigidly aligned to the aDW image of template using the six-parameter rigid registration function in the automated image registration (AIR) package (Woods et al., 1998a; Woods et al., 1998b). The derived rigid transformation was then applied to the T2-weighted and tensor data. The orientation of each tensor was aligned using the methods described by Xu et al. (Xu et al., 2003).
Segmentation, volume, and cross-sectional area measurements and fiber tract reconstruction
Major brain compartments, e.g., neocortex, hippocampus, and cerebellum, were manually segmented in the rigidly aligned MR data using Amira (Visage Imaging, Inc. San Diego, CA, USA), and their volumes were calculated based on the segmentation results. We followed the definition of compartmental boundaries in the Paxinos' atlases (Paxinos and Franklin, 2003; Paxinos et al., 2006). The cross-sectional area of major white matter tracts was measured at selected locations along each white matter tract in the DEC images. Using DtiStudio, we first located the tract in the DEC image, then rotated the image so that the viewing plane was orthogonal to the tract, and finally defined the area by tracing the boundary of the tract in the rotated DEC image. This operation was repeated three times for each tract, and the average area was saved as the cross-sectional area of the tract. Three-dimensional tract reconstruction was performed using the FACT method (DtiStudio) (Mori et al., 1999; Xue et al., 1999) with an FA threshold of 0.2 and fiber angles less than 40° between two connected pixels. A multi-ROI approach was used to reconstruct tracts of interest (Conturo et al., 1999; Huang et al., 2004).
Quantification of anatomical structures during development
Changes in the volumes of major brain compartments and cross-sectional areas of major white matter tracts during development were fitted to a sigmoidal model, originally proposed by Wingert (Wingert, 1969).
The sigmoidal model characterizes the growth of brain structures using three parameters: a, x0, and b. The first parameter, a, represents the estimated volume or cross-sectional area of a structure in the adult mouse brain. The second parameter, x0, is the time to reach half the estimated adult volume (a). The third parameter, b, describes the growth curve; the smaller the b value, the faster the growth. The variable x is the gestational age (days since conception). Volume data was fitted to the above sigmoidal model using nonlinear regression (SigmaPlot 10.0, Systat Software, San Jose, CA, USA) with a threshold for significance of p < 0.0001.
Generation of population averaged brains and individual variation maps
Ex vivo images acquired from mouse brains at 7 postnatal stages (P7, P11, P15, P21, P28, P42, and P60, n=6 at each stage) were used to generate population averaged mouse brain images at each stage. In order to minimize differences in aDW image intensity caused by different receiver gain values used during acquisition, the intensity values of the rigidly aligned aDW images from each specimen were adjusted using a multiplicative scaling factor to the representative brain with median brain volume at each stage. The multiplicative scaling factor was obtained by comparing the intensity values of several gray matter structures (the cortex, thalamus, and striatum) and white matter structures (the genu and splenium of the corpus callosum, anterior commissure, and cerebral peduncle). The intensity values of the aDW images were then scaled uniformly to a 0 to 255 range, while the intensity values in the FA images, which range from 0 to 1, were multiplied by 255. For generation of population averaged mouse brain images, the iterative procedure previously described in (Kovacevic et al., 2005) was adopted. Image registration was performed using dual contrast (aDW + FA) Large Deformation Diffeomorphic Metric Mapping (LDDMM) with equal weighting for aDW and FA signals, which can accurately register images from two subjects based on the tissue contrasts in the aDW and FA images (Ceritoglu et al., 2009). The transformation of the tensor field was carried out by the method described in (Xu et al., 2003). The results included population averaged T2-weighted, diffusion weighted, and diffusion tensor data and the deformation maps between individual mouse brain images and the population averaged images, from which the covariance tensor at each pixel, x0, in the population averaged images was computed as
where Dcov is the covariance tensor, n is the number of subjects, xk is the coordinates in the image of subject k that corresponds to pixel x0 in the population averaged image according to the deformation map, and is the average of all xk, k = 1, 2, …, n. The trace value of the covariance tensor characterizes the magnitude of anatomical variation at each pixel.
Results
The MRI / DTI-based atlas of the developing mouse brain
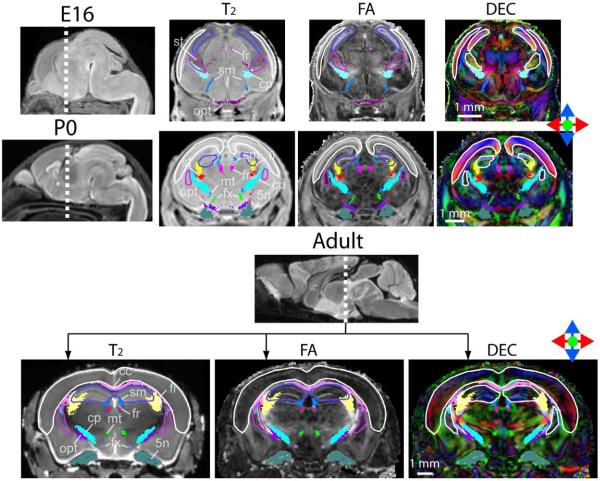
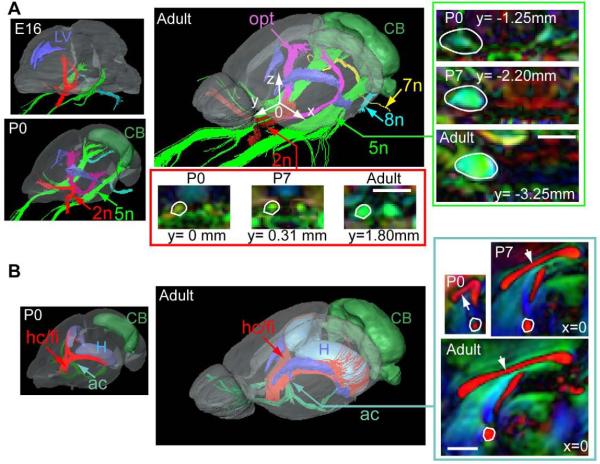
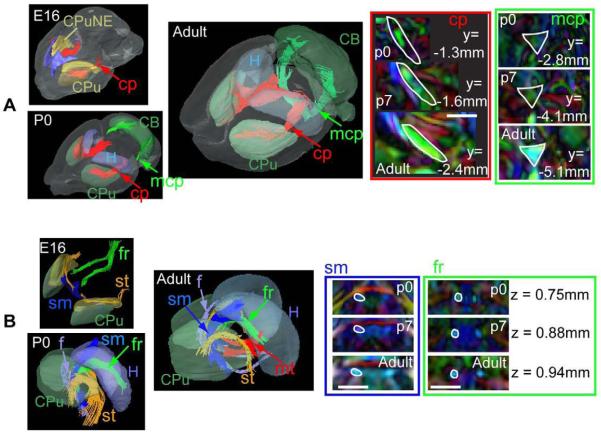
Figure 1 shows coronal T2-weighted, FA, and direction-encoded color-map (DEC) images of an E16, a P0 (postnatal day 0), and an adult (P60) mouse brains, overlaid with segmented gray and white matter structures. The atlas focuses on the major white matter tracts in the mouse brains at embryonic, postnatal and adult stages. Figure 2 and figure 3 show three-dimensional views of the segmented structures in the E16, P0, and adult mouse brains. The spatial relationships among various neural structures and anatomical changes during development are clearly demonstrated. White matter structures, with their intermediate to high fractional anisotropy (FA) and unique orientations (as distinctive colors in the DEC image) dictated by the trajectory of each tract, can be identified in the embryonic and neonatal mouse brains. For example, the trigeminal nerve (5n), vestibulococlear nerve (8n), optic nerve (2n), and optic tract are already visible in the images of an E16 mouse brain (Fig. 2A). The three major commissural structures, the anterior commissure, the hippocampal commissure, and the corpus callosum, are barely perceptible in our MR data at E16 (not shown), but become visible by P0 in the mid-sagittal plane (Fig. 2B). In the thalamic region, the cerebral peduncles, the stria terminalis, and the fasciculus retroflexus are visible at E16 (Fig. 3B). Tracts related to the cerebellum are difficult to detect in our MR data at E16. For example, the middle cerebellar peduncle first becomes visible at P0 (Fig. 3A). Examination of the cross-sectional areas of white matter tracts at comparable locations during development suggests that they have different temporal profiles of growth. For example, the cross-sectional area of the fasciculus retroflexus (fr inFig. 3B) only shows a small increase from P0 to adult, while the cerebral peduncle (cp in Fig. 3A), middle cerebellar peduncle (mcp in Fig. 3A), and corpus callosum (indicated by white arrows in Fig. 2B) show dramatic growth during the same period.
Figure 1.
Single subject coronal images of the E16, P0, and adult mouse brains from the mouse brain database. At each stage, the brain specimen with the median total brain volume was shown. The images are overlaid with reconstructed white and gray matter structures currently included in the atlas. The color schemes for gray matter structures in the T2 and FA images are: white: neocortex (cortical plates in the E16 mouse brain); blue: hippocampus (hippocampal neuroepithelium in the E16 mouse brain); purple: striatum. The color schemes for White matter structures are: yellow: fimbria (fi); pink: corpus callosum (cc); magenta: fasciculus retroflexus (fr); red: stria terminalis (st); light green: fornix (fx); dark green: the trigeminal nerve (5n); light blue: cerebral peduncle (cp); dark blue: stria medularis (sm); light purple: mammillothalamic tract (mt); purple: optic tract (opt).
Figure 2.
Development of the cranial nerves and commissural tracts in the mouse brain. At each stage,single subject images from the brain specimen with the median total brain volume was shown. A) The trigeminal nerve (5n, green), facial nerve (7n, yellow), vestibulocochlear nerve (8n, light blue), optic nerve (red), and optic tracts (purple), together with the lateral ventricles (LV, blue) and cerebellum (CB, green). To facilitate comparison between different specimens, we define the mid-sagittal anterior commissure as the origin, with the X, Y, and Z axes along the left-right, anterior-posterior, superior-inferior axes as shown by the white arrows in the 3D rendering of the adult mouse brain. Time-related changes in cross-sectional areas of the 5th nerves and optic nerves are examined in coronal sections (inside the green and red boxes, respectively). The locations of the coronal sections along the Y axis (anterior-posterior) are marked under each image, and also indicated by the green and red arrows in the 3D images. B) The anterior commissure (ac, green), hippocampal commissure and fimbria (hc/fi, red), with the lateral ventricles (blue), hippocampus (H, light blue), and cerebellum (CB, green) in P0 and adult mouse brains. The white arrows indicate the corpus callosum. Time-related changes in cross-sectional areas of the commissural fibers are examined at the mid-sagittal level. No embryonic data are shown in panel B because commissural and cerebellar tracts were not visible in MR images until late embryonic stages. The locations of the sagittal sections along the X axis (the left-right axis, x=0 for the mid-sagittal section) are indicated by the dark green arrows in the 3D images Scale bars = 1 mm.
Figure 3.
Development of the white matter tracts related to the cerebellum and thalamus. At each stage,single subject images from the brain specimen with the median total brain volume was shown. A) The middle cerebellar peduncle (mcp, green), the internal capsule and cerebral peduncle (cp, red), together with the caudate putamen (CPu), neuroepithelium of the caudate putamen (CPuNE, only at E16), hippocampus (H), and cerebellum (CB). Time-related changes in the cross-sectional areas of the cerebral peduncle and middle cerebellar peduncle are examined at selected coronal sections (inside the red and green boxes, respectively). The locations of the coronal sections along the Y axis (the anterior-posterior axis) are marked in each image, and also indicated by the red and green arrows in the 3D images. B) White matter tracts that pass through the thalamus, including the fasciculus retroflexus (fr), the stria medularis (sm, blue), the fornix (f, light blue), the stria terminalis (st, orange), and the mammalothalamic tract (mt, red), together with the caudate putamen (CPu), and the hippocampus (H). Time-related changes in the cross-sectional areas of the stria medularis (sm) and the fasciculus retroflexus (fr) are examined at selected horizontal sections, as indicated by the green and blue arrows in the 3D images. In the 2D images, boundaries of the white matter tracts in the adult brains are defined and overlaid on the images from P0 and P7 mouse brains. Scale bars = 1 mm.
Volumetric Development
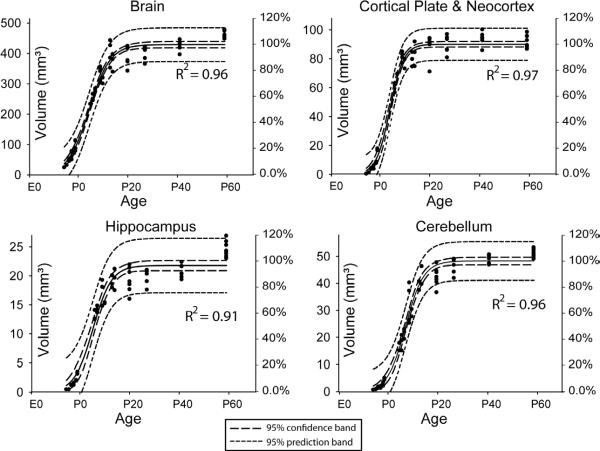
In Figure 4, the volume of the whole brain, neocortex, hippocampus, and cerebellum were examined from E14 (gestational day 14) to adult. We observed sigmoidal growth patterns in our MR-based volume measurements and the estimated parameters are summarized in Table 2. These measurements establish the normal growth curve of C57BL/6 mice and the normal range of individual variability measured by ex vivo MRI (via the 95% prediction bands). Close inspection of the growth of individual structures reveals delayed (greater x0) growth of the cerebellum, which coincides with the known delayed anatomical development of the cerebellum (Table 2).
Figure 4.
Volumes of the whole brain, neocortex, hippocampus, and cerebellum (left vertical axis) and their percentage of adult volumes (right vertical axis). The volume measurements were fitted to a sigmoidal model with 95% confidence band (long dash lines) and 95% prediction band (short dash lines). The right vertical axis shows the percentile of the estimated adult values (parameter a in Table 2).
Table 2.
Estimated parameters and standard errors of the sigmoidal model that characterize the volumetric development of the whole brain, neocortex, cerebellum, and hippocampus in the C57BL/6 mouse. The parameters for the C57BL/6 mice were estimated, based on MRI data. Notice that gestational age was used in this model, so that 0 day equals E0 in other places in the paper, 18.5 days equals P0, and 38.5 days equals P20, and so on.
| C57BL/6 (MRI) | Estimated parameters | ||
|---|---|---|---|
| Structures | a (mm3) | b (day) | x0 (gestational day) |
| whole brain | 429 ± 5.3 | 4.2 ± 0.2 | 24.0 ± 0.3 |
| neocortex | 90 ± 1.0 | 2.7 ± 0.2 | 23.4 ± 0.3 |
| cerebellum | 48 ± 0.7 | 3.6 ± 0.3 | 27.0 ± 0.3 |
| hippocampus | 22 ± 0.4 | 3.7 ± 0.4 | 24.7 ± 0.5 |
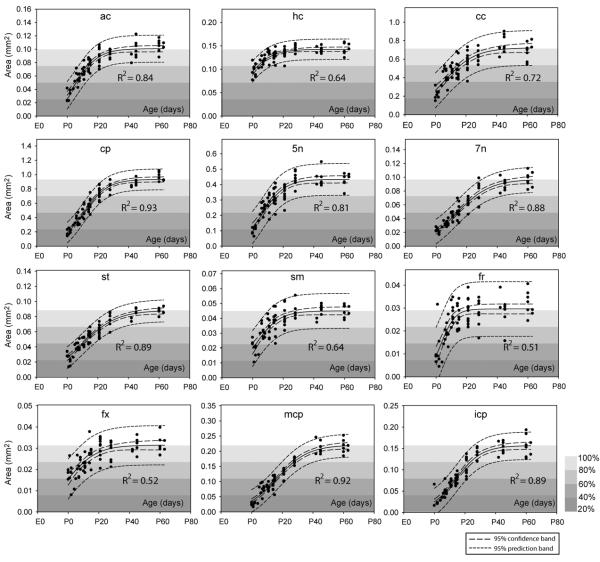
White Matter Fiber Tract Cross-Sectional Areas
Figure 5 shows the changes in cross-sectional areas from birth to adult in twelve white matter tracts. The measurements were fitted to the sigmoidal model, and the estimated parameters and standard errors for each tract are tabulated in Table 3. At birth (18.5 days after conception, P0), the cross-sectional areas of most tracts were less than 40% of their estimated adult values (a), except the hippocampal commissure, whose cross-sectional area at P0 was approximately 60% of its estimated adult value (a = 21.11 ± 0.36 mm2). The estimated x0s (time to reach 50% of the estimated adult values), based on the cross-sectional areas, suggested that these white matter tracts can be classified into three groups: an early group (x0 < 18.5 days (P0), before birth) that includes the hipppocampal commissure (hc); a middle group (x0 between 18.5 days (P0) and 26.5 days (P8), the first week after birth) that includes the anterior commissure (ac), corpus callosum (cp), fornix (fx), stria medularis (sm), fasciculus retroflexus (fr), and the trigeminal nerve (5n); and a late group (x0 larger than 26.5 days or P8) for the rest. In terms of the growth rate of the cross-sectional area, the fasciculus retroflexus showed the highest growth rate (the smallest b).
Figure 5.
Changes in white matter cross-sectional areas in twelve major white matter tracts. The area data were fitted to a sigmoidal model and the 95% confidence intervals are shown in the figures, with figure background indicative of the percentile of the estimated adult values (parameter a in Table 3). Structural abbreviations are: ac: anterior commissure; hc: hippocampal commissure; cc: corpus callosum; cp: cerebral peduncle; 5n: the trigeminal nerve; 7n: the facial nerve; st: stria terminalis; sm: stria medularis; fr: fasciculus retroflexus; fx: fornix; mcp: middle cerebellar peduncle; icp: inferior cerebellar peduncle.
Table 3.
Estimated parameters and standard erros of the sigmoidal model that characterize the postnatal development of white matter tract cross-sectional areas in the C57BL/6 mouse brains. Structural abbreviations are: ac: anterior commissure; hc: hippocampal commissure; cc: corpus callosum; cp: cerebral peduncle; 5n: the trigeminal nerve; 7n: the facial nerve; opt: the optic nerve; sm: stria medularis; fr: fasciculus retroflexus; fx: fornix; mcp: middle cerebellar peduncle; icp: inferior cerebellar peduncle; st: stria terminalis; fi: fimbria. All the estimated parameters have a p < 0.0001. Notice that gestational age was used in this model.
| Area | |||
|---|---|---|---|
| a (mm3) | b (day) | x0 (gestational day) | |
| ac | 0.100 ± 0.002 | 7 ± 0.8 | 25 ± 0.7 |
| hc | 0.143 ± 0.002 | 8 ± 1.5 | 13 ± 1.7 |
| cc | 0.72 ± 0.03 | 10 ± 1.6 | 24 ± 1.2 |
| cp | 0.93 ± 0.02 | 8 ± 0.6 | 29 ± 0.5 |
| 5n | 0.43 ± 0.01 | 7 ± 0.9 | 25 ± 0.8 |
| 7n | 0.097 ± 0.003 | 11 ± 1.0 | 32 ± 1.0 |
| st | 0.088 ± 0.002 | 11 ± 1.0 | 29 ± 0.8 |
| sm | 0.045 ± 0.001 | 8 ± 1.4 | 21 ± 1.1 |
| fr | 0.030 ± 0.001 | 4 ± 1.0 | 22 ± 1.0 |
| fx | 0.031 ± 0.001 | 9 ± 2.0 | 19 ± 1.7 |
| mcp | 0.220 ± 0.006 | 10 ± 0.9 | 35 ± 1.0 |
| icp | 0.157 ± 0.004 | 8 ± 0.8 | 33 ± 0.8 |
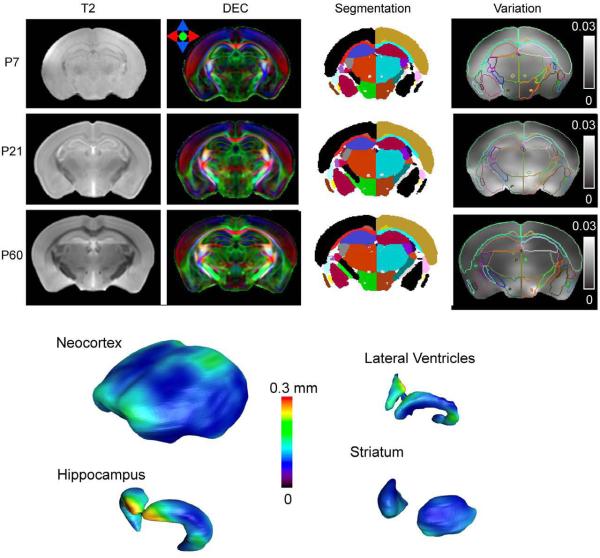
Population-Averaged Mouse Brain Atlas
Population averaged T2-weighted and Direction-encoded colormap (DEC) images of the mouse brain at several stages are shown in Figure 6 with segmentation of major gray and white matter structures. The average mouse brain images have high signal-to-noise ratio and sharply defined boundaries of major white matter tracts. Furthermore, with the transformations generated by LDDMM, the morphological variation in adult mouse brains was quantitatively characterized using a covariance tensor at each pixel. Figure 6 shows the amount of anatomical variation in the P7, P21, and P60 mouse brain with respect to the population averaged mouse brain images using the trace of the covariance tensor (Fig. 6, Variation). The distribution of anatomical variation in the brain is not uniform and the amount of variation varies at different developmental stages. Figure 6 also illustrates the amounts of variation on the surfaces of the cortex, hippocampus, lateral ventricles, and striatum in the adult mouse brain. The frontal and posterior dorsal regions of the cortex display variation at the level of 0.1 mm. The anterior part of the hippocampus, which is adjacent to the lateral ventricles, displayed a high degree of variation (~ 0.3 mm). The striatum has a relatively low degree of variation.
Figure 6.
Top: Population average T2, directional encoded colormap (DEC) images of the P7, P21, and P60 mouse brain and maps of segmentation and maps of anatomical variation overlaid with outlines of structural segmenations. The unit in the variation maps is mm2. Bottom: anatomical variations in the P60 mouse brain rendered on the surfaces of the neocortex, hippocampus, lateral ventricles, and striatum.
Discussion
Compared to existing histology or MR-based atlases, the atlas presented here is unique in its characterization of developing mouse brains using high resolution DTI technique. DTI has become a useful phenotyping tool in neuroscience research, especially at early developmental stages. While conventional T1 and T2 based MRI show poor tissue contrast in immature mouse brains, probably due to lack of myelin, DTI was able to provide satisfactory and relatively consistent tissue contrasts in the developing mouse brains. Ex vivo mouse brain DTI has been used to examine the white matter structures and their development in wild-type and mutant strains (Andrews et al., 2006; Tyszka et al., 2006; Verma et al., 2005; Wang et al., 2006; Zhang et al., 2005; Zhang et al., 2003). With these developments, this atlas will be a useful guide for locating anatomical structures in mouse DTI data, and our database may serve as a baseline for the study of abnormal mouse brain development.
The information in the atlas is presented in 2D and 3D formats, and the atlas is simple to use. The 2D atlas contains images from developing brains with structural labels, and can be used in the same ways as current histology-based atlases. We followed the structural nomenclature used in the two atlases as closely as possible. The 3D atlas provides visualization of the segmented white and gray matter structures of interest, thus allowing exploration of the 3D spatial relationships between these structures, e.g., the 3D trajectories of a particular white matter tract. By coregistering the atlas and MRI/DTI data to CT data for the external skull features, the 3D atlas may also be useful for surgical planning among other interventions (Aggarwal et al., 2009). It is necessary to note that even though the images in the atlas have been rotated to the orientation defined in the Paxinos atlas (Paxinos and Franklin, 2003) and Foster atlas (Foster, 1998) for postnatal and embryonic mouse brains, respectively, small discrepancies in slice orientation may still present because accurate matching of our 3D MRI based atlases to the 2D section based atlases is difficult.
To avoid tissue deformation or damage during dissection and imaging, the images were acquired with the brains preserved in the skull. The disadvantages associated with imaging brains within the skull are in-skull susceptibility artifacts near the ear drum, and reduced SNR and resolution due to the use of larger, less sensitive RF coils. Y. Ma et al. (Ma et al., 2005) reported significant differences in brain shapes between images obtained from brain specimens preserved inside and outside the skull, which was attributed to tissue relaxation and stretching after dissection. The volume measurements reported here are comparable with data from previous reports based on histology and/or MRI. A previous histology-based study on albino mice showed that the volumetric growth of the brain, neocortex, and cerebellum follow a sigmoidal model (Wingert, 1969), with relatively slow growth in the late embryonic period (E16 ~ E18), followed by a rapid increase during the neonatal and early postnatal period (P0 ~ P20), subsequently slowly approaching their adult values. The data showed that the development of different brain compartments is not uniform. For example, while the volumetric growth of the neocortex and hippocampus followed the volumetric growth of the whole brain closely, the growth of the cerebellum is slower than the growth of the whole brain (Fig. 4). The estimated x0 for the cerebellum is around P8 to P9, compared to an estimated x0s of P5 to P6 for the whole brain, neocortex, and hippocampus. The sigmoidal model, however, is a general model used here to characterize the brain growth throughout the embryonic and postnatal periods. For more detailed analysis on volume development, it is necessary to focus on a particular period with more sophisticated models (Uylings et al., 1990) than the model adopted here.
While volume changes are prevalent in various types of human brain diseases, e.g., Huntington's disease (Nopoulos et al., 2007; Rosas et al., 2004) and Alzheimer's disease (Csernansky et al., 2005; Thompson et al., 2003) brain morphology has rarely been used as a phenotypic marker in mouse disease models. With the increasing availability of small animal imaging instruments, this situation is rapidly changing. The normal growth curves of C57BL/6 with 95% prediction bands will be a useful reference for future morphological phenotype studies.
In this study, population-averaged mouse brain images at several postnatal stages were generated by normalizing individual mouse brain image to a template using linear (rigid) and non-linear (LDDMM) transformations. After the transformations, we could obtain information about anatomical variability among individual mice. Recently, several groups have reported measurements of anatomical variation in mouse brains based on segmentation of MRI data (Badea et al., 2007; Dorr et al., 2008; Kovacevic et al., 2005; Ma et al., 2005). In this study, anatomical variation in adult mouse brains was characterized without segmentation. Instead, we relied on LDDMM, driven by the rich anatomical information in DTI data, to generate measurements of anatomical variation for each pixel. Figure 6 demonstrates that certain brain regions have greater variation than other regions. In addition, if the atlases are used for stereotactic operations, the variability data provide a level of confidence for the delivery of cells, chemicals, or electrodes, depending on the experiment. This type of information is difficult to obtain from histology-based morphology studies.
In this study, the maturation process of various white matter tracts was also quantitatively characterized in terms of their sizes. DTI can monitor only the macroscopic sizes of axonal bundles, which by no means provides precise cellular information, such as axonal counts, axonal calibers, and myelination. However, macroscopic axonal degeneration or misguidance is prevalent in various types of mutants, which are sometimes difficult to comprehend from serial histology sections. We believe that our atlases and quantitative growth curves of each white matter tract provide anatomical information in 0.1 – 1 mm scales, that is complementary to histological information (which is typically on a μm scale) and will serve as important references for axonal tract development studies.
Currently, the main limitation of our atlas is a lack of precise gray matter assignment based on correlations with histology. Although it is rather straightforward to identify major white matter tracts based on their trajectory information provided by DTI, assignment of gray matter structures is not always straightforward. MR-histology correlation studies will be an important future effort to establish more comprehensive atlases. Other limitations of our atlas are mainly DTI related. First, DTI is inherently a technique with low-signal-to-noise ratio, and therefore, it is more difficult to achieve a high spatial resolution image and is more time-consuming than conventional MRI and other 3D imaging modalities. The resolution achieved in this atlas ranges from 80 μm to 125 μm isotropic resolution. Previous reports based on T1- and T2/T2*-weighted MRI were able to reach up to 20 ~ 30 μm resolution (Badea et al., 2007; Dorr et al., 2008; Ma et al., 2005), while episcopic and microCT can achieve even higher resolution (Johnson et al., 2006; Weninger and Mohun, 2002). Second, DTI is based on a simplified model of water diffusion. When the anatomical structures are complex, for example in regions where two or more populations of fibers cross each other, DTI oversimplifies the underlying anatomy. Contrasts for such problematic areas could be improved by more sophisticated models, such as diffusion spectrum imaging, Q-ball imaging, and HARD (Frank, 2001; Tuch et al., 2002; Tuch et al., 2003; Wedeen et al., 2000).
In summary, an MRI-based atlas and a database of the developing mouse brain have been established. The atlas and database will be useful for the quantitative examination of brain phenotypes and their development in wildtype and mutant mouse strains. The atlas and database can also be used to study abnormal anatomy and lesions in mouse models of neurological diseases, and as a dataset for the development of novel analysis tools. In the future, we will continue to collect high-resolution data to add to our current database and pursue correlations between information provided by histology and MRI.
Research Highlights.
Mouse brain development studied using MRI/DTI
MRI/DTI atlas and database of developing mouse brain
Characterization of anatomical changes in mouse brain development
Acknowledgments
This study was supported by National Institutes of Health AG20012, EB003543, NS059529, and ES012665. Support was also provided, in part, by P41 RR013642 and U24 RR021760 MouseBIRN to A. W. Toga and NIH R21 NS04584 to S. Mori, L. J. Richards, and Frank L. Margolis (University of Maryland).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal M, Zhang J, Miller MI, Sidman RL, Mori S. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience. 2009;162(4):1339–50. doi: 10.1016/j.neuroscience.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Embryonic development of the rat cerebellum. I. Delineation of the cerebellar primordium and early cell movements. J Comp Neurol. 1985;231:1–26. doi: 10.1002/cne.902310103. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the rat thalamus: I. Mosaic organization of the thalamic neuroepithelium. J Comp Neurol. 1988;275:346–377. doi: 10.1002/cne.902750304. [DOI] [PubMed] [Google Scholar]
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- Badea A, Ali-Sharief AA, Johnson GA. Morphometric analysis of the C57BL/6J mouse brain. Neuroimage. 2007;37(3):683–93. doi: 10.1016/j.neuroimage.2007.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-dfifusion-tensor MRI. J Magn Reson B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J. Neocortical Development. Raven Press; New York: 1991. [Google Scholar]
- Benveniste H, Kim K, Zhang L, Johnson GA. Magnetic resonance microscopy of the C57BL mouse brain. Neuroimage. 2000;11:601–611. doi: 10.1006/nimg.2000.0567. [DOI] [PubMed] [Google Scholar]
- Ceritoglu C, Oishi K, Li X, Chou MC, Younes L, Albert M, Lyketsos C, van Zijl PC, Miller MI, Mori S. Multi-contrast large deformation diffeomorphic metric mapping for diffusion tensor imaging. Neuroimage. 2009;47:618–627. doi: 10.1016/j.neuroimage.2009.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E, Kovacevic N, Ho SK, Henkelman RM, Henderson JT. Development of a high resolution three-dimensional surgical atlas of the murine head for strains 129S1/SvImJ and C57Bl/6J using magnetic resonance imaging and microcomputed tomography. Neuroscience. 2007;144:604–615. doi: 10.1016/j.neuroscience.2006.08.080. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Proc. Natl. Acad. Sci. USA. 1999;96:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC. Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005;25:783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Foster GA. Chemical Neuroanatomy of the Prenatal Rat Brain: A Developmental Atlas. 1st ed. Oxford University Press; Oxford: 1998. [Google Scholar]
- Frank LR. Anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2001;45:935–939. doi: 10.1002/mrm.1125. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang J, van Zijl PC, Mori S. Analysis of noise effects on DTI-based tractography using the brute-force and multi-ROI approach. Magn Reson Med. 2004;52:559–565. doi: 10.1002/mrm.20147. [DOI] [PubMed] [Google Scholar]
- Jacobs RE, Ahrens ET, Dickinson ME, Laidlaw D. Towards a microMRI atlas of mouse development. Comput Med Imaging Graph. 1999;23:15–24. doi: 10.1016/s0895-6111(98)00059-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed. 2006;81:106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Gewalt SL, Hedlund LW. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology. 2002;222:789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Johnson JT, Hansen MS, Wu I, Healy LJ, Johnson CR, Jones GM, Capecchi MR, Keller C. Virtual histology of transgenic mouse embryos for high-throughput phenotyping. PLoS Genet. 2006;2:e61. doi: 10.1371/journal.pgen.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic N, Henderson JT, Chan E, Lifshitz N, Bishop J, Evans AC, Henkelman RM, Chen XJ. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cereb Cortex. 2005;15:639–645. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hof PR, Grant SC, Blackband SJ, Bennett R, Slatest L, McGuigan MD, Benveniste H. A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience. 2005;135:1203–1215. doi: 10.1016/j.neuroscience.2005.07.014. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, Hu G, Jacobs RE, Toga AW. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J Anat. 2004;204:93–102. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PCM. Three dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annal. Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, Itoh R, Zhang J, Kaufmann WE, van Zijl PCM, Solaiyappan M, Yarowsky P. Diffusion tensor imaging of the developing mouse brain. Magn Reson Med. 2001;46:18–23. doi: 10.1002/mrm.1155. [DOI] [PubMed] [Google Scholar]
- Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Natt O, Watanabe T, Boretius S, Radulovic J, Frahm J, Michaelis T. High-resolution 3D MRI of mouse brain reveals small cerebral structures in vivo. J Neurosci Methods. 2002;120:203–209. doi: 10.1016/s0165-0270(02)00211-x. [DOI] [PubMed] [Google Scholar]
- Nopoulos P, Magnotta VA, Mikos A, Paulson H, Andreasen NC, Paulsen JS. Morphology of the cerebral cortex in preclinical Huntington's disease. Am J Psychiatry. 2007;164:1428–1434. doi: 10.1176/appi.ajp.2007.06081266. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2nd ed. Academic Press; Scan Diego: 2003. [Google Scholar]
- Paxinos G, Halliday GM, Watson C, Koutcherov Y, Wang H. Atlas of the Developing Mouse Brain at E17.5, P0 and P6. Academic Press; New York: 2006. [Google Scholar]
- Petiet AE, Kaufman MH, Goddeeris MM, Brandenburg J, Elmore SA, Johnson GA. High-resolution magnetic resonance histology of the embryonic and neonatal mouse: a 4D atlas and morphologic database. Proc Natl Acad Sci U S A. 2008;105:12331–12336. doi: 10.1073/pnas.0805747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas HD, Feigin AS, Hersch SM. Using advances in neuroimaging to detect, understand, and monitor disease progression in Huntington's disease. NeuroRx. 2004;1:263–272. doi: 10.1602/neurorx.1.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer's disease. J Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- Tyszka JM, Readhead C, Bearer EL, Pautler RG, Jacobs RE. Statistical diffusion tensor histology reveals regional dysmyelination effects in the shiverer mouse mutant. Neuroimage. 2006;29:1058–1065. doi: 10.1016/j.neuroimage.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uylings H, Van Eden C, Parnavelas J, Kalsbeek A. The prenatal and Postnatal Development of Rat Cerebral Cortex. In: Kolb B, Tees R, editors. The Cerebral Cortex of the Rat. MIT Press; Boston, MA: 1990. pp. 35–76. [Google Scholar]
- Verma R, Mori S, Shen D, Yarowsky P, Zhang J, Davatzikos C. Spatiotemporal maturation patterns of murine brain quantified by diffusion tensor MRI and deformation-based morphometry. Proc Natl Acad Sci U S A. 2005;102:6978–6983. doi: 10.1073/pnas.0407828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Mori S, Nathans J. Axonal growth and guidance defects in Frizzled3 knock-out mice: a comparison of diffusion tensor magnetic resonance imaging, neurofilament staining, and genetically directed cell labeling. J Neurosci. 2006;26:355–364. doi: 10.1523/JNEUROSCI.3221-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen V, Reese T, Tuch DS, Wieigel MR, Dou JG, Weisskoff R, Chessler D. Mapping fiber orientation spectra in cerebral white matter with Fourier-transform diffusion MRI. Annual meeting of the international society of Magnetic Resonance in Medicine (ISMRM); Denver, Colorodo, USA. 2000. p. 82. [Google Scholar]
- Weninger WJ, Mohun T. Phenotyping transgenic embryos: a rapid 3-D screening method based on episcopic fluorescence image capturing. Nat Genet. 2002;30:59–65. doi: 10.1038/ng785. [DOI] [PubMed] [Google Scholar]
- Wingert F. Biometrical analysis of growth functions of cerebral component parts and body weight. J Hirnforsch. 1969;11:133–197. [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998a;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998b;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- Xu D, Mori S, Shen D, van Zijl PC, Davatzikos C. Spatial normalization of diffusion tensor fields. Magn Reson Med. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- Xue R, van Zijl PCM, Crain BJ, Solaiyappan M, Mori S. In vivo three-dimensional reconstruction of rat brain axonal projections by diffusion tensor imaging. Magn. Reson. Med. 1999;42:1123–1127. doi: 10.1002/(sici)1522-2594(199912)42:6<1123::aid-mrm17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chen YB, Hardwick JM, Miller MI, Plachez C, Richards LJ, Yarowsky P, van Zijl P, Mori S. Magnetic resonance diffusion tensor microimaging reveals a role for Bcl-x in brain development and homeostasis. J Neurosci. 2005;25:1881–1888. doi: 10.1523/JNEUROSCI.4129-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Richards LJ, Yarowsky P, Huang H, van Zijl PC, Mori S. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20:1639–1648. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]