Abstract
Central to the pathogenesis of atypical hemolytic uremic syndrome (aHUS) is over-activation of the alternative pathway of complement. Following the initial discovery of mutations in the complement regulatory protein, factor H, mutations have been described in factor I, membrane cofactor protein and thrombomodulin, which also result in decreased complement regulation. Autoantibodies to factor H have also been reported to impair complement regulation in aHUS. More recently, gain of function mutations in the complement components C3 and Factor B have been seen. This review focuses on the genetic causes of aHUS, their functional consequences, and clinical effect.
Keywords: Hemolytic uremic syndrome, Transplantation, Complement, Factor H, Factor I, Membrane cofactor protein, Thrombomodulin, Thrombotic thrombocytopenic purpura
Introduction
Hemolytic uremic syndrome (HUS) is characterized by the triad of microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure. It has been classified as either diarrhoeal-associated (D+ve) or non-diarrhoeal/atypical (aHUS) [1].
Pathologically, glomerular capillary wall thickening is seen due to endothelial cell swelling and accumulation of material between the endothelial cell and the basement membrane. This narrowing of the vessel lumen, in addition to the platelet and fibrin thrombi, results in occlusion of the glomerular capillaries. Fibrinoid necrosis of the afferent arteriole associated with thrombosis may also be seen. Mesangiolysis occurs early in the disease process and is subsequently replaced by sclerotic changes. Early arterial changes are variable, ranging from only mild endothelial swelling to fibrinoid necrosis with occlusive thrombus formation. Later in the disease process there is mucoid intimal hyperplasia with narrowing of the vessel lumen. Immunofluorescence demonstrates deposition of fibrin or fibrinogen in the glomeruli and in the mesangium as well as within the vessel walls. Granular deposits of complement and immunoglobulins along the capillary loops of glomeruli are occasionally seen [2].
D+ve HUS accounts for the majority of cases and is usually caused by a preceding illness with vero cytotoxin-producing bacteria, most commonly E. coli O157:H7 [3]. aHUS is rare, accounting for ∼10% of HUS and has a poor prognosis with up to 50% of patients requiring ongoing dialysis [4]. aHUS occurs at any age and may be sporadic or familial. Guidelines on the diagnosis of aHUS have recently been published [5].
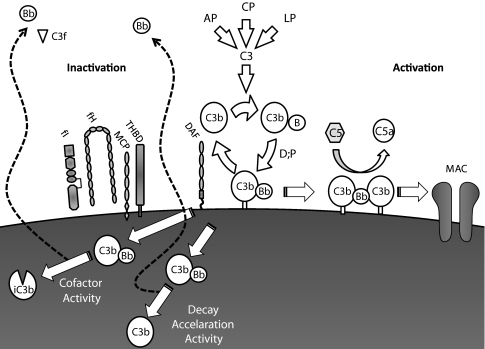
The past decade has revealed aHUS to be a disease characterized by over-activation of the alternative complement pathway (AP) (Fig. 1). Since the initial description of mutations in the gene (CFH) encoding the complement regulator factor H in aHUS, mutations in other complement proteins and the presence of autoantibodies against complement regulators have been demonstrated in many cohorts of aHUS patients. In this review, we will discuss the genetic and autoimmune factors which predispose to aHUS and their functional consequences.
Fig. 1.
Complement regulation. The alternative complement pathway (AP) is a positive-feedback amplification loop. The AP constantly undergoes tick over but can also be primed by the classical (CP) and lectin (LP) pathways. The C3b that is formed interacts with factor B (B), which is then cleaved by factor D (D) to form the AP C3 convertase (C3bBb). This enzyme complex is attached to the target covalently via C3b while Bb is the catalytic serine protease subunit. It is stabilized by binding properdin (P). Because C3 is the substrate for this convertase, a powerful feedback loop is created. Unchecked, this will lead to activation of the terminal complement pathway with generation of the effector molecules; the anaphylatoxin C5a and the membrane attack complex (MAC). To limit damage to host cells, the AP is down-regulated by cofactor activity (CA) and decay accelerating activity (DAA). CA results in the permanent inactivation of C3b to iC3b, such that it is no longer capable of binding B and thus cannot form the AP convertase. CA requires both a cofactor protein (factor H [fH] or membrane cofactor protein [MCP]) and a protease, factor I (fI). DAA is the dissociation of the C3/C5 convertases. The decay accelerator protein, in this example decay accelerating factor (DAF), displaces the catalytic Bb from target-bound C3b. However, this C3b can bind another B and then reform the convertase
Role of complement factor H
Factor H is an abundant serum glycoprotein (∼500 µg/ml) which is composed of 20 complement control protein modules (CCPs) [6]. It is the most important fluid-phase regulator of the AP. It acts as a cofactor for factor I-mediated proteolytic inactivation of C3b, competes with factor B for C3b binding, and accelerates the decay of the C3 convertase into its components [7]. These functions are mediated through the N-terminal four CCPs (CCP 1–4) [8]. Factor H also regulates complement on host surfaces by binding to the glycosaminoglycans of endothelial cells and exposed basement membranes via its carboxyl-terminal domain (CCPs 19–20) [9].
Mutations in CFH associated with aHUS were first demonstrated by Warwicker et al. [10] and have been demonstrated to be the commonest genetic abnormality found in many cohorts of aHUS patients [11–16]. The majority of mutations are heterozygous and are located in the exons encoding the C-terminal domain of the protein which is responsible for mediating complement protection on the cell surface (Fig. 2). These mutations do not usually result in a quantitative deficiency of factor H.
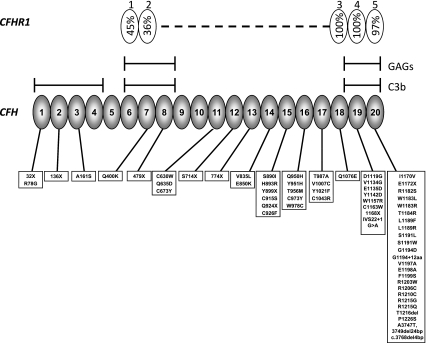
Fig. 2.
Complement factor H and complement factor H-related 1. The figure demonstrates the 20 CCP modules of factor H (bottom) and the five CCP modules of factor H-related protein 1 (top). The homology of each factor H-related protein 1 CCP to the corresponding factor H CCP is given as a percentage in the factor H-related protein 1 figure. Note the homology between CCP18 of factor H and CCP 3 of factor H-related protein 1 is given for the basic isoform. An acidic isoform differs by three amino acids [62]. The glycosaminoglycan and C3b binding sites of factor H are indicated on the diagram. Mutations in CFH reported in aHUS are listed below the figure
CFH lies in the RCA gene cluster at chromosome 1q32 and is in close proximity to the genes (CFHR1–5) encoding the five factor H-related proteins. The CFHRs show a very high degree of sequence identity to CFH (Fig. 2) and are thought to have arisen from several large genomic duplications. This homology predisposes to both gene conversion and genomic rearrangements through non-allelic homologous recombination (NAHR). Heinen et al. [17] showed that the aHUS-associated factor H mutations S1191L, V1197A, and combined S1191L/V1197A had arisen through gene conversion between CFHR1 and CFH. Venables et al. showed that a hybrid (fusion) gene comprising the 21 N-terminal exons of CFH and the 2 C terminal exons of CFHR1 has arisen through NAHR and is associated with aHUS [18].
The functional consequences of aHUS-associated factor H mutations have been studied, particularly those which cluster in the C-terminal domain of the protein. Structural analysis has shown that all such mutants are folded and have only very localized structural perturbations [19]. Functional analysis has shown varied consequences on the binding to heparin, C3b, and endothelial cells (Table 1). However, all aHUS-associated factor H mutants show impaired complement regulation at the cell surface using erythrocyte lysis assays [19–21]. Thus it is hypothesized that these C-terminal mutants fail to control complement activation at the glomerular endothelium particularly where basement membrane is exposed by the fenestrated endothelium. Renal biopsy data from an aHUS patient with a C-terminal mutant showed reduced factor H binding to renal endothelium compared to wild-type, in keeping with this hypothesis [22]. Additionally, it has been demonstrated that aHUS-associated C-terminal factor H mutants have reduced ability to bind to platelets, resulting in complement activation on the surface of platelets. This in turn causes platelet activation with aggregation and release of tissue-factor-expressing micro-particles [23]. Thus complement activation on the glomerular vasculature and on platelets is thought to result in the pro-coagulant phenotype which leads to aHUS.
Table 1.
Structural and functional consequences of mutations in aHUS
| Mutant | CCP | Structural changes | C3b/d binding | Heparin binding | Endothelial cell binding | Hemolysis assay | Reference |
|---|---|---|---|---|---|---|---|
| D1119G | 19 | Local | ↓e | ↔ | ↔a | ↓c | [19, 86] |
| Y1142C | 19 | ND | ND | ND | ND | ↓d | [21] |
| W1157R | 19 | ND | ↓ | ↓ | ND | ND | [87] |
| E1172X | 20 | ND | ↓ | ↓ | ND | ND | [88, 89] |
| R1182S | 20 | Local | ↓ | ↓ | ND | ↓c,d | [19, 20] |
| W1183R | 20 | Local | ↑ | ↑ | ND | ↓c | [19] |
| W1183L | 20 | Local | ↓ | ↓e | ↓a,b,e | ↓c | [19, 86, 87, 90] |
| T1184R | 20 | Local | ↑e | ↑ | ↑a | ↓c | [19, 86] |
| L1189R | 20 | Local | ↑ | ↑ | ↑a | ↓c,d | [19, 20, 86] |
| L1189F | 20 | Local | ↑ | ↑ | ND | ↓c,d | [19, 20] |
| S1191W | 20 | ND | ND | ND | ND | ↓d | [20] |
| S1191L | 20 | Local | ↑e | ↔ | ND | ↓c,d | [17, 19] |
| S1191L/V1197A | 20 | Local | ↑e | ↔ | ND | ↓c,d | [17, 19] |
| V1197A | 20 | ND | ↓e | ↓e | ND | ↓d | [17, 20, 87, 90] |
| E1198K | 20 | ND | ND | ND | ↓b | ↓d | [22] |
| R1210C | 20 | Local | ↓ | ↓e | ↓b | ↓c | [19, 20, 87, 89, 90] |
| R1215G | 20 | Local | ↓ | ↓ | ↓b | ↓c | [19, 87, 89] |
| R1215Q | 20 | ND | ↔ | ↓ | ↔a | ND | [86] |
| P1226S | 20 | ND | ↓ | ↓ | ND | ND | [87] |
ND not done; Endothelial cell binding relates to either amGEnC-1 [86] or bHUVEC [22, 88, 89] binding. Hemolysis assay using crecombinant proteins to compete with full-length CFH on human erythrocytes [19] or dusing patient serum on sheep erythrocytes [20]. To amalgamate these different hemolysis assays, the arrows indicate the effect on cell surface complement regulation produced by the mutation. eIndicates contradictory results
As most CFH mutations associated with aHUS are heterozygous, it has been postulated that these mutations may exert a dominant negative effect [24]. Recent studies have suggested that factor H may exist in monomer-dimer equilibrium, but that up to 95% will be monomeric in isolation in serum [25]. However, oligomerization of factor H via glycosaminoglycans [26] or C3d [27] on cell surfaces has been described. The extent to which oligomerization of mutant factor H with wild-type may interfere with the complement regulatory function has yet to be established.
The CFH knockout mouse (Cfh-/-) and a transgenic mouse lacking the C-terminal region of factor H (Cfh-/-Δ16–20) have proved illuminating [28]. The Cfh-/- mouse has very low C3 levels due to uncontrolled turnover of the alternative pathway and has a renal phenotype similar to membranoproliferative glomerulonephritis (MPGN) [29]. This is similar to the phenotype of the factor H-deficient Norwegian–Yorkshire pig [30]. In contrast, the Cfh-/-Δ16–20 mouse has higher plasma C3 levels than the Cfh-/- mouse, and spontaneously develops aHUS, not MPGN [31]. Thus, this mouse model provides the first in vivo evidence that the CFH mutations seen in aHUS impair endothelial cell surface recognition, resulting in local complement dysregulation, while controlling the alternative pathway in plasma. Goicoechea et al. [32] have also crossed the Cfh-/-Δ16–20 with a C5-deficient mouse to investigate the role of C5 activation in the pathogenesis of aHUS. These C5-/-CFH-/-Δ16–20 mice do not develop aHUS, suggesting a critical role downstream of C3b generation in aHUS.
Mutations in membrane cofactor protein (MCP: CD46)
Membrane cofactor protein (MCP: CD46) is a membrane glycoprotein present on the surface of all cells with the exception of erythrocytes [33]. It consists of an extracellular segment comprising 4 CCPs, an alternatively spliced STP region and a group of 12 amino acids of unknown function. This is followed by a transmembrane domain and an alternatively spliced cytoplasmic tail [33] (Fig. 3). As with CFH and the CFHRs, the gene (MCP, CD46) encoding MCP is located in the RCA cluster. MCP acts as a cofactor for factor I in the proteolytic inactivation of C3b and C4b bound to host cells.
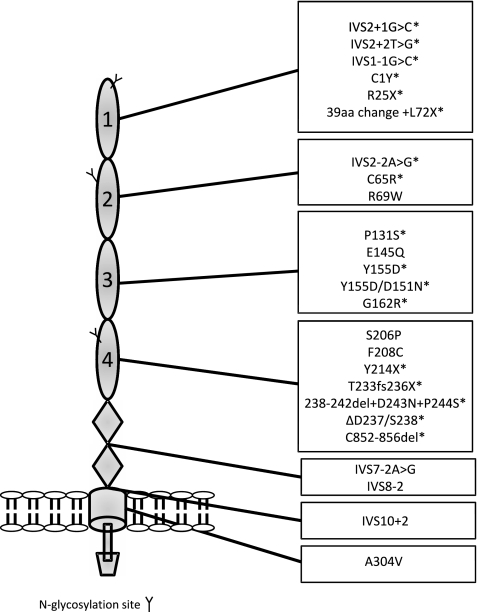
Fig. 3.
Membrane cofactor protein. The figure demonstrates clustering of mutations in the four extracellular CCP domains of membrane cofactor protein in aHUS. * denotes mutations resulting in decreased cell surface expression of membrane cofactor protein
Mutations in MCP have been demonstrated in up to 15% of cohorts of aHUS patients [13, 34–37]. The majority of MCP mutations in aHUS are heterozygous (∼75%), although homozygous or compound heterozygous mutations are reported [33]. Most MCP mutations cluster in the four extracellular complement control protein domains of MCP (Fig. 3), the region critical of complement regulation [33].
Around 75% of MCP mutations result in decreased cell surface expression and a quantitative deficiency in complement regulation [33]. Functional analysis of the remaining mutations has demonstrated a qualitative defect in complement regulation. Ligand binding and cofactor assays of two mutations with normal expression (S206P; F208C) demonstrated reduced C3b binding and cofactor activity [13, 33, 34]. These assays of complement regulatory activity failed to reveal any defect in two further normally expressed variants (R69W; A304V), however further analysis of function using cell surface assays demonstrated deficient control of the alternative pathway [38]. Only one mutation (E145Q) so far described has affected only C4b cofactor activity and the surface expression of this mutant was increased [33, 35].
Complement factor I
Factor I is a highly specific serine protease that acts through its proteolytic activity to control complement activation. It consists of a catalytic light-chain disulfide bonded to a heavy chain of unknown function. Factor I cleaves the α chains of C3b and C4b in the presence of its cofactor proteins: factor H for C3b [39, 40]; C4-binding protein (C4BP) for C4b [41, 42]; and MCP [43] and complement receptor 1 (CR1; CD35) [44, 45] for both. By inactivating C3b and C4b through limited proteolytic cleavage, factor I prevents the formation of the C3 and C5 convertases and thus down-regulates the AP and classical pathway (CP). Factor I is a serum glycoprotein predominantly synthesized by the liver. Unlike many other complement regulatory genes CFI does not reside in the RCA cluster at 1q32 but is located at chromosome 4q25.
CFI mutations have been reported in 2–12% of aHUS patients [13, 46–50]. The mutations are seen throughout the molecule but are most commonly seen the serine protease domain (Fig. 4).
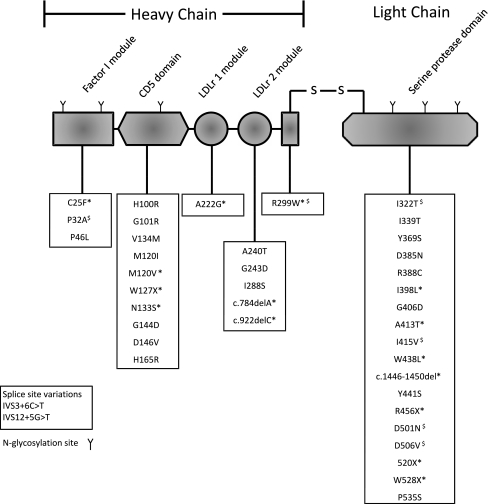
Fig. 4.
Complement factor I. The figure demonstrates the modular structure of factor I with aHUS-associated mutations below. $ denotes factor I mutants that are secreted but have been demonstrated to have decreased activity. * denotes factor I mutants that have impaired secretion
In vitro analysis of aHUS-associated mutations has demonstrated that the majority of mutations in CFI result in a quantitative defect. Analysis has shown that even in those mutations which have been shown to decrease secretion, the serum factor I level can be within the normal range [47]. Factor I is an acute-phase protein and this is probably responsible for the large variation seen in factor I levels in normal individuals (39–100 µg/ml) [51]. A minority of mutations in CFI result in a qualitative defect in complement regulation. Functional analysis of these mutations demonstrated a loss of alternative and classical cofactor activity, both in the fluid phase and on cell surfaces [50, 52]. However, analysis of some CFI variants, using recombinant proteins, has failed to demonstrate a functional consequence (e.g. G243D [53]). It is possible that they are not involved in the pathogenesis of disease or alternatively, the defect may be too subtle to be detected by currently available assays. This emphasizes the difficulty in attributing disease causality to mis-sense mutations.
Thrombomodulin
Thrombomodulin (THBD) is a key component of the protein C anticoagulation pathway and facilitates the activation of protein C by thrombin [54]. Additionally, it enhances thrombin-mediated activation of plasma procarboxypeptidase B (TAFI) an inhibitor of fibrinolysis that also inactivates the complement-derived anaphylatoxins C3a and C5a. THBD has recently been shown to down-regulate the AP of complement by accelerating factor I-mediated inactivation of C3b in the presence of co-factors [55]. Mutations in the gene (THBD) encoding thrombomodulin have recently been shown to predispose to atypical HUS. These mutations resulted in a loss of cofactor activity [55].
Factor B (CFB) mutations
Mutations have also recently been described in factor B (CFB) although they are rare in the cohorts so far described (0–3%) [56–58]. Factor B carries the catalytic site of the complement AP convertase (C3bBb) and as such, unlike previously described mutants associated with aHUS, acts as an activator of complement, not a regulator. Mutations in CFB associated with aHUS, are therefore activating mutations. Goicoechea de Jorge et al. [56] elegantly described two mechanisms through which these separate mutations led to increased complement activation. One mutant (F286L) showed enhanced formation of the C3bB proenzyme, which will result in a more active enzyme in vivo. The other mutant (K323E) formed a C3bBb enzyme more resistant to decay by the complement regulators decay accelerating factor (DAF; CD55) and factor H. This also caused increased enzyme activity [56]. The two mutations described in the French cohort were also located in the von-Willebrand type A domain, suggesting that this is a hotspot for mutations in aHUS. These mutations were both demonstrated to be more active than the wild-type protein, resulting in increased complement deposition on human glomerular endothelial cells [58].
C3 mutations
C3 is the central component of the complement cascade, critical to activation by the classical, lectin, and alternative pathways. C3 is cleaved to form the anaphylatoxin C3a and C3b, which is highly reactive and can bind to cell surfaces via its reactive thioester. C3b can then interact with factor B in the presence of factor D to form the alternative pathway convertase, thus introducing a positive-feedback amplification loop.
Recently mutations in C3 have been described in aHUS patients [59, 60]. Functional analysis of five of the nine mutations so far described has revealed decreased binding to MCP with a consequent decrease in the ability of MCP to act as a cofactor to inactivate C3b [60]. Thus, in vivo these will act as gain of function mutations. In two of the mutations, no functional impairment could be demonstrated and in another two, the mutations resulted in a decreased secretion of C3. How impaired secretion of C3 fits into the current model of complement over-activation in aHUS is yet to be resolved.
Autoantibodies to factor H/factor H-related 1
In addition to the genetic abnormalities described in aHUS, autoantibodies to factor H have also been linked to disease in 5–10% of aHUS patients [61–66]. Available data would suggest that the onset of disease or disease recurrence correlates with the presence of factor H autoantibodies. The titre of these antibodies may also spontaneously decline with time.
Jozsi et al. demonstrated using recombinant fragments of factor H that the autoantibodies in five patients bound to the C-terminus of the molecule [65]. This is an area of factor H that is responsible for cell surface protection and is a hotspot for mutations. Factor H autoantibodies have been demonstrated to impair the binding of factor H to C3b and are associated with increased hemolysis of sheep erythrocytes in patient plasma [65]. In the Newcastle cohort, the majority of the autoantibodies also bound to CCPs19–20 [61].
An 80-kb-long genomic deletion of CFHR1 and CFHR3 has been associated with an increased risk of aHUS [66]. It was subsequently demonstrated that this complete deficiency of CFHR1 and CFHR3 was strongly associated with factor H autoantibodies [64]. More detailed analysis has subsequently revealed aHUS patients with CFHR1 deficiency resulting from point mutations in CFHR1 [62] or from a deletion incorporating CFHR1 and CFHR4 [61, 62]. Thus the association between the CFHR1/CFHR3 deletion and the presence of autoantibodies in aHUS is probably related to the absence of CFHR1. It should be noted, however, that deficiency of CFHR1 is not a prerequisite for formation of autoantibodies as Moore et al. describe three patients with no evidence of deficiency of CFHR1 or CFHR3 and high titres of autoantibodies [61].
The two C-terminal CCPs (4 and 5) of CFHR1 are almost identical to CCPs19–20 of factor H (Fig. 2) and it is not surprising therefore that autoantibodies to CCPs19–20 of factor H also bind to CCPs 4–5 of CFHR1 [61]. It has been suggested that the antibodies generated in the absence of CFHR1 are different to those in its presence [62] and it is interesting that one of the patients with two copies of CFHR1 had antibodies against factor H CCPs1–4 [61], the region of the molecule responsible for cofactor and decay accelerating activity. Although most individuals with autoantibodies and CFHR1 deficiency do generate autoantibodies to CCP19–20, antibodies to this epitope are seen in the presence of CFHR1 [61].
Other complement genes
A mutation (Q433P) in the gene (Clu) encoding clusterin has been described in one family with aHUS [67]. Clusterin is a serum regulator of the terminal pathway of complement, it is predominantly produced by the liver, but is also released by activated platelets [68, 69]. In addition to the Clu mutation, the patient also had a functionally significant MCP mutation. This was of paternal origin while the Clu mutation came through the maternal line. Functional analysis of the mutant clusterin revealed decreased binding to C5b-7 and reduced complement regulation in a hemolytic assay. Serum from this patient also induced complement deposition on platelets and their activation. Although this mutation has been demonstrated to be functionally significant, aHUS only occurred in the presence of an MCP mutation known to predispose to aHUS. This is in keeping with the hypothesis that multiple concurrent factors may be necessary in individual patients for disease manifestation [70]. Mutation screening of other complement genes has been undertaken in aHUS. DAF is another complement regulatory gene located in the RCA cluster. It encodes a widely expressed membrane bound regulatory protein which accelerates the decay of either C4bC2a or C3bBb and the corresponding C5 convertases. Mutation screening of DAF in two cohorts has shown only one mutation which did not impair regulatory activity [56, 71]. Complement receptor 1 (CR1) is a cell surface complement regulator with cofactor and decay accelerating activity for both the AP and CP. In the one reported cohort of patients, no mutations were detected [36]. Screening of the genes (CFHR1–5) encoding the factor H-related proteins in one cohort of aHUS individuals failed to reveal mutations [37] while no causative mutations in CFHR5 were demonstrated in a separate cohort [72]. However, Abarrategui-Garrido have shown that deficiency of factor H-related protein 1 is in some aHUS patients secondary to point mutations in CFHR1. They also described a novel variant of CFHR1, which is probably a result of gene conversion between CFH and CFHR1. This variant is strongly associated with aHUS [62].
Incomplete penetrance
Incomplete penetrance has been reported for all the genes associated with aHUS. For mutations in CFH, CFI, MCP, and CFB penetrance is ∼50% while in the limited number of C3 mutations described to date, the penetrance is lower [59].
It has now been demonstrated that for disease to manifest in an individual a combination of mutations, risk haplotypes, and single nucleotide polymorphisms (SNPs) must be present [13, 31, 64, 66, 73–77]. The CFH risk haplotype for aHUS (CFH–H3) contains a SNP in the region of CFH responsible for cofactor activity. Functional analysis has demonstrated that the risk variant, CFH-Val62, has a subtle decrease in cofactor activity compared to the protective variant, in keeping with the minor structural differences between these SNPs [8, 78].
Similar risk haplotypes have also been described in MCP [73, 74] with a haplotype termed MCPggaac conferring a two-fold increased risk of aHUS compared with controls [74]. This contains two SNPs in the MCP promoter and reporter gene assays suggests that these haplotype differences may reduce transcriptional activity in the risk haplotypes by 25% [74, 75]. In vivo differences have not been demonstrated between the haplotypes, however, and it is possible that this is only revealed at times of cellular activation [74, 75].
C4b binding protein is the predominant regulator of the classical pathway of complement in the fluid phase [79, 80]. In addition to classical pathway activity, C4BP is also a weak regulator of the alternative pathway, acting as a cofactor for the factor I mediated cleavage of C3b. It is comprised of 7 identical α chains and a single β chain. C4BP is encoded by two genes, C4BPA and C4BPA in the RCA cluster. A SNP in C4BP (R240H) was associated with aHUS in cohorts from the UK and France [77]. Functional analysis of the change demonstrated normal secretion of the protein and normal ability to regulate classical and lectin pathways. The R240H SNP was, however, unable to down regulate the alternative pathway as efficiently as wild-type [77]. Many of the aHUS patients with this polymorphism also carried mutations in other complement genes associated with aHUS, and it is possible that this change is an additive risk factor for aHUS. However, this association was not confirmed in a separate Spanish cohort of patients [81].
The CFHR1/CFHR3 deletion is another risk factor for the development of aHUS as described previously.
Thus, there are now described a number haplotypes and SNPs which act in concert with mutations in complement genes and inhibitory autoantibodies. Only when an unfavorable group of risk factors co-segregate will aHUS develop. Even then the disease may not manifest until middle age, suggesting that a trigger is required to reveal the latent complement regulatory deficiency. These precipitating events are hypothesized to be the endothelial cell insults which have historically been associated with aHUS [82]. In an Italian cohort of patients with MCP mutations, infection precipitated the onset of disease in all individuals. In those with factor H mutations, 70% of cases were preceded by infection, and pregnancy and drugs each accounted for 4%. For CFI mutations 40% were preceded by pregnancy and 60% were precipitated by infection [13].
Thus, the environmental trigger initiates the positive feedback loop of complement and in susceptible individuals unable to control complement turnover, disease will manifest.
Genotype: phenotype effects
The prognosis of patients correlates to a certain extent with the genetic defect. Those with CFH mutations carry the worst outcome. Within a year after disease onset, 60–70% [13, 49] of patients with CFH mutations die or reach ESRF. This genotype: phenotype is not absolute, however. This was best illustrated by analysis of 13 individuals carrying the same mutation (R1210C) [76]. As a whole the group did poorly, eight patients developed end-stage renal failure and one patient had chronic kidney disease, but there was one individual with this mutation who had a single episode of aHUS with no long-term sequelae. It is noteworthy that of this group only this individual did not have additional genetic risk factors for aHUS. Thus, although mutations in CFH are a poor prognostic indicator, it appears that other genetic risk factors do modify disease outcome.
In comparison to those with CFH mutations, the outcome of patients with MCP mutations is good, with ∼80% remaining dialysis-independent [13, 49]. Recurrence of disease in MCP-associated aHUS is frequent however. In the minority of cases with MCP mutations that do go onto develop ESRF, it is likely that additional genetic modifiers are present.
In those with CFI-associated aHUS, the prognosis is intermediate between CFH and MCP-associated aHUS. A report from an Italian cohort of patients suggested that in those with CFI-associated aHUS, >60% of patients developed ESRF [13]. In the French cohort, in those with mutations in CFI, around 50% of patients died or progressed to ESRF within 2 years of the initial episode of aHUS, while ∼30% of individuals recovered with no disease recurrences [49]. Bienaime et al. demonstrated that the severity of disease was influenced by additional genetic factors. In particular, those with an associated deletion of CFHR1 had a significantly worse outcome [50].
To date, all those individuals with factor H autoantibodies are children [61, 63] and less than 50% have gone on to develop ESRF [61, 63].
Definite clinical correlations have yet to emerge in those with CFB and C3 mutations due to the limited number of cases so far reported. The outcome of patients with CFB mutations would appear poor, however, with the majority of patients developing ESRF. Of the 14 individuals reported with C3-associated aHUS, eight developed ESRF and one developed chronic renal impairment.
In addition to pre-determining the outcome of aHUS in the native kidneys, the genetic predisposition also affects the aHUS recurrence rate after renal transplantation. The complement regulatory defect in those with mutations in the membrane bound, MCP, is corrected by an allograft bearing wild-type MCP and so the recurrence rate is low. In those with mutations in CFH and CFI, which are produced in the liver, a renal transplant does not correct the defect so the recurrence rate is high [83–85].
Summary
Although many different alterations in complement genes have been reported to predispose to aHUS, the downstream consequence of all is over-activation of the alternative pathway of complement on the glomerular vasculature. It is increasingly becoming clear that a combination of mutations, SNPs, and haplotypes are required for disease to manifest upon exposure to an environmental trigger. The genetic predisposition also determines the prognosis after the initial episode and following renal transplantation.
Acknowledgements
DK is funded by Kidney Research UK, The Academy of Medical Science, and the Mason Medical Research Trust.
Abbreviations
- HUS
Hemolytic uremic syndrome
- D+ve HUS
Diarrhoeal-associated hemolytic uremic syndrome
- aHUS
Atypical hemolytic uremic syndrome
- AP
Alternative pathway
- CP
Classical pathway
- CCPs
Complement control protein modules
- RCA
Regulators of complement activation
- ESRF
End-stage renal failure
- SNPs
Single nucleotide polymorphisms
Questions
(Answers appear following the reference list)
- Familial atypical HUS
- Is completely penetrant.
- Has been causally associated with Eculizumab treatment.
- Is predisposed to by mutations in complement factor H.
- Always presents in childhood.
- Never recurs following renal transplantation.
- In atypical HUS
- Complement factor B mutations are the commonest genetic cause.
- Individuals with membrane cofactor mutations never have disease recurrence.
- Individuals who present during pregnancy never have complement mutations.
- Complement factor H mutations are the rarest genetic cause.
- Autoantibodies to factor H have been associated with disease.
- Atypical hemolytic uremic syndrome is associated with mutations in complement regulatory genes. A low recurrence rate of aHUS post renal transplantation is associated with mutations in which of the following genes?
- Complement factor I
- Complement factor H
- Membrane cofactor protein
- Complement factor B
- C3
- In aHUS, autoantibodies to complement factor H
- Do not result in impaired complement regulation at host cell surfaces.
- Are associated with a deletion of the CFHR1 and CFHR3 genes.
- Always bind to the C-terminal region of complement factor H.
- Never cross react with the complement factor H-related 1 protein.
- Cannot be removed by plasma exchange.
Answers:
c
e
c
b
References
- 1.Kavanagh D, Richards A, Atkinson J. Complement regulatory genes and hemolytic uremic syndromes. Annu Rev Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110. [DOI] [PubMed] [Google Scholar]
- 2.Fogo A, Kashgarian M. Diagnostic atlas of renal pathology. Amsterdam: Elsevier Science; 2005. [Google Scholar]
- 3.Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 4.Noris M, Remuzzi G. Hemolytic uremic syndrome. J Am Soc Nephrol. 2005;16:1035–1050. doi: 10.1681/ASN.2004100861. [DOI] [PubMed] [Google Scholar]
- 5.Ariceta G, Besbas N, Johnson S, Karpman D, Landau D, Licht C, Loirat C, Pecoraro C, Taylor CM, Kar N, Vandewalle J, Zimmerhackl LB. Guideline for the investigation and initial therapy of diarrhea-negative hemolytic uremic syndrome. Pediatr Nephrol. 2009;24:687–696. doi: 10.1007/s00467-008-0964-1. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. The human complement factor H: functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Richards A, Kavanagh D, Atkinson JP. Inherited complement regulatory protein deficiency predisposes to human disease in acute injury and chronic inflammatory states: the examples of vascular damage in atypical hemolytic uremic syndrome and debris accumulation in age-related macular degeneration. Adv Immunol. 2007;96:141–177. doi: 10.1016/S0065-2776(07)96004-6. [DOI] [PubMed] [Google Scholar]
- 8.Hocking HG, Herbert AP, Kavanagh D, Soares DC, Ferreira VP, Pangburn MK, Uhrin D, Barlow PN. Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations. J Biol Chem. 2008;283:9475–9487. doi: 10.1074/jbc.M709587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt CQ, Herbert AP, Kavanagh D, Gandy C, Fenton CJ, Blaum BS, Lyon M, Uhrin D, Barlow PN. A new map of glycosaminoglycan and C3b binding sites on factor H. J Immunol. 2008;181:2610–2619. doi: 10.4049/jimmunol.181.4.2610. [DOI] [PubMed] [Google Scholar]
- 10.Warwicker P, Goodship TH, Donne RL, Pirson Y, Nicholls A, Ward RM, Turnpenny P, Goodship JA. Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int. 1998;53:836–844. doi: 10.1111/j.1523-1755.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- 11.Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH. Factor H mutations in hemolytic uremic syndrome cluster in exons 18–20, a domain important for host cell recognition. Am J Hum Genet. 2001;68:485–490. doi: 10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragon-Durey MA, Fremeaux-Bacchi V, Loirat C, Blouin J, Niaudet P, Deschenes G, Coppo P, Herman Fridman W, Weiss L. Heterozygous and homozygous factor h deficiencies associated with hemolytic uremic syndrome or membranoproliferative glomerulonephritis: report and genetic analysis of 16 cases. J Am Soc Nephrol. 2004;15:787–795. doi: 10.1097/01.ASN.0000115702.28859.A7. [DOI] [PubMed] [Google Scholar]
- 13.Caprioli J, Noris M, Brioschi S, Pianetti G, Castelletti F, Bettinaglio P, Mele C, Bresin E, Cassis L, Gamba S, Porrati F, Bucchioni S, Monteferrante G, Fang CJ, Liszewski MK, Kavanagh D, Atkinson JP, Remuzzi G. Genetics of HUS: the impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood. 2006;108:1267–1279. doi: 10.1182/blood-2005-10-007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann HP, Salzmann M, Bohnert-Iwan B, Mannuelian T, Skerka C, Lenk D, Bender BU, Cybulla M, Riegler P, Konigsrainer A, Neyer U, Bock A, Widmer U, Male DA, Franke G, Zipfel PF. Haemolytic uraemic syndrome and mutations of the factor H gene: a registry-based study of German-speaking countries. J Med Genet. 2003;40:676–681. doi: 10.1136/jmg.40.9.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Caballero D, Gonzalez-Rubio C, Gallardo ME, Vera M, Lopez-Trascasa M, Rodriguez de Cordoba S, Sanchez-Corral P. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet. 2001;68:478–484. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westra D, Volokhina E, Heijden E, Vos A, Huigen M, Jansen J, Kaauwen E, Velden T, Kar N, Heuvel L. Genetic disorders in complement (regulating) genes in patients with atypical haemolytic uraemic syndrome (aHUS) Nephrol Dial Transplant. 2010 doi: 10.1093/ndt/gfq010. [DOI] [PubMed] [Google Scholar]
- 17.Heinen S, Sanchez-Corral P, Jackson MS, Strain L, Goodship JA, Kemp EJ, Skerka C, Jokiranta TS, Meyers K, Wagner E, Robitaille P, Esparza-Gordillo J, Rodriguez de Cordoba S, Zipfel PF, Goodship TH. De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Hum Mutat. 2006;27:292–293. doi: 10.1002/humu.9408. [DOI] [PubMed] [Google Scholar]
- 18.Venables JP, Strain L, Routledge D, Bourn D, Powell HM, Warwicker P, Diaz-Torres ML, Sampson A, Mead P, Webb M, Pirson Y, Jackson MS, Hughes A, Wood KM, Goodship JA, Goodship TH. Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med. 2006;3:e431. doi: 10.1371/journal.pmed.0030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira VP, Herbert AP, Cortes C, McKee KA, Blaum BS, Esswein ST, Uhrin D, Barlow PN, Pangburn MK, Kavanagh D. The binding of factor H to a complex of physiological polyanions and C3b on cells is impaired in atypical hemolytic uremic syndrome. J Immunol. 2009;182:7009–7018. doi: 10.4049/jimmunol.0804031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Corral P, Gonzalez-Rubio C, Rodriguez de Cordoba S, Lopez-Trascasa M. Functional analysis in serum from atypical hemolytic uremic syndrome patients reveals impaired protection of host cells associated with mutations in factor H. Mol Immunol. 2004;41:81–84. doi: 10.1016/j.molimm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Abarrategui-Garrido C, Melgosa M, Pena-Carrion A, Jorge EG, Cordoba SR, Lopez-Trascasa M, Sanchez-Corral P. Mutations in proteins of the alternative pathway of complement and the pathogenesis of atypical hemolytic uremic syndrome. Am J Kidney Dis. 2008;52:171–180. doi: 10.1053/j.ajkd.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 22.Vaziri-Sani F, Holmberg L, Sjoholm AG, Kristoffersson AC, Manea M, Fremeaux-Bacchi V, Fehrman-Ekholm I, Raafat R, Karpman D. Phenotypic expression of factor H mutations in patients with atypical hemolytic uremic syndrome. Kidney Int. 2006;69:981–988. doi: 10.1038/sj.ki.5000155. [DOI] [PubMed] [Google Scholar]
- 23.Stahl AL, Vaziri-Sani F, Heinen S, Kristoffersson AC, Gydell KH, Raafat R, Gutierrez A, Beringer O, Zipfel PF, Karpman D. Factor H dysfunction in patients with atypical hemolytic uremic syndrome contributes to complement deposition on platelets and their activation. Blood. 2008;111:5307–5315. doi: 10.1182/blood-2007-08-106153. [DOI] [PubMed] [Google Scholar]
- 24.Heinen S, Jozsi M, Hartmann A, Noris M, Remuzzi G, Skerka C, Zipfel PF. Hemolytic uremic syndrome: a factor H mutation (E1172Stop) causes defective complement control at the surface of endothelial cells. J Am Soc Nephrol. 2007;18:506–514. doi: 10.1681/ASN.2006091069. [DOI] [PubMed] [Google Scholar]
- 25.Nan R, Gor J, Perkins SJ. Implications of the progressive self-association of wild-type human factor H for complement regulation and disease. J Mol Biol. 2008;375:891–900. doi: 10.1016/j.jmb.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Pangburn MK, Rawal N, Cortes C, Alam MN, Ferreira VP, Atkinson MA. Polyanion-induced self-association of complement factor H. J Immunol. 2009;182:1061–1068. doi: 10.4049/jimmunol.182.2.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okemefuna AI, Li K, Nan R, Ormsby RJ, Sadlon T, Gordon DL, Perkins SJ. Multimeric interactions between complement factor H and its C3d ligand provide new insight on complement regulation. J Mol Biol. 2009;391:119–135. doi: 10.1016/j.jmb.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Richards A, Kavanagh D. Pathogenesis of thrombotic microangiopathy: insights from animal models. Nephron Exp Nephrol. 2009;113:e97–e103. doi: 10.1159/000235253. [DOI] [PubMed] [Google Scholar]
- 29.Pickering MC, Cook HT, Warren J, Bygrave AE, Moss J, Walport MJ, Botto M. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat Genet. 2002;31:424–428. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- 30.Hegasy GA, Manuelian T, Hogasen K, Jansen JH, Zipfel PF. The molecular basis for hereditary porcine membranoproliferative glomerulonephritis type II: point mutations in the factor H coding sequence block protein secretion. Am J Pathol. 2002;161:2027–2034. doi: 10.1016/S0002-9440(10)64481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickering MC, Jorge EG, Martinez-Barricarte R, Recalde S, Garcia-Layana A, Rose KL, Moss J, Walport MJ, Cook HT, Cordoba SR, Botto M. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J Exp Med. 2007;204:1249–1256. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goicoechea de Jorge E, Paixao-Cavalcante D, Rose K, Cook H, Botto M, Pickering M. C5 activation is required for the development of atypical haemolytic uraemic syndrome in Cfh-/-FH Delta 16–20 mice. Mol Immunol. 2008;44:4100. [Google Scholar]
- 33.Richards A, Kathryn Liszewski M, Kavanagh D, Fang CJ, Moulton E, Fremeaux-Bacchi V, Remuzzi G, Noris M, Goodship TH, Atkinson JP. Implications of the initial mutations in membrane cofactor protein (MCP; CD46) leading to atypical hemolytic uremic syndrome. Mol Immunol. 2007;44:111–122. doi: 10.1016/j.molimm.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Richards A, Kemp EJ, Liszewski MK, Goodship JA, Lampe AK, Decorte R, Muslumanoglu MH, Kavukcu S, Filler G, Pirson Y, Wen LS, Atkinson JP, Goodship TH. Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc Natl Acad Sci USA. 2003;100:12966–12971. doi: 10.1073/pnas.2135497100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fremeaux-Bacchi V, Moulton EA, Kavanagh D, Dragon-Durey MA, Blouin J, Caudy A, Arzouk N, Cleper R, Francois M, Guest G, Pourrat J, Seligman R, Fridman WH, Loirat C, Atkinson JP. Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2006;17:2017–2025. doi: 10.1681/ASN.2005101051. [DOI] [PubMed] [Google Scholar]
- 36.Noris M, Brioschi S, Caprioli J, Todeschini M, Bresin E, Porrati F, Gamba S, Remuzzi G. Familial haemolytic uraemic syndrome and an MCP mutation. Lancet. 2003;362:1542–1547. doi: 10.1016/S0140-6736(03)14742-3. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan M, Erlic Z, Hoffmann MM, Arbeiter K, Patzer L, Budde K, Hoppe B, Zeier M, Lhotta K, Rybicki LA, Bock A, Berisha G, Neumann HP. Epidemiological approach to identifying genetic predispositions for atypical hemolytic uremic syndrome. Ann Hum Genet. 2010;74:17–26. doi: 10.1111/j.1469-1809.2009.00554.x. [DOI] [PubMed] [Google Scholar]
- 38.Fang CJ, Fremeaux-Bacchi V, Liszewski MK, Pianetti G, Noris M, Goodship TH, Atkinson JP. Membrane cofactor protein mutations in atypical hemolytic uremic syndrome (aHUS), fatal Stx-HUS, C3 glomerulonephritis, and the HELLP syndrome. Blood. 2008;111:624–632. doi: 10.1182/blood-2007-04-084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pangburn MK, Schreiber RD, Muller-Eberhard HJ. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein beta1H for cleavage of C3b and C4b in solution. J Exp Med. 1977;146:257–270. doi: 10.1084/jem.146.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whaley K, Ruddy S. Modulation of C3b hemolytic activity by a plasma protein distinct from C3b inactivator. Science. 1976;193:1011–1013. doi: 10.1126/science.948757. [DOI] [PubMed] [Google Scholar]
- 41.Nagasawa S, Stroud RM. Mechanism of action of the C3b inactivator: requirement for a high molecular weight cofactor (C3b-C4bINA cofactor) and production of a new C3b derivative (C3b′) Immunochemistry. 1977;14:749–756. doi: 10.1016/0019-2791(77)90345-7. [DOI] [PubMed] [Google Scholar]
- 42.Shiraishi S, Stroud RM. Cleavage products of C4b produced by enzymes in human serum. Immunochemistry. 1975;12:935–939. doi: 10.1016/0019-2791(75)90256-6. [DOI] [PubMed] [Google Scholar]
- 43.Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- 44.Medof ME, Iida K, Mold C, Nussenzweig V. Unique role of the complement receptor CR1 in the degradation of C3b associated with immune complexes. J Exp Med. 1982;156:1739–1754. doi: 10.1084/jem.156.6.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross GD, Lambris JD, Cain JA, Newman SL. Generation of three different fragments of bound C3 with purified factor I or serum. I. Requirements for factor H vs. CR1 cofactor activity. J Immunol. 1982;129:2051–2060. [PubMed] [Google Scholar]
- 46.Kavanagh D, Kemp EJ, Mayland E, Winney RJ, Duffield JS, Warwick G, Richards A, Ward R, Goodship JA, Goodship TH. Mutations in complement factor I predispose to development of atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2005;16:2150–2155. doi: 10.1681/ASN.2005010103. [DOI] [PubMed] [Google Scholar]
- 47.Kavanagh D, Richards A, Noris M, Hauhart R, Liszewski MK, Karpman D, Goodship JA, Fremeaux-Bacchi V, Remuzzi G, Goodship TH, Atkinson JP. Characterization of mutations in complement factor I (CFI) associated with hemolytic uremic syndrome. Mol Immunol. 2008;45:95–105. doi: 10.1016/j.molimm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Fremeaux-Bacchi V, Dragon-Durey MA, Blouin J, Vigneau C, Kuypers D, Boudailliez B, Loirat C, Rondeau E, Fridman WH. Complement factor I: a susceptibility gene for atypical haemolytic uraemic syndrome. J Med Genet. 2004;41:e84. doi: 10.1136/jmg.2004.019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sellier-Leclerc AL, Fremeaux-Bacchi V, Dragon-Durey MA, Macher MA, Niaudet P, Guest G, Boudailliez B, Bouissou F, Deschenes G, Gie S, Tsimaratos M, Fischbach M, Morin D, Nivet H, Alberti C, Loirat C. Differential impact of complement mutations on clinical characteristics in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2007;18:2392–2400. doi: 10.1681/ASN.2006080811. [DOI] [PubMed] [Google Scholar]
- 50.Bienaime F, Dragon-Durey MA, Regnier CH, Nilsson SC, Kwan WH, Blouin J, Jablonski M, Renault N, Rameix-Welti MA, Loirat C, Sautes-Fridman C, Villoutreix BO, Blom AM, Fremeaux-Bacchi V. Mutations in components of complement influence the outcome of factor I-associated atypical hemolytic uremic syndrome. Kidney Int. 2010;77:339–349. doi: 10.1038/ki.2009.472. [DOI] [PubMed] [Google Scholar]
- 51.Paula PF, Barbosa JE, Junior PR, Ferriani VP, Latorre MR, Nudelman V, Isaac L. Ontogeny of complement regulatory proteins—concentrations of factor h, factor I, c4b-binding protein, properdin and vitronectin in healthy children of different ages and in adults. Scand J Immunol. 2003;58:572–577. doi: 10.1046/j.1365-3083.2003.01326.x. [DOI] [PubMed] [Google Scholar]
- 52.Nilsson SC, Kalchishkova N, Trouw LA, Fremeaux-Bacchi V, Villoutreix BO, Blom AM. Mutations in complement factor I as found in atypical hemolytic uremic syndrome lead to either altered secretion or altered function of factor I. Eur J Immunol. 2010;40:172–185. doi: 10.1002/eji.200939280. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson SC, Karpman D, Vaziri-Sani F, Kristoffersson AC, Salomon R, Provot F, Fremeaux-Bacchi V, Trouw LA, Blom AM. A mutation in factor I that is associated with atypical hemolytic uremic syndrome does not affect the function of factor I in complement regulation. Mol Immunol. 2007;44:1835–1844. doi: 10.1016/j.molimm.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Weiler H, Isermann BH. Thrombomodulin. J Thromb Haemost. 2003;1:1515–1524. doi: 10.1046/j.1538-7836.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 55.Delvaeye M, Noris M, Vriese A, Esmon CT, Esmon NL, Ferrell G, Del-Favero J, Plaisance S, Claes B, Lambrechts D, Zoja C, Remuzzi G, Conway EM. Thrombomodulin mutations in atypical hemolytic-uremic syndrome. N Engl J Med. 2009;361:345–357. doi: 10.1056/NEJMoa0810739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, Carreras L, Arranz EA, Garrido CA, Lopez-Trascasa M, Sanchez-Corral P, Morgan BP, Rodriguez de Cordoba S. Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci USA. 2007;104:240–245. doi: 10.1073/pnas.0603420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kavanagh D, Kemp EJ, Richards A, Burgess RM, Mayland E, Goodship JA, Goodship TH. Does complement factor B have a role in the pathogenesis of atypical HUS? Mol Immunol. 2006;43:856–859. doi: 10.1016/j.molimm.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 58.Roumenina LT, Jablonski M, Hue C, Blouin J, Dimitrov JD, Dragon-Durey MA, Cayla M, Fridman WH, Macher MA, Ribes D, Moulonguet L, Rostaing L, Satchell SC, Mathieson PW, Sautes-Fridman C, Loirat C, Regnier CH, Halbwachs-Mecarelli L, Fremeaux-Bacchi V. Hyperfunctional C3 convertase leads to complement deposition on endothelial cells and contributes to atypical hemolytic uremic syndrome. Blood. 2009;114:2837–2845. doi: 10.1182/blood-2009-01-197640. [DOI] [PubMed] [Google Scholar]
- 59.Lhotta K, Janecke AR, Scheiring J, Petzlberger B, Giner T, Fally V, Wurzner R, Zimmerhackl LB, Mayer G, Fremeaux-Bacchi V. A large family with a gain-of-function mutation of complement C3 predisposing to atypical hemolytic uremic syndrome, microhematuria, hypertension and chronic renal failure. Clin J Am Soc Nephrol. 2009;4:1356–1362. doi: 10.2215/CJN.06281208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fremeaux-Bacchi V, Miller EC, Liszewski MK, Strain L, Blouin J, Brown AL, Moghal N, Kaplan BS, Weiss RA, Lhotta K, Kapur G, Mattoo T, Nivet H, Wong W, Gie S, Ligny BH, Fischbach M, Gupta R, Hauhart R, Meunier V, Loirat C, Dragon-Durey MA, Fridman WH, Janssen BJ, Goodship TH, Atkinson JP. Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood. 2008;112:4948–4952. doi: 10.1182/blood-2008-01-133702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore I, Strain L, Pappworth I, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Staniforth S, Holmes L, Ward R, Morgan L, Goodship TH, Marchbank K. Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4 and with mutations in CFH, CFI, CD46 and C3 in patients with atypical haemolytic uraemic syndrome. Blood. 2010 doi: 10.1182/blood-2009-05-221549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abarrategui-Garrido C, Martinez-Barricarte R, Lopez-Trascasa M, Rodriguez de Cordoba S, Sanchez-Corral P. Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood. 2009;114:4261–4271. doi: 10.1182/blood-2009-05-223834. [DOI] [PubMed] [Google Scholar]
- 63.Dragon-Durey MA, Loirat C, Cloarec S, Macher MA, Blouin J, Nivet H, Weiss L, Fridman WH, Fremeaux-Bacchi V. Anti-Factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2005;16:555–563. doi: 10.1681/ASN.2004050380. [DOI] [PubMed] [Google Scholar]
- 64.Jozsi M, Licht C, Strobel S, Zipfel SL, Richter H, Heinen S, Zipfel PF, Skerka C. Factor H autoantibodies in atypical hemolytic uremic syndrome correlate with CFHR1/CFHR3 deficiency. Blood. 2008;111:1512–1514. doi: 10.1182/blood-2007-09-109876. [DOI] [PubMed] [Google Scholar]
- 65.Jozsi M, Strobel S, Dahse HM, Liu WS, Hoyer PF, Oppermann M, Skerka C, Zipfel PF. Anti factor H autoantibodies block C-terminal recognition function of factor H in hemolytic uremic syndrome. Blood. 2007;110:1516–1518. doi: 10.1182/blood-2007-02-071472. [DOI] [PubMed] [Google Scholar]
- 66.Zipfel PF, Edey M, Heinen S, Jozsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, Goodship JA, Licht C, Goodship TH, Skerka C. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stahl AL, Kristoffersson A, Olin AI, Olsson ML, Roodhooft AM, Proesmans W, Karpman D. A novel mutation in the complement regulator clusterin in recurrent hemolytic uremic syndrome. Mol Immunol. 2009;46:2236–2243. doi: 10.1016/j.molimm.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 68.Jenne DE, Tschopp J. Clusterin: the intriguing guises of a widely expressed glycoprotein. Trends Biochem Sci. 1992;17:154–159. doi: 10.1016/0968-0004(92)90325-4. [DOI] [PubMed] [Google Scholar]
- 69.Tschopp J, Jenne DE, Hertig S, Preissner KT, Morgenstern H, Sapino AP, French L. Human megakaryocytes express clusterin and package it without apolipoprotein A-1 into alpha-granules. Blood. 1993;82:118–125. [PubMed] [Google Scholar]
- 70.Rodriguez de Cordoba S. aHUS: a disorder with many risk factors. Blood. 2010;115:158–160. doi: 10.1182/blood-2009-11-252627. [DOI] [PubMed] [Google Scholar]
- 71.Kavanagh D, Burgess R, Spitzer D, Richards A, Diaz-Torres ML, Goodship JA, Hourcade DE, Atkinson JP, Goodship TH. The decay accelerating factor mutation I197V found in hemolytic uraemic syndrome does not impair complement regulation. Mol Immunol. 2007;44:3162–3167. doi: 10.1016/j.molimm.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 72.Monteferrante G, Brioschi S, Caprioli J, Pianetti G, Bettinaglio P, Bresin E, Remuzzi G, Noris M. Genetic analysis of the complement factor H-related 5 gene in haemolytic uraemic syndrome. Mol Immunol. 2007;44:1704–1708. doi: 10.1016/j.molimm.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 73.Fremeaux-Bacchi V, Kemp EJ, Goodship JA, Dragon-Durey MA, Strain L, Loirat C, Deng HW, Goodship TH. The development of atypical haemolytic-uraemic syndrome is influenced by susceptibility factors in factor H and membrane cofactor protein: evidence from two independent cohorts. J Med Genet. 2005;42:852–856. doi: 10.1136/jmg.2005.030783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Esparza-Gordillo J, Goicoechea de Jorge E, Buil A, Carreras Berges L, Lopez-Trascasa M, Sanchez-Corral P, Rodriguez de Cordoba S. Predisposition to atypical hemolytic uremic syndrome involves the concurrence of different susceptibility alleles in the regulators of complement activation gene cluster in 1q32. Hum Mol Genet. 2005;14:703–712. doi: 10.1093/hmg/ddi066. [DOI] [PubMed] [Google Scholar]
- 75.Esparza-Gordillo J, Jorge EG, Garrido CA, Carreras L, Lopez-Trascasa M, Sanchez-Corral P, Cordoba SR. Insights into hemolytic uremic syndrome: segregation of three independent predisposition factors in a large, multiple affected pedigree. Mol Immunol. 2006;43:1769–1775. doi: 10.1016/j.molimm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 76.Martinez-Barricarte R, Pianetti G, Gautard R, Misselwitz J, Strain L, Fremeaux-Bacchi V, Skerka C, Zipfel PF, Goodship T, Noris M, Remuzzi G, Cordoba SR. The complement factor H R1210C mutation is associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2008;19:639–646. doi: 10.1681/ASN.2007080923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blom AM, Bergstrom F, Edey M, Diaz-Torres M, Kavanagh D, Lampe A, Goodship JA, Strain L, Moghal N, McHugh M, Inward C, Tomson C, Fremeaux-Bacchi V, Villoutreix BO, Goodship TH. A novel non-synonymous polymorphism (p.Arg240His) in C4b-binding protein is associated with atypical hemolytic uremic syndrome and leads to impaired alternative pathway cofactor activity. J Immunol. 2008;180:6385–6391. doi: 10.4049/jimmunol.180.9.6385. [DOI] [PubMed] [Google Scholar]
- 78.Tortajada A, Montes T, Martinez-Barricarte R, Morgan BP, Harris CL, Cordoba SR. The disease-protective complement factor H allotypic variant Ile62 shows increased binding affinity for C3b and enhanced cofactor activity. Hum Mol Genet. 2009;18:3452–3461. doi: 10.1093/hmg/ddp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scharfstein J, Ferreira A, Gigli I, Nussenzweig V. Human C4-binding protein. I. Isolation and characterization. J Exp Med. 1978;148:207–222. doi: 10.1084/jem.148.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fujita T, Gigli I, Nussenzweig V. Human C4-binding protein. II. Role in proteolysis of C4b by C3b-inactivator. J Exp Med. 1978;148:1044–1051. doi: 10.1084/jem.148.4.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinez-Barricarte R, Goicoechea de Jorge E, Montes T, Layana AG, Rodriguez de Cordoba S. Lack of association between polymorphisms in C4b-binding protein and atypical haemolytic uraemic syndrome in the Spanish population. Clin Exp Immunol. 2009;155:59–64. doi: 10.1111/j.1365-2249.2008.03798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kavanagh D, Goodship TH, Richards A. Atypical haemolytic uraemic syndrome. Br Med Bull. 2006;77–78:5–22. doi: 10.1093/bmb/ldl004. [DOI] [PubMed] [Google Scholar]
- 83.Loirat C, Fremeaux-Bacchi V. Hemolytic uremic syndrome recurrence after renal transplantation. Pediatr Transplant. 2008;12:619–629. doi: 10.1111/j.1399-3046.2008.00910.x. [DOI] [PubMed] [Google Scholar]
- 84.Kavanagh D, Richards A, Goodship TH, Jalanko H (2010) Transplantation in atypical hemolytic uremic syndrome. Semin Thromb Hemost. doi [DOI] [PubMed]
- 85.Bresin E, Daina E, Noris M, Castelletti F, Stefanov R, Hill P, Goodship TH, Remuzzi G. Outcome of renal transplantation in patients with non-Shiga toxin-associated hemolytic uremic syndrome: prognostic significance of genetic background. Clin J Am Soc Nephrol. 2006;1:88–99. doi: 10.2215/CJN.00050505. [DOI] [PubMed] [Google Scholar]
- 86.Lehtinen MJ, Rops AL, Isenman DE, Vlag J, Jokiranta TS. Mutations of factor H impair regulation of surface-bound C3b by three mechanisms in atypical hemolytic uremic syndrome. J Biol Chem. 2009;284:15650–15658. doi: 10.1074/jbc.M900814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jozsi M, Heinen S, Hartmann A, Ostrowicz CW, Halbich S, Richter H, Kunert A, Licht C, Saunders RE, Perkins SJ, Zipfel PF, Skerka C. Factor H and atypical hemolytic uremic syndrome: mutations in the C-terminus cause structural changes and defective recognition functions. J Am Soc Nephrol. 2006;17:170–177. doi: 10.1681/ASN.2005080868. [DOI] [PubMed] [Google Scholar]
- 88.Jokiranta TS, Cheng ZZ, Seeberger H, Jozsi M, Heinen S, Noris M, Remuzzi G, Ormsby R, Gordon DL, Meri S, Hellwage J, Zipfel PF. Binding of complement factor H to endothelial cells is mediated by the carboxy-terminal glycosaminoglycan binding site. Am J Pathol. 2005;167:1173–1181. doi: 10.1016/S0002-9440(10)61205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HP, Remuzzi G, Zipfel PF. Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J Clin Invest. 2003;111:1181–1190. doi: 10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sanchez-Corral P, Perez-Caballero D, Huarte O, Simckes AM, Goicoechea E, Lopez-Trascasa M, Cordoba SR. Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am J Hum Genet. 2002;71:1285–1295. doi: 10.1086/344515. [DOI] [PMC free article] [PubMed] [Google Scholar]