Abstract
The real-time, personalized and highly sensitive early-stage diagnosis of disease remains an important challenge in modern medicine. With the ability to interact with matter at the nanoscale, the development of nanotechnology architectures and materials could potentially extend subcellular and molecular detection beyond the limits of conventional diagnostic modalities. At the very least, nanotechnology should be able to dramatically accelerate biomarker discovery, as well as facilitate disease monitoring, especially of maladies presenting a high degree of molecular and compositional heterogeneity. This article gives an overview of several of the most promising nanodevices and nanomaterials along with their applications in clinical practice. Significant work to adapt nanoscale materials and devices to clinical applications involving large interdisciplinary collaborations is already underway with the potential for nanotechnology to become an important enabling diagnostic technology.
Many diseases, including cancer, originate from mutations and alterations to normal cellular regulatory and metabolic pathways at molecular level.1,2 Accurate and sensitive diagnosis has been constrained by the lack of biosensors and molecular probes capable of rapidly recognizing the distinct molecular features of these diseases. The ability of nanomaterials and nanopatterned devices to directly interact with biologically significant molecules, and to convert those interactions into directly transduced or significantly amplified electrical or electromagnetic signals, has enabled a new generation of early-stage diagnostic techniques.
The greater the detail with which the molecular components of a specific disease can be determined the more specifically the therapeutic regime can be tailored to the individual.3 The development of microfluidics and ‘lab on a chip’ systems with specifically designed nanoscale features enables a number of complex diagnostic procedures to be combined into one simple device for point-of-care diagnosis. Many products have emerged for laboratory clinical and diagnostic uses including separation technologies for blood into its components, the fractionation of complex biofluidic mixtures into, e.g., its protein and nucleic acid digest sub-populations, DNA amplification strategies via PCR-on-chip, precise fluidic dispensation technologies for automated high throughput analyses, multiplexed analyte sensors for point-of-care diagnostics, and many more. Despite the enormous potential of nanotechnology as it relates to diagnosis, many important concerns must still be addressed including the toxicological effects of the in vivo use of the relevant nanomaterials, the identification of appropriate target molecules and biomarkers to be screened for, the proper protocols for sample preparation, and the complete interpretation of diagnostic results obtained from both animal models and human trials. The ability to address these concerns will ultimately determine how extensively nanotechnology-driven diagnostic techniques can percolate into the clinic, but as you will see in this review article, initial preclinical efforts are quite promising and the array of possible applications are quite broad.
Currently, conventional radiological diagnostic techniques focus on detecting the physical manifestations of disease. Given the high correlation between survival rate and early detection,4 it is highly advantageous to identify abnormal cellular function before significant physiological modifications become apparent. Mammography is the process of using low energy X-rays to examine the human breast for characteristic masses or microcalcifications.5 It is a widely used technique because of its relatively low cost and operational simplicity allowing for population-wide screening. Regular mammographic screening can detect between 80 and 90% of breast cancers in asymptomatic individuals, resulting in reduced breast cancer mortality.4 However, for women with dense breasts, the sensitivity of mammography is low (45.8–55%).6 Furthermore, successful screening and diagnosis rely too heavily on the skill and experience of the radiological practitioner. Finally, this imaging-based technology requires a critical mass of localized accumulated tumor cells for effective neoplasm identification and is therefore not capable of detecting tumors with masses or densities below this critical dimension. The diagnosis of breast cancer can be also accomplished by directly analyzing tumor cells through techniques such as the cytomorphology of exfoliated cells.7 Direct visualization of tumor cells does not lend itself to regular population-wide screening of asymptomatic women, however, because of the subjective nature of tumor cell identification and the labor-intensiveness of the process. Several studies have recognized that diagnostic biomarkers isolated from blood, such as circulating proteins and nucleic acids, may offer the greatest potential for the reliable and earliest possible screening of diseases.8–11 The assistance of nanomaterials and nanotechnologies will greatly enhance the throughput and sensitivity of the identification and screening of potential biomarkers. For example, the breakthroughs in using inorganic nanoparticles for contrast enhance in imaging biomarkers provide a robust framework for biomedical application.12–16 The marriage of biology and micro- and nanofabrication has revolutionized biosensing by allowing for the integration of biological recognition elements into devices that will significantly impact the commercial availability of detection and diagnostic technologies at the genome, proteome, and metabalome levels. This in turn should dramatically decrease the time between disease onset and the initiation of tailored medical intervention and thus greatly increase the likelihood of a positive clinical outcome.
In this review, we will discuss disease diagnosis through molecular recognition in addition to several of the most promising and advanced nanotechnologies for achieving the goal of the earliest possible detection of abnormal cellular function. This technology review will first cover nanopatterned devices, including mesoporous silica chips, for rapid proteomic and genomic screening of blood and plasma. Next, we will look at the nanomaterials used in diagnostics, including the DNA-based nanobarcode, quantum dots (QDs), and gold nanoparticles. In this section, we will also discuss magnetic nanoparticles, predominantly as contrast agents for magnetic resonance imaging (MRI). Finally, we will conclude with a discussion of future trends, including the movement toward theranostics whereby the functional flexibility of many nanotechnologies allows for the coupling of both the personalized diagnostic and therapeutic modalities within the same construct.
MOLECULAR RECOGNITION IN DIAGNOSTICS
Origin of Biomarkers
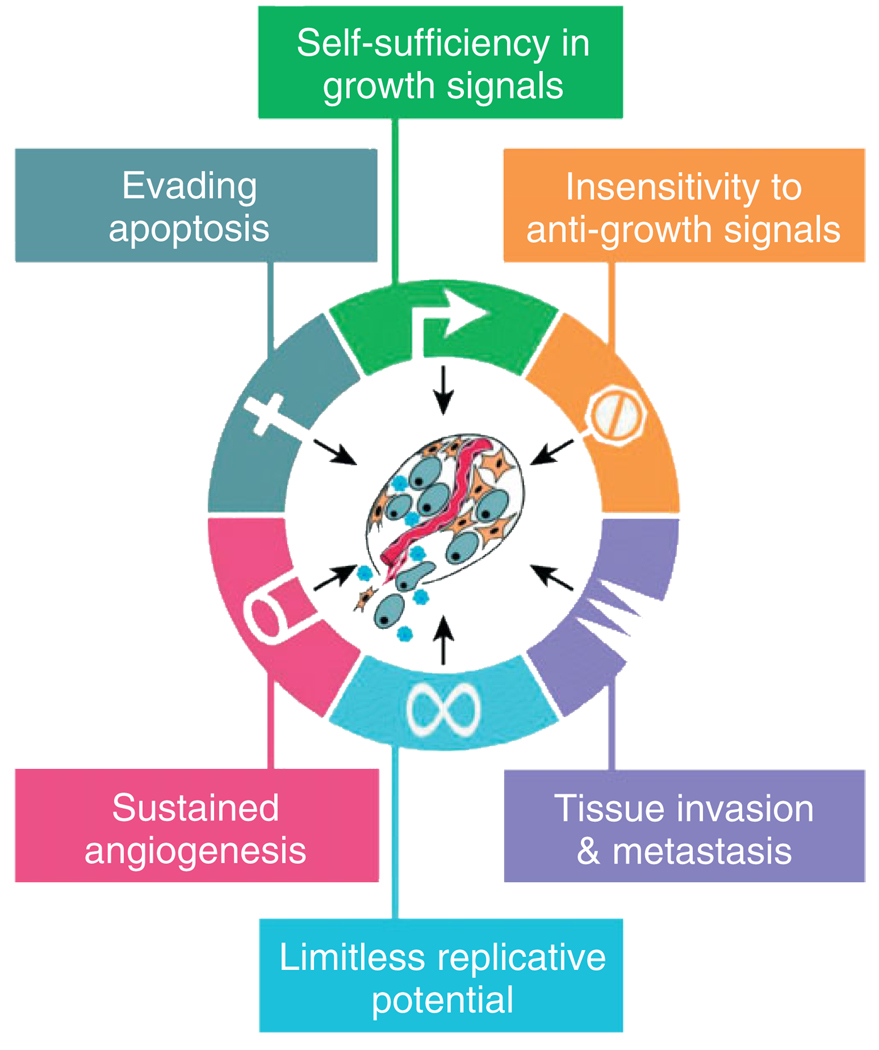
Increasing knowledge of the molecular pathways driving disease development and understanding the mechanism beyond the specific pathways malfunction can provide scientists, pharmacists, and clinicians with possible molecular targets that can be used to generate new diagnostic strategies. In case of tumor progression, Hanahan and Weinberg have defined six important subcellular regulatory systems whose malfunctioning is required for most cancers (Figure 1). These malfunctions include the acquisition of self-sufficiency in growth signals, insensitivity to anti-growth signals, limitless replication, evading apoptosis, sustained angiogenesis, and tissue invasion and metastasis.17 Cancer represents a class of diseases containing more than 100 subtypes whose distinguishing characteristics are related to the tissue of origin and the type and nature of the specific molecular pathways alterations.18 Current diagnostic and prognostic classifications still suffer from major limitations and do not reflect interpatient and intratumoral heterogeneity (which contain benign, cancerous, and stromal cells) impeding precise histological diagnosis of the pathology, raising the need for new molecular markers to improve diagnosis, prognosis, as well as the prediction of the therapeutic response.19,20
FIGURE 1.
The six hallmarkers of cancer. It has been suggested that most if not all cancers must acquired the common set of functional capabilities depicted here during their development, albeit through a variety of possible mechanistic strategies (reprinted, with permission, from Ref 17. Copyright 2000 Cell).
Biomarkers can be altered genes, RNA products, proteins, or other metabolites that reflect the pathological state of the patient. Molecular diagnostics is based on the detection of abnormal nucleic acids sequences, proteins, and other biomolecules derived from biological fluids (serum, urine, saliva, cerebrospinal fluid), which comprise the machinery, or in some cases the by-products, of these regulatory signaling pathways and can therefore be indicative of diseased states.
Genomics and Proteomics in Disease Diagnosis
The spectacular success in sequencing the complete human genomes has dramatically advanced the possibility of realizing personalized medicine.21,22 With the development of gene chips and microarrays, genomics has moved to a functional phase where gene expression can be determined by detecting gene-specific mRNA sequences.
The use of gene expression patterns for molecular profiling of disease has provided new opportunities for molecular diagnosis and prediction of response to therapies.23,24 Fully understanding the molecular processes leading from health to disease, however, is considerably more involved than identifying active gene sequences. Attention is increasingly focusing on proteins and enzymes and their interactions that determine cellular architecture and function.25–27
Proteomics, in contrast to genomics, refers to the systematic study of the total protein complement expressed by the genome of particular cells or tissues, both healthy and diseased.28,29 Proteomic studies to determine the structure and function of each protein and the complexities of protein–protein interactions will be important for developing accurate, effective, and timely diagnostic and therapeutic modalities. Currently, a number of techniques, including western blots,30 immunohistochemical analysis,31 enzyme-linked immunosorbent assays (ELISA),32 or mass spectrometry (MS),33 allow us to test for specific proteins and thus diagnose a particular disease. There is no doubt, however, that completely defining the human proteome is going to involve a much different set of challenges than the sequencing the human genome. As an example, it is essential to know the concentration of a particular protein and not just its presence to determine its functional activity. The accurate quantification of low abundance proteins is therefore one of the biggest challenges in the study of proteomics. Fortunately with the aid of advanced protein-based nanotechnology methods, capable of detecting zeptogram quantities, this problem can be overcome and will be of great use for specific and sensitive diagnosis.34–36
NANOPATTERNED DEVICES
Realizing the full potential of nanotechnology as it pertains to disease diagnosis requires the ability to fabricate nanoscale devices and materials with a high degree of precision and accuracy. Reproducibility is of crucial importance to ensure that device and material variation can be eliminated as a source of diagnostic variability. Fabrication of such nanoscale constructs can proceed from either a ‘top-down’ or ‘bottom-up’ approach. In the ‘top-down’ approach, micro-and nanofabrication techniques, much of which were originally developed for the semiconductor industry, are used to produce biosensing devices with micro- and nanoscale features. The ‘bottom-up’ approach uses the thermodynamically driven accumulation of atoms or molecules by means of directed or self-assembly under tightly controlled conditions to create materials with internal or external nanoscale structure, such as mesoporous thin films or gold nanocrystals.37,38
In this section, we will investigate nanoscale architectures that are produced predominantly from a ‘top-down’ approach, although the fabrication protocols of several of these constructs possess elements of both fabrication methodologies. These architectures also benefit from their ability to identify biological species without the need for florescent or radiological prelabeling, a valuable characteristic for achieving target molecule quantification. All of the nanoscale biosensing devices in this section are included in this review based on their potential to significantly impact point-of-care diagnosis because of their high sensitivity and capability for high throughput screening of biological samples.
Nanoporous Silica Chips
Several studies have indicated that the composition of the low molecular weight proteome (LMWP) extracted from serum may reflect ongoing pathological conditions and thus can be used not only to identify and screen for circulating biomarkers but also to monitor therapeutic efficiency in real time as well.39,40 The LMWP (see Figure 2) is composed of small proteins shed from tissues and cells or peptide fragments derived from the proteolytic degradation of larger proteins. Despite its potential in clinical applications, profiling of the LMWP has proven to be a significant technical challenge because of the extremely high dynamic range of protein concentrations in serum and plasma. This ultimately limits the resolution of most protein screening methodologies in the range of protein sizes of diagnostic interest. Without proper sample preparation, the constituents in the LMWP will be obscured by signals generated from the larger more highly abundant proteins such as albumin. Furthermore, many common enzymes (i.e., thrombin, plasmin, and complement proteins) also circulate in the blood that may initialize endoprotolytic cleavage and cause the LMWP signature to be altered during sample processing. For this reason, sample collection, storage, and processing must be carefully standardized and documented, and occur within a specifically controlled duration. Traditional proteomic-based approaches, such as two-dimensional polyacrylamide gel electrophoresis (2D–PAGE), are labor-intensive procedures with limited dynamic range (>7000 kDa) and with a library of available reagents for targeting proteins that operate over a very narrow pH range, making it difficult to detect small proteins and peptides. 2D–PAGE also requires a large amount of starting material and suffers from low throughput and low sensitivity ultimately limiting its clinical diagnostic value.41 Innovative technologies that can address the issues of the intrinsic complexity of the plasma proteome and the rapid degradation of proteins in sampled blood are necessary to realize effective biomarker discovery. Progress in this area depends on the development of fast, reliable, and highly sensitive and specific sample analysis methods.
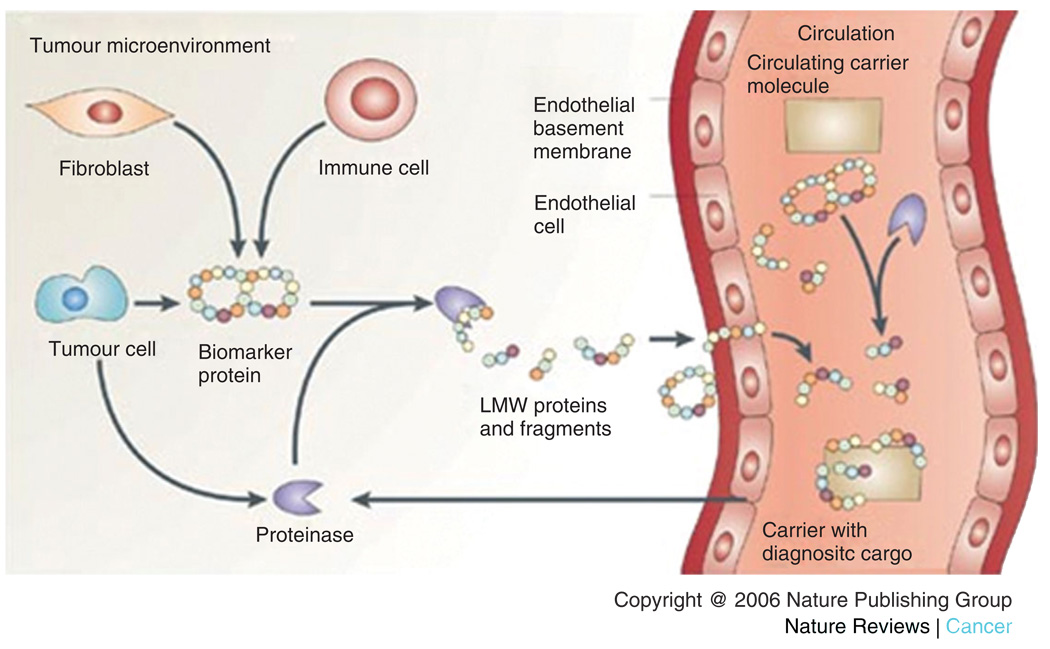
FIGURE 2.
Circulating peptide and protein fragments are shed from all cell types within the tissue microenvironment. Proteolytic cascades within the tissue generate peptide and protein fragments that diffuse into the circulatory system. The identity and cleavage pattern of the peptides provide two important types of diagnostic information (reprinted, with permission, from Ref 40. Copyright 2006. Nature Reviews Cancer).
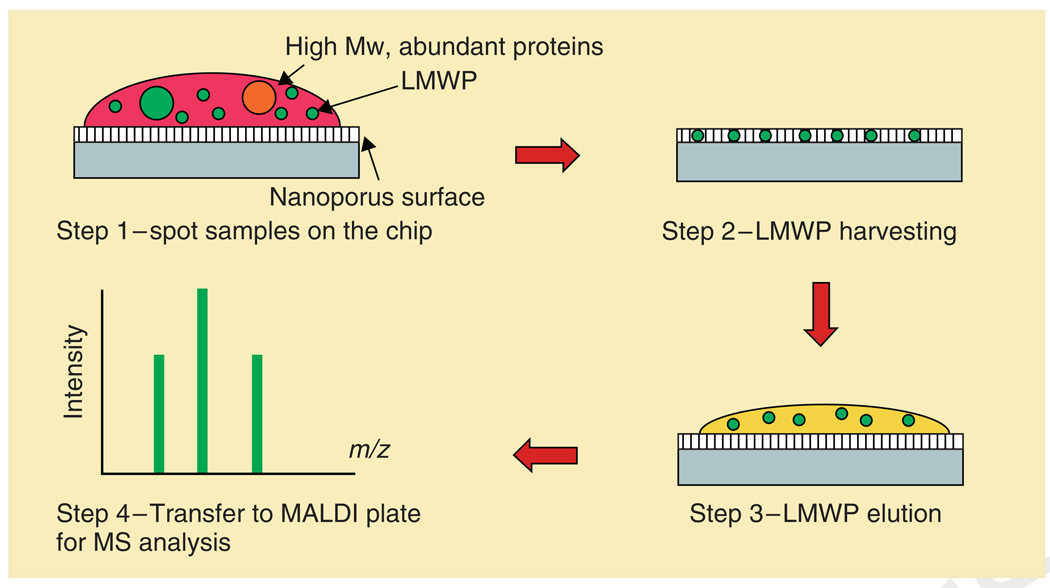
A promising nanotechnology-based technique combining mesoporous silica thin films with spectrographic analysis has been developed, which has demonstrated the ability to efficiently deplete large proteins and selectively enrich and analyze the LMWP in serum samples.42–44 These mesoporous silica chips, with pore sizes in the nanoscale, were synthesized using a process that began with the self-assembly of a mixture of a triblock copolymer [poly(ethylene oxide) (PEO)-poly(propylene oxide) (PPO)- poly(ethylene oxide) (PEO)] and hydrolyzed silicate precursors.45,46 The preferential evaporation of the solvent after dip or spin coating drives the self-assembly of this mixture into a uniform thin-film nanophase by increasing the concentration of polymer in the solution until it exceeds the critical micelle concentration. Mesoporous silica thin films with narrow nanoscale pore size distributions and high surface area to pore volume ratios were formed after removing the organic template through calcination. Serum fractionation using the mesoporous silica thin films was carried out by a rapid four-step on-chip strategy shown in Figure 3. The sample is first spotted on the chip surface (step 1) and incubated to allow the low molecular weight (LMW) proteins to be trapped in the pores (step 2). Next, the chip surface is washed to remove the larger protein species that remained outside the pores. The enriched small molecules are then eluted from the nanopores (step 3) and subjected to further MS analysis (step 4). To validate this approach in the context of complex biological samples, the MS profiles of human serum before and after fractionation using the mesoporous chips were compared, shown in Figure 4. After washing using deionized water, the larger proteins were shown to have been thoroughly removed as demonstrated by a dramatic reduction in signal intensity compared with that of the LMW proteins and peptides. The proteomic profiles depicted in Figure 4(e,f) clearly demonstrate the efficiency of this fractionation strategy in eliminating most of the abundant high molecular weight (HMW) proteins and enriching the otherwise undetectable LMW species present in the serum. An important characteristic of mesoporous silica thin films is the wide variety of achievable pore morphologies. By controlling the pore’s nanoscale characteristics, such as pore size, pore structure, porosity, and the chemical functionalization of the pore surface,47,48 mesoporous silica chips can be tailored to selectively enrich desired components of the LMWP. Thus, mesoporous thin films chips provide a platform with the capability for low-cost production, simple and rapid sample collection, and greatly reduced sample volumes required for analysis. Furthermore, the evaluation of the differential recovery of peptides and proteins from human serum is performed from statistical analysis of spectrographic profiles obtained using MS, a well-established spectrographic technique. Finally, the fabrication of arrays of mesoporous thin films chips with a range of carefully tailored characteristics interfaced through microfluidic channels is also being investigated, which should provide even greater sensitivity and functionality. This proteomic profiling platform allows for rapid, efficient, and flexible collection and analysis of the LMWP from human serum and should therefore prove quite useful in the fields of proteomic biomarker research and clinical proteomic assessment.
FIGURE 3.
Schematic of the harvesting protocol consisting of (1) the deposition of plasma directly onto the chip surface; (2) the washing away of unbound substances; (3) the extraction of bound molecules; and (4) mass spectrometry analysis (reprinted, with permission, from Ref 44. Copyright 2006. American Chemical Society).
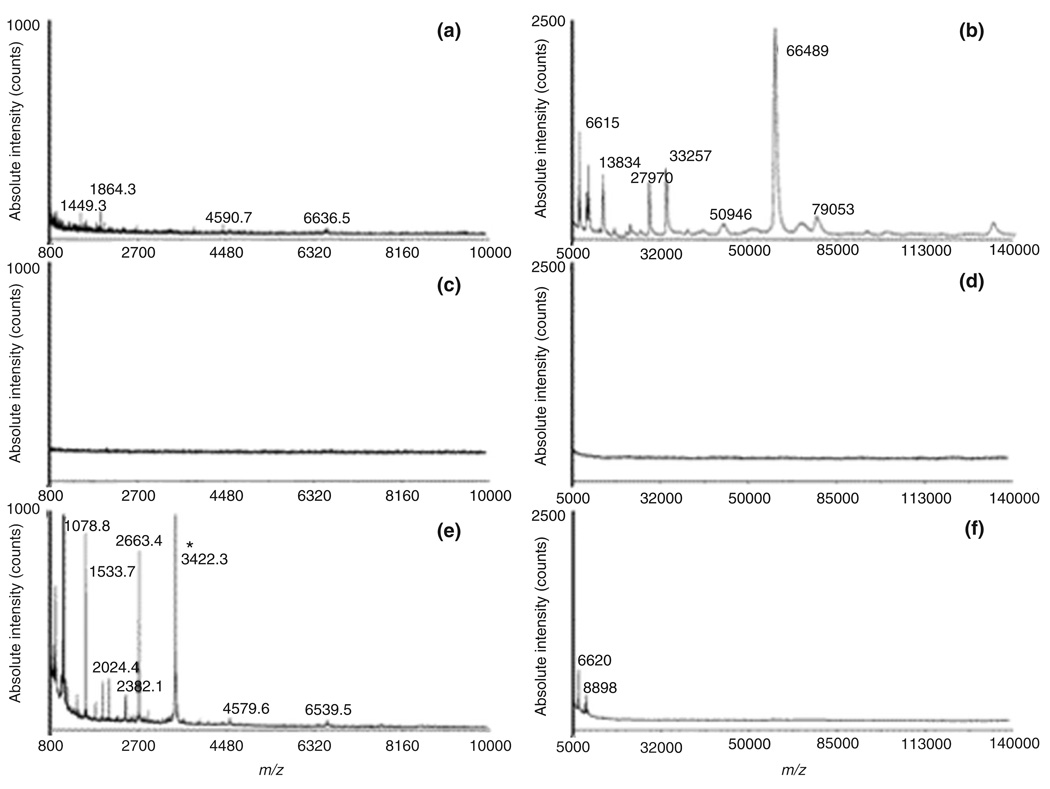
FIGURE 4.
MALDI-TOF profiles of human plasma using as matrixes Alpha-cyano-4-hydroxy-cinnamic acid (CHCA) (left panels) and Sinapinic acid (SA) (right panels). (a), (b) Control experiment without chip incubation [direct mass spectrometry (MS) analysis with no pretreatment]; a plasma aliquot was diluted 100-fold with matrix for MS analysis. (c), (d) Control experiment using a solid silica surface (nonporous). (e), (f) Analysis of human plasma proteins after exposure to a nanoporous silica chip. All experiments used 5 µL of human plasma spiked with calcitonin at a concentration of 1 µg/mL. For obtaining spectra (c) and (e), chip surfaces were extracted directly with matrix solution; for spectra (d) and (f), the extract was instead mixed with matrix in a subsequent step (matrix/extract ratio 3:1). The calcitonin peak, only visible in (e), is marked with a star (reprinted, with permission, from Ref 40. Copyright 2006. American Chemical Society).
Nanowire Biosensors
The field-effect transistor (FET), the basic three terminal logic device found in almost all integrated circuits and which has been in development for over 50 years, can be fabricated using a number of different semiconductor materials, such as silicon, germanium, and gallium arsenide.49 This device can be configured as a sensor by eliminating the metal gating electrode and directly modifying the gate oxide with molecular receptors capable of binding to charged species of interest resulting in a direct change to the channel conductance. The FET channel conductance indicates the amount of current that can flow through the channel for a given bias voltage.50,51 Boron-doped silicon nanowires (SiNWs) have been used to create extremely sensitive, realtime, electrically based sensors for the quantitative detection of biological species.52 The sensitivity of FETs whose active areas are comprised of nanowires is greatly enhanced over conventional bulk semiconductor FETs because of their small diameters, between 10 and 20 nm, and high surface area to volume ratios, which allow the binding of a target molecule to cause accumulation or depletion of carriers throughout a much larger percentage of the channel cross-section. Furthermore, a very large percentage of the nanowire surface is available for functionalization with chemical or biological molecular recognition units that provide selective analyte detection.53–55 Devices based on chemically functionalized or antigen-conjugated SiNW have been demonstrated to successfully transduce biological binding events in real time at sensitivities approaching 0.9 pg/mL.
Because FETs respond to changes in surface charge, careful consideration must be given to maintaining the low ionic strength of the buffer solution while performing measurements. The reason for this is that increased ionic strength compresses the electrical double layer around the wires; the spatial extent to which the electric field of a charged and bound target can interact with the nanowire is limited by the Debye screening length.56 Physiological systems such as serum have relatively high ionic strengths resulting in low nanowire FET sensitivity. This has been overcome by either diluting or desalting samples before analysis to increase the Debye length and allow the detection of the charge on the wire surface.57,58
The accurate identification and categorization of diseases are essential for public health, and multiplexed testing is very often required for individualized screening and diagnostics because of the several subtypes of diseases and conditions which present similar symptoms.59,60 MicroRNA (miRNA), an 18- to 24-nucleotide (nt) noncoding RNA molecule in the genes of humans, plants, and animals, is emerging as a key player in gene regulation, and many applications within and beyond the realm of cancer have been proposed and developed in recent years.61–63 An SiNWs device has been developed using peptide nucleic acids as baits for the direct recognition of unlabelled, target miRNAs. Resistance change measured before and after hybridization correlates directly to the concentrations of hybridized target miRNA. This technique enables the identification of fully matched versus mismatched miRNA sequences with sensitivity as low as 1 fM in total RNA extracted from cancer cells. This approach is a promising example of label-free, early detection of miRNA as a biomarker in cancer diagnostics with very high sensitivity and good specificity.64
Another interesting approach was based on the real-time, label-free, electrical detection of vascular endothelial growth factor (VEGF) for cancer diagnosis using an anti-VEGF aptamer-modified SiNW-FET. In this system, the target VEGF molecules consistently act on the gate dielectrics, and their recognition to the anti-VEGF aptamers induces changes in the detection currents. The detection limit of VEGFs in this study was determined as 104 pM, a sensitivity treshold compatible with the natural concentration of this protein in blood.65 Recent results suggest the possibility of incorporating significant numbers of nanowire FETs into large-scale arrays with complex hierarchical structure for high-density biosensor, electronic, and optoelectronic applications.66 A patterning methodology, referred to as superlattice nanowire pattern transfer (SNAP), has been developed to fabricate high-density, highly ordered nanowire arrays with wire diameters and center-to-center distances as small as 8 nm and up to 106 of aspect ratios.67 Multiplexed biosensing can be achieved by postfabrication and functionalization of nanowires with a range of different biorecognition elements. The multiplexing system should be capable of simultaneous detection of multiple biomolecular targets, such as nucleic acids or proteins, by correlating the change in their respective conductance with the configuration of electrical charges contained within the bound molecules.
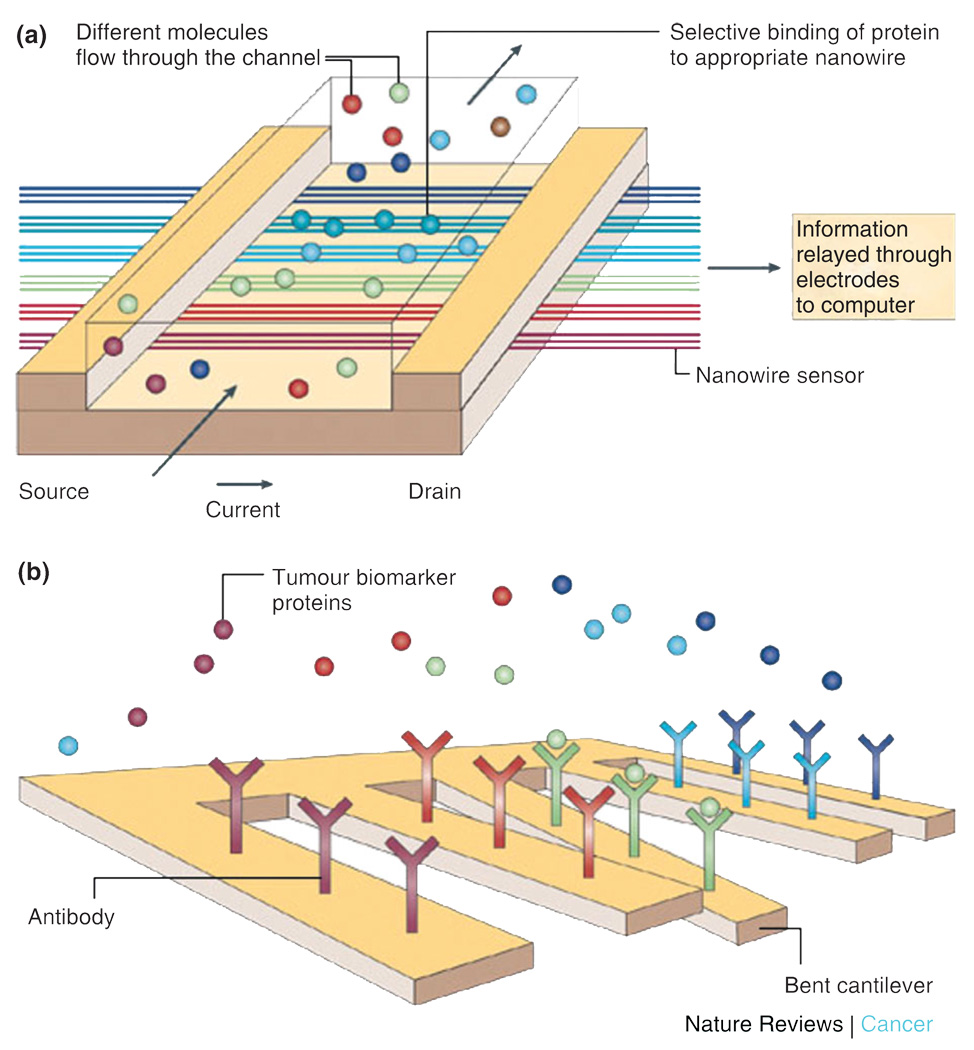
To realize the considerable potential of sensors based on electrical detection, arrays of independent nanodevices must be fully integrated with on-chip computation and wireless communication. Nanowire assemblies with microchip technology, as shown in Figure 5(a), are emphasized as a key step toward the ultimate goal of multiplexed detection at the point-of-care using portable, low power, electronic biosensor chips.68
FIGURE 5.
(a) Nanowires deployed within a microfluidic system. The different colors represent that different molecules adsorb or affinity-bind to different nanowire sensors. The binding causes a change in conductance of the wires, which can be electronically and quantitatively detected in real time. The working principle is that of a (biologically gated) transistor and is illustrated in the insert. The charges of the binding protein disrupt electrical conduction in the underlying nanowire. The nanosize of the wire is required to attain high signal-to-noise ratios. (b) Nanocantilever array. The biomarker proteins are affinity-bound to the cantilevers and cause them to deflect. The deflections can be directly observed with lasers. Alternatively, the shift in resonant frequencies caused by the binding can be electronically detected. As for nanowire sensors, the breakthrough potential in nanocantilever technology is the ability to sense a large number of different proteins at the same time, in real time (reprinted, with permission, from Ref 68. Copyright 2005. Nature Reviews Cancer).
Micro- and Nanocantilever Arrays
One widely investigated architecture to realize this label-free strategy is the micro- or nanocantilever, with the nomenclature indicating the length scale of the cantilever beam thickness. These cantilevers, usually microfabricated in silicon or a piezoelectric material such as quartz, can operate either statically, by measuring absolute cantilever deflection, or dynamically, by measuring resonance frequency shifts.69 The static mode is based on the bending of the cantilever because of surface stress induced when ligands or target single-stranded DNA (ssDNA) bind or hybridize to their conjugate receptor or probe ssDNA immobilized to the cantilever surface. The micro- and nanocantilevers measure the absolute beam deflection using optical reflection from the cantilever surface.70 The dynamic mode operates by measuring a change in the cantilever resonance frequency, which occurs because of a change in adsorbed mass.71–73 Micro- and nanocantilevers, in addition to quantifying DNA hybridization with single base pair resolution, have used been to detect a variety of biomarkers (Figure 5(b)) using both the static74 and dynamic75 measurement modes. One such biomarker that has been rigorously studied is prostate-specific antigen (PSA), a 33–34 kDa glycoprotein exhibiting chymotrypsin-like protease activity. Micro- and nanocantilevers have been used to detect concentrations of PSA between 0.2 ng/mL and 60 µg/mL in solution also containing human serum albumin (HSA) and human plasminogen (HP) at concentrations of 1 mg/mL. The useful clinical range for PSA is 4–10 ng/ml which falls within this range. Perhaps of even greater diagnostic interest, cantilevers surface functionalized with the appropriate polyclonal anti-PSA antibody have also been used to determine the ratio of complexed (bound) to free (unbound) conformations of PSA whose ratio should be more indicative of malignancy than absolute PSA levels.76 PSA illustrates, however, the inherent difficulty of biomarker discovery, and thus the urgency for high throughput screening methodologies, as there is still significant controversy as to its ultimate effectiveness for early prostate cancer detection.77 Other emerging biomarkers include the angiogenic factors VEGF and fibroblast growth factor (bFGF) and the anti-angiogenic factor endostatin.78 Changes in the concentration of these components in sampled blood have been demonstrated to be predictive of the extent and risk of recurrence in patients with soft tissue sarcoma. The ability fabricate assays arrays of differentially functionalized cantilevers allow for the simultaneous monitoring of low concentrations. Micro- and nanocantilevers impart significant advantages over other molecular-based diagnostic and screening technologies because of their sensitivity, compatibility with silicon technology, and capacity for microfluidic integration. Through multiplexed testing using arrays of differentially functionalized cantilevers, this device architecture demonstrates great potential for high throughput protein screening, a crucial component for effective proteomic profiling.
Molecularly Gated Single Electron Transistors
As previously noted, FETs can be converted to effective biosensors by eliminating the gate metal and using chemically functionalized ligand or DNA probes directly on the gate oxide to capture charged species of interest and thereby cause a change in the channel conductance. Nanowire FETs extend this principle but with reduced channel cross-section and increased surface area to volume ratio allowing for the generation of much greater channel conductance changes for a given amount of immobilized surface charge (accumulated through probe-target binding events).
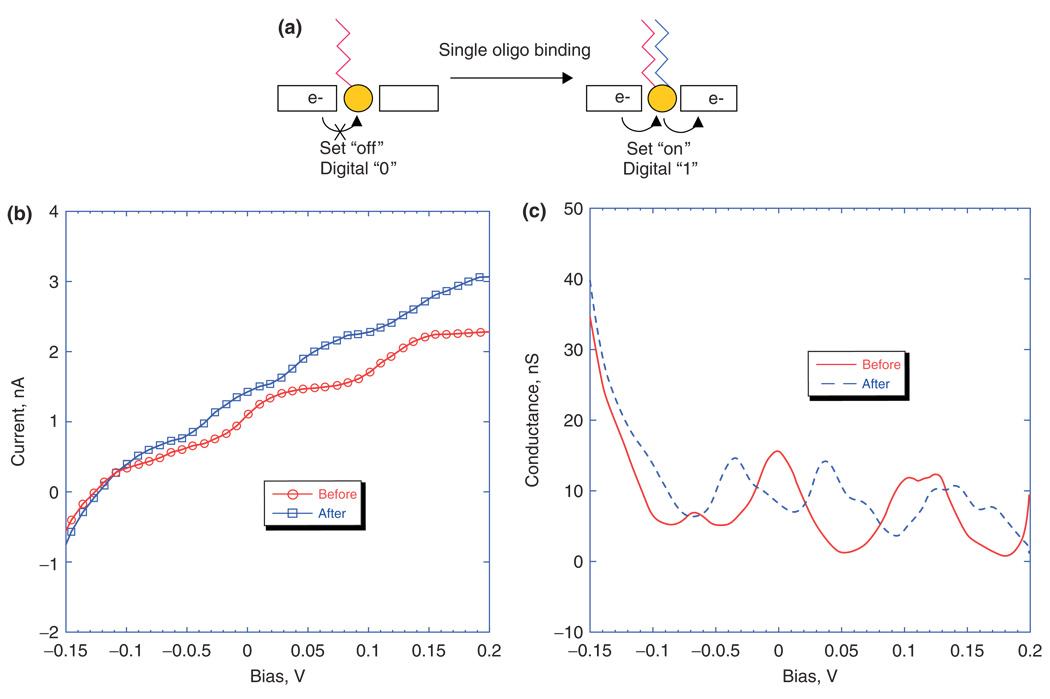
Efforts are currently underway to develop a third configuration of the FET for achieving highly sensitive label-free detection of proteins, DNA, RNA, and other biologically relevant components. The device is called a single electron transistor (SET) and operates based on the quantum mechanical phenomenon of tunneling. When an electron or hole (also called a carrier) is confined spatially at the nanoscale in three dimensions, the energy levels available to that carrier become discrete, similar to electron orbital occupation in atoms.79 The energy level spacing represents that the energy required to add another carrier to this nanoscale island is equal to e2/2C, where e is the charge of an electron or hole and C is the total capacitance of the island. In an SET, a schematic representation of which is shown in Figure 6(a), the nanoscale island is separated from the source and drain contact electrodes by nanoscale dielectric gaps. These dielectric gaps help the island to maintain the necessary three-dimensional (3D) quantum mechanical confinement of carriers to ensure discrete energy levels. Such devices have been fabricated using a top-down approach for producing the source and drain contacts and a bottom-up approach for the incorporation of nanoparticles, which provide the 3D quantum confinement, through self-assembly with an appropriate linker whose molecular chain length comprises the dielectric gap width.80 Although the dielectric gaps ordinarily represent barriers to conduction, in quantum mechanical systems it is possible for carriers to tunnel through dielectric barriers, provided they are thin enough and there are available energy states on either side which are at exactly the same energy.81 Conduction can occur only when the energy imparted by the external source to drain bias (eVsd) exceeds the energy level separation on the island, provided the energy separation is less than the thermal energy available at a given ambient temperature.79 Under these conditions, conduction is quantized and therefore only increases when the source to drain bias energy becomes large enough to reach the next energy level and incorporate another conductive channel. Nanoparticles synthesized using a host of conductive and semi-conductive materials with diameters of <10 nm have been shown to have total capacitances of less than an attofarad (10−18 farad), sufficient to achieve quantized conduction at room temperature.82 The quantization of conductance in SETs is revealed by observing plateaus in plots of current versus source/drain bias voltage and peaks in plots of conductance versus source/drain bias voltage,80 as depicted in Figure 6(b,c). The energy levels on the nanoscale island can also be shifted through the application of an external electric field, commonly referred to as a gating voltage.81 As the active area of these devices has such low capacitance, only a single externally coupled charge is required to significantly shift the energy levels and thus the conductance peaks. As many biologically relevant molecules have either a net charge or large domains of differential charge, a single binding event can be used to gate the device once an appropriate receptor or probe has been bound to the nanoscale island.80
FIGURE 6.
(a) Illustration of the concept of ‘digital detection’ of oligonucleotide hybridization, and plots of I/V (b) and G/V (c) for positive binding of complementary oligo target to probe. For positive binding, ΔV decreases by approximately half (reprinted, with permission, from Ref 80. Copyright 2006. American Chemical Society).
SETs have thus far been fabricated, using gold nanoparticles as the nanoscale island and self-assembled monolayers to define the dielectric gaps, and been demonstrated to be capable of successfully and accurately measuring single molecule DNA hybridization at femtomolar concentrations and to distinguish between single base pair mismatches in oligionucleotide strands as long as 35 base pairs in real time. These devices have also been able to distinguish homogeneous and heterogeneous mixtures of 25 mer and 35 mer chains of oligionucleotides in distilled water, citrate-stabilized saline, phosphate buffer solution, quanidinium thiocyanate lysis buffer, and human serum.80 The merging of top-down and bottom-up fabrication should also allow for the fabrication of large arrays of SETs capable of multiplexed biosensing. The SET’s capability for direct electrical signal transduction significantly reduces the complexity of the required measurement apparatus, which should dramatically increase its portability, flexibility, and applicability for real-time point-of-care diagnosis.
The aforementioned nanopatterned biosensing architectures represent promising avenues for identifying important molecular components from clinically relevant samples with greater sensitivity and/or less lead time than many current screening and diagnostic methodologies. The next section will transition to nanomaterials used for molecular labeling, imaging, and identification for in vitro studies. These probes provide a powerful tool in determining not only the presence of a particular molecular species but also its distribution within a cell or an animal model.
FREE STANDING NANOPARTICLES
Nanomaterials are defined as materials comprised of basic components, which have at least one dimension in the nanoscale (<100 nm). This confined dimensionality yields a host of unique properties not present in the bulk material, which are being investigated for an array of clinical applications.83 In this section, we will limit our discussion to materials whose basic building blocks have all three dimensions in the nanoscale. These nanoscale structures represent powerful diagnostic tools as they can be surface functionalized with a range of specific targeting agents or amplification modalities and then systemically circulated in vivo to locate and monitor specific biological targets. They can also be used for in vitro assays and experiments to increase the sensitivity of a particular assay or label important subcellular or molecular features of cells. Nanomaterials are typically fabricated using a bottom-up approach in which molecular components self-assemble into more complex structures.
We will first cover the use of magnetic iron oxide nanoparticles as MRI contrast agents. We will then investigate gold nanoparticles that can absorb light through the excitation of a surface plasmon resonance (SPR), a phenomenon with wide ranging applicability. The next topic will be a discussion on bio-barcodes, an assay amplification strategy that incorporates both gold and magnetic nanoparticles. This will be followed by a look at aptamers, a targeting modality which, when conjugated to the surface of gold and magnetic nanomaterials, can be used for both targeting as well as for assay amplification. The use of QDs in a variety of labeling strategies based on the ability to tune their optical emission spectra to almost any visible wavelength will follow next. Finally, this section will conclude with a treatment of multiplex dendrimers whereby carbon lattices are used to encapsulate contrast agents allowing for very tight control over their nanoscale structure.
Magnetic Nanoparticles
Since the early 1980s, superparamagnetic nanoparticles and their derivatives have been developed and commercialized to enable the purification, separation, and detection of important components within biological samples.84,85 More recently, the use of this class of nanomaterials has been extended in vivo to drug delivery and to the development of molecular contrast agents for MRI.86 MRI is a diagnostic imaging technique, which uses a strong external magnetic field to align the nuclear magnetization of hydrogen atoms incorporated on water or fat molecules within the body and then uses radiofrequency (RF) waves to excite these aligned magnetizations out of equilibrium.87 The excited hydrogen nuclei then relax back into their aligned equilibrium positions with two characteristic relaxation times, the longitudinal relaxation time T1 and the transverse relaxation time T2, resulting in the emission of the excitation energy absorbed from the RF waves.87 Stronger external magnetic field strength leads to shorter relaxation times and thus better image contrast and resolution. An MRI with an external magnetic field of 7 Tesla is capable of a minimum resolution of 50–100 microns.88 By imaging techniques weighted toward either T1 or T2, different tissue contrasts can be realized. Furthermore, the relaxation times are also highly dependent on the chemical composition of the tissue yielding enhanced soft tissue contrast. This ability to better image soft tissue and the use of nonionizing radiation in the imaging process endow MRI with many important advantages as compared with other imaging modalities such as computed tomography (CT). Colloidal superparamagnetic iron oxide (SPIO) have been investigated as MRI contrast agents because of their ability to reduce T2 proton relaxation times of specific tissues.89 The tissue is targeted through the surface functionalization of the colloidal nanoparticles with recognition elements, such as antibodies or ligands, leading to an improvement of the MRI resolution down to the level of single cells and allowing for the specific recognition of the molecular components of important cellular features and processes.89
The SPIO nanoparticles are synthesized using a variety of techniques, the easiest and most common being an aqueous co-precipitation process initiated by mixing an iron salt with polymer surfactants under alkaline conditions.89 The precise pH value in the solution and the amount and structure of the surfactant-coating materials play the predominant role in tuning the nanoparticle properties.90,91 High-temperature decomposition of organometallic precursors has also been used to improve nanoparticle size control. This technique is capable of producing uniform spherical Fe3O4 nanoparticles with a size variation of <2 nm and diameters ranging from 4 to 20 nm.92
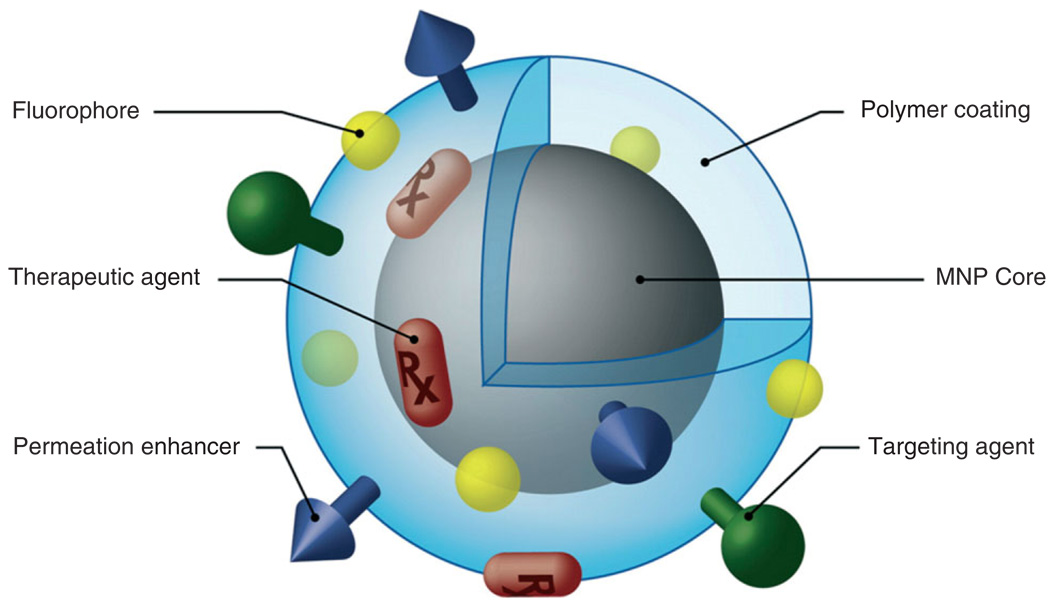
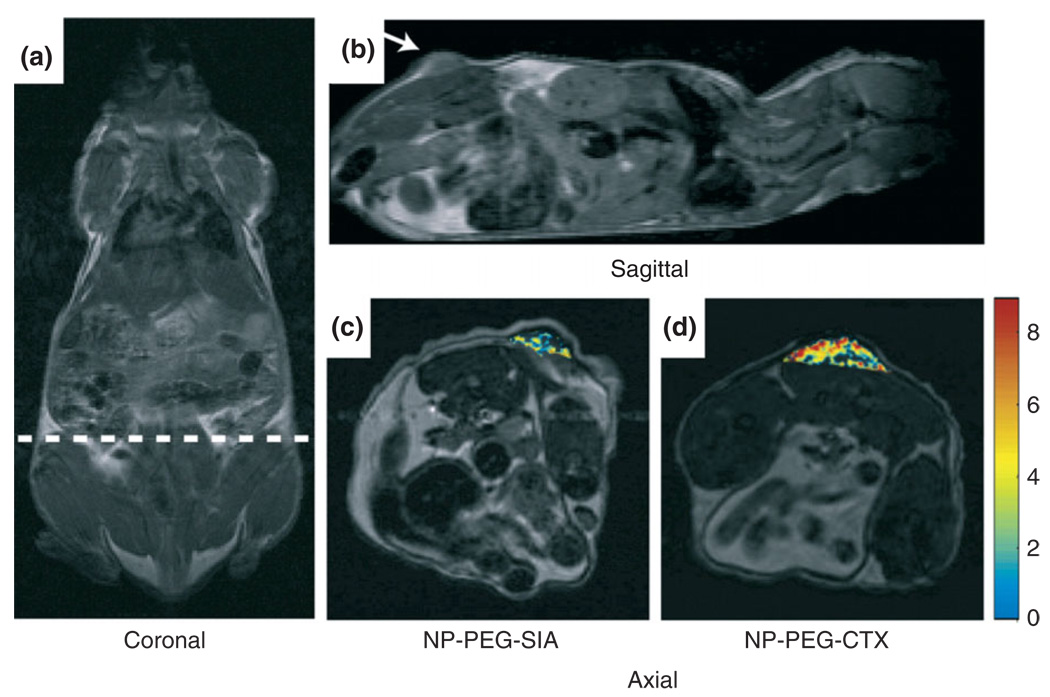
Once the magnetic nanoparticles have been synthesized, they are coated with various types of chemical modifiers. These modifiers can include polymers to prevent nanoparticle aggregation and functional ligands, organic dyes, permeation enhancers, or antibodies (see Figure 7) to imbue them with greater biological functionality. SPIO nanoparticles can be coated with poly(ethylene glycol) (PEG) to avoid nanoparticle uptake by macrophages allowing for extended blood circulation time in vivo.94,95 CTX-targeted iron oxide nanoparticles have been demonstrated by Sun et al. to specifically accumulate in 9L glioma flank xenografts in vivo, resulting in more thorough contrast enhancement of these tumors in comparison with non-targeted control nanoparticles, as shown in Figure 8. Furthermore, Artemov et al.97 has used streptavidin-conjugated SPIO nanoparticles for magnetic resonance molecular imaging of Her-2/neu receptors expressed by breast cancer cells in vitro. The receptors were tagged with biotinylated monoclonal antibodies, which allowed for the binding of streptavidin-conjugated nanoparticles. The contrast level of the resulting image was proportional to the level of expression of Her-2/neu receptors.
FIGURE 7.
Magnetic nanoparticle possessing various ligands to enable multifunctionality from a single nanoparticle platform (reprinted, with permission, from Ref 93. Copyright 2008. Elsevier B.V.).
FIGURE 8.
Magnetic resonance imaging (MRI) anatomical image of a mouse in (a). Coronal plane with the dotted line displaying the approximate location of the axial cross-sections displayed in (c) and (d). Anatomical image in (b). Sagittal plane displaying the location of the 9L xenograft tumor. Change in R2 relaxation values for the tumor regions (superimposed over anatomical MR images) for mouse receiving (c) non-targeting PEG-coated iron oxide nanoparticles and (d) CTX-targeted PEG-coated iron oxide nanoparticles 3 h post nanoparticle injection (reprinted, with permission, from Ref 96. Copyright 2008. Wiley-VCH Verlag GmbH & Co.).
Clustering of superparamagnetic iron oxide nanoparticles inside the hydrophobic core of a micelle further increases the T2 relaxivity more than 10 times over single SPIO particles at the same Fe concentration.98 Selective imaging with these micelles was demonstrated by surface functionalizing them with a cyclic arginine-glycine-aspartic acid (RGD) ligand targeted to vβ 3 integrins on the surface of tumor endothelial cells, which subsequently induced receptor-mediated endocytosis of the micelles.99
Quantum Dots
QDs consist of a semiconductor core encapsulated by another semiconductor shell with a typical diameter of 2–10 nm. Because of their tunable nanoscale dimensions, high photostability, broad absorption spectra, and narrow emission bands, QDs have been used as florescent labels to optically image a host of biological structures and processes, ranging from DNA, small organelles, and tumors to cell–cell interactions and cell signaling processes.100–102 The quantum confinement effect allows for careful control of the emission properties of QDs by varying their size and material composition.103 The fact that multiple QDs may be excited by a single excitation wavelength, thanks to their aforementioned broad absorption spectra, allows for them to facilitate multiplexed diagnosis.104–107 Figure 9 illustrates the impressive multiplexing capability of QD tags in live animals, comparing with the detection sensitivity and spectral features of encoded fluorescent proteins (green fluorescent protein, GFP).13 Although QDs themselves are insoluble in water, their active surface can be conjugated by a layer of functionalized silica or any number of linkers, including mercaptoacetic acid, dihydrolipoic acid, or modified polyacrylic acid, thus rendering them biocompatible.108 In addition, QDs can be adapted for specific target recognition by coating them with antibodies, streptavidin, or oligonucleotides.
FIGURE 9.
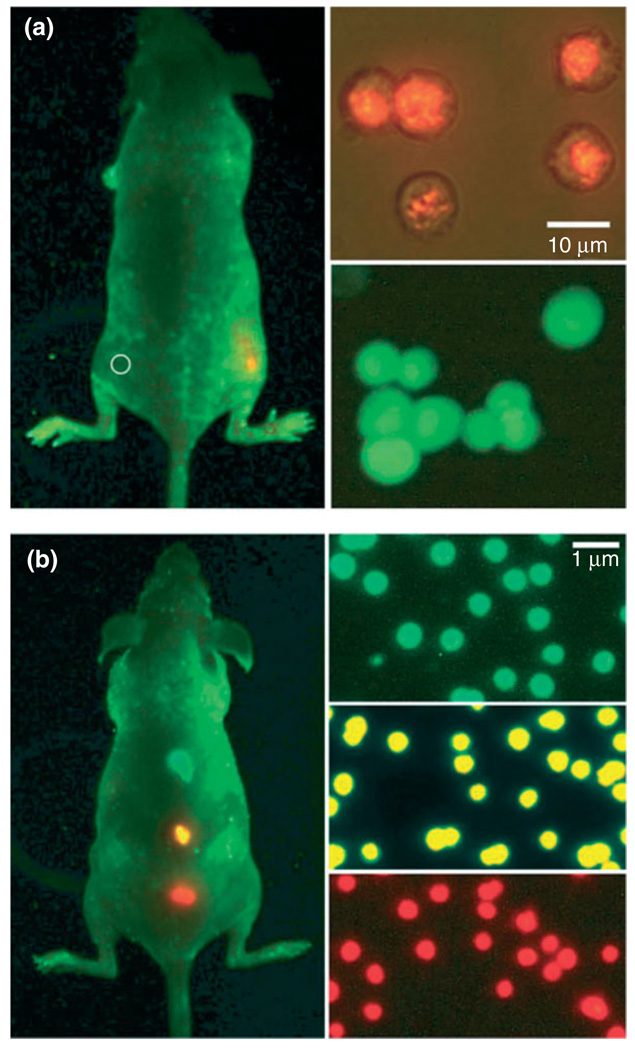
Sensitivity and multicolor capability of quantum dot (QD) imaging in live animals. (a) Sensitivity and spectral comparison between QD-tagged and green fluorescent protein (GFP)-transfected cancer cells and (b) simultaneous in vivo imaging of multicolor QD-encoded microbeads. The right-hand images in (a) show QD-tagged cancer cells (orange, upper) and GFP-labeled cells (green, lower). Approximately 1000 of the QD-labeled cells were injected on the right flank of a mouse, whereas the same number of GFP-labeled cells was injected on the left flank (circle) of the same animal. Similarly, the right-hand images in (b) show QD-encoded microbeads (0.5 m diameter) emitting green, yellow, or red light. Approximately 1–2 million beads in each color were injected subcutaneously at three adjacent locations on a host animal. In both (a) and (b), cell and animal imaging data were acquired with tungsten or mercury lamp excitation, a filter set designed for GFP fluorescence and true color digital cameras. Transfected cancer cell lines for high level expression of GFP were developed using retroviral vectors, but the exact copy numbers of GFP per cell have not been determined quantitatively (reprinted, with permission, from Ref 13. Copyright 2006. Nature Biotechnology).
A multiplex immunoassay has been developed by Goldman et al.109 for the simultaneous and sensitive detection of cholera toxin, ricin, shiga-like toxin 1, and staphylococcal enterotoxin B using the relevant antibodies conjugated to QDs with different sizes, which lie in different colors. In addition, a multiplex diagnostic system using a microfluidic chip and antigen-coated QDs embedded in polystyrene microbeads was developed by Klostranec et al.110 The integrated device is able to detect antibodies against hepatitis B virus, hepatitis C virus, and HIV in serum samples with sensitivity at the level of near picomolar. The microfluidic system enables the approach to be more automatic, accurate, and efficient, rendering it 50 times more sensitive than the currently available methodology using the same antigen and antibodies.
Gold Nanoparticles
Gold nanoparticles represent another promising diagnostic technology as their optical properties can easily be tuned and their surfaces functionalized using a variety of well-characterized chemical moieties (thiols, disulfides, amines).111 These nanoscale constructs can be fabricated either as solid gold spherical nanoparticles with diameters in the range of 0.8–250 nm, asthin gold shells surrounding a dielectric core (i.e., silica) or as high aspect ratio nanorods.111 Au nanoparticles, unlike their QD counter parts, do not emit light but absorb and scatter it in a process called SPR.111 SPR is a phenomenon whereby a coherent oscillation of electrons at the surface of a gold nanoparticle (or thin film) is excited by incident electromagnetic radiation of a particular frequency causing the incident radiation to be either absorbed, scattered, or both.112 This oscillation, or plasmon, can only be sustained in materials (including the noble metals gold, silver, and copper) that possess a dielectric constant that is complex-valued with a negative real part and a slightly positive imaginary part.111,112 The size and shape of the Au nanoparticles determine their optical resonance and can be tuned to achieve absorption and scattering at electromagnetic wavelengths from visible light to the mid infrared.113,114 The resonance frequency of the excited plasmon is also highly sensitive to the local refractive index of the solution in which the nanoparticles are dissolved, but only at small distances (nanometers) because of the exponential decay of the evanescent field normal to the surface.112,114 This property can be used to determine both the presence and binding kinetics of molecular targets adsorbed to the particle surface.
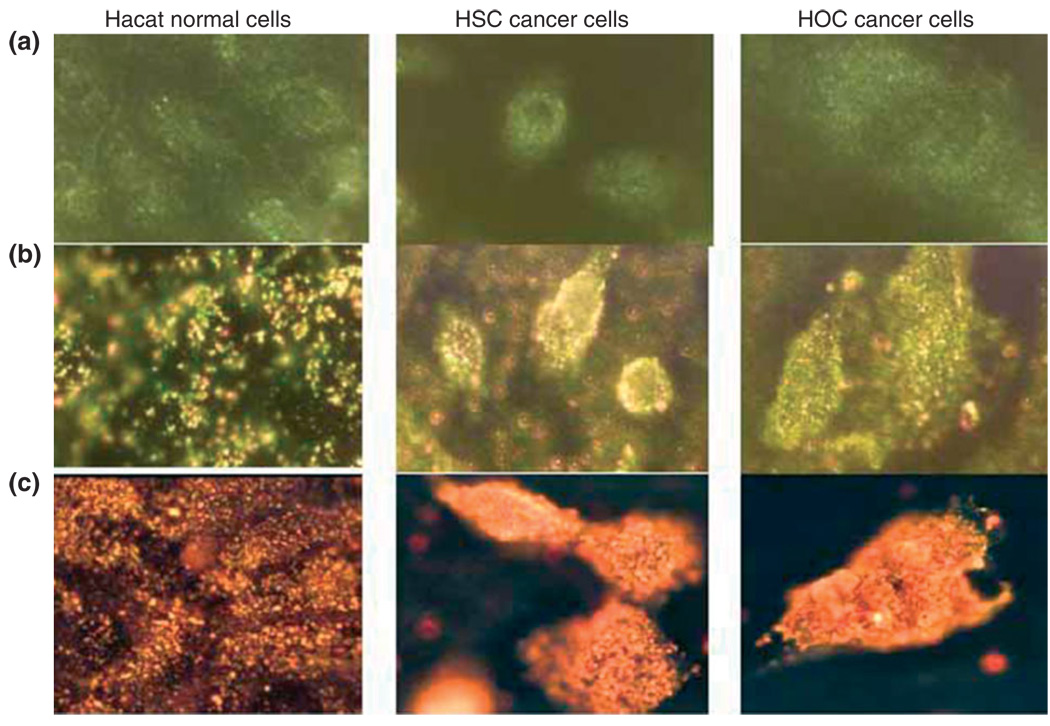
When used for in vitro assays, the binding of Au nanoparticle-labeled recognition elements to their respective targets leads to aggregation of the nanoparticles resulting in a color change in the optical signal as compared with that for the unbound monodispersed gold nanoparticle solution.111,115 Figure 10 shows anti-endothelial growth factor receptor (EGFR)-conjugated gold nanospheres (Figure 10(b)) and nanorods (Figure 10(a)) bound to cancer cells in an organized fashion, as compared with their random distribution around normal cells, thus enabling the optical differentiation and detection of the cancer cells.116 Hirsch et al. has developed an immunoassay capable of quickly detecting (within 10–30 min) subnanogram/mL quantities of various analytes using gold nanoshells optimized for the near-infrared, including rabbit lgG in different media.117 For in vivo imaging, nanoparticles optimized for a subset of the near-infrared (650–900 nm) are frequently used, as the body and biological tissue are highly transmissive in this wavelength range.111
FIGURE 10.
(a) Light-scattering images of normal and cancer cells without nanoparticles.
(b) Light-scattering images of normal and cancer cells after incubation with anti-endothelial growth factor receptor (EGFR) antibody-conjugated gold nanospheres.
(c) Light-scattering images of normal and cancer cells after incubation with anti-EGFR antibody-conjugated gold nanorods. HOC, human osteocalcin; HSC, hematopoietic stem cells (reprinted, with permission, from Ref 116. Copyright 2006. American Chemical Society).
Aptamer-Conjugated Nanoparticles
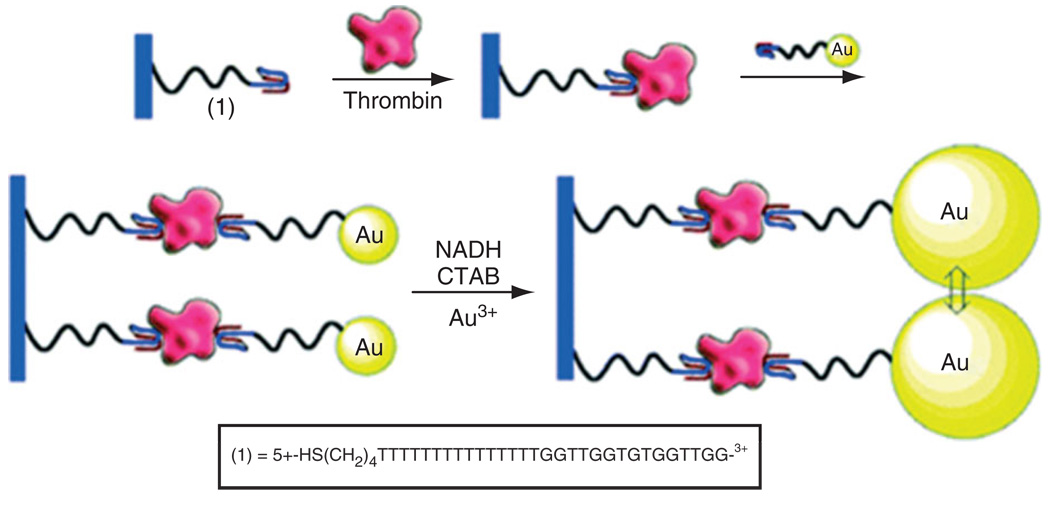
The in vitro detection of specific mRNA and DNA sequences originating in vivo from circulating cancer cells and micrometastases has proven to be a very useful diagnostic technique, because their extracellular instability leads to low extracellular lifetimes and prevents significant circulation.118 Thus, there is a high correlation between the presence of these specific sequences and the presence of diseased cells in biological samples. Some of the most widely used techniques for detecting these DNA and mRNA sequences are based on the use of the polymerase chain reaction (PCR) to amplify these biomarkers in blood and tissue samples by six to eight orders of magnitude.118,119 These PCR-based techniques have been demonstrated to yield a detection limit of one diseased cell in 1–10 million normal cells.118 This extreme sensitivity can also increase the number of false positives, however, because of the combination of the small probability that non-target sequences will initiate the amplification process and the small number of total molecules necessary to achieve significant amplification. Thus, the clinical effectiveness of this technique is limited.118,120,121 Fluorescent and magnetic nanoparticles surface functionalized with a variety of biological recognition elements, such as aptamers and antibodies, have been used to screen for specific cells, including circulating cancer cells, in buffer, blood, and fetal bovine serum, and to extract those cells from the samples using magnetic fields.122 Aptamers are receptors constituted of short DNA or RNA sequences, which have been selected in vitro from a large library of random sequences to bind a host of biological components in a manner similar to antibodies.123,124 The apatamer selection process, referred to as systematic evolution of ligands by exponential enrichment (SELEX), was first reported in 1990 by Ellington and Szostak.125,126 The unique features of aptamers, making them superior to antibodies in clinical use, include the ability to produce them from repeatable chemical synthesis, as opposed to antibodies which must be biologically synthesized in an animal model, and their ability to fold into complex 3D structures with distinct molecular binding motifs. Gold nanoparticles functionalized with thiolated aptamers (about 80 aptamers per particle) were used for the optical transduction of aptamer–protein interactions (see Figure 11).127 The binding of aptamers to a thrombin protein target resulted in aggregation of the gold nanoparticles. Removal of the aggregates from solution with the use of a centrifuge precipitation technique led to a decrease in the solution’s plasmon absorbance as compared with a sample with an equal concentration of unbound dispersed gold nanoparticles.
FIGURE 11.
Amplified detection of thrombin on surfaces by the catalytic enlargement of thrombin aptamer-functionalized Au nanoparticles (reprinted, with permission, from Ref 127. Copyright 2006. American Chemical Society).
Nanoparticle-Based Bio-Bar Codes
Effective clinical biomarker screening requires three crucial capabilities to be effective, the ability to look for several targets simultaneously, the ability to detect small concentrations of proteins in samples containing a complex mixture of biological constituents, and the ability to operate using minimal sample sizes. A variety of multiplex assays have already been developed, such as microarrays128,129 and microsphere-based flow cytometry,130 capable of high throughput screening for a wide range of different molecular species simultaneously. Greater assay sensitivity has also been achieved through either directly amplifying the quantity of low abundance targets, such as the replication of DNA and RNA sequences using PCR, for use with a technique such as microarray analysis, or by developing more sensitive detection methodologies, several of which are described in this review. Finally, microfluidic systems are being developed to achieve a lot of these techniques, such as flow cytometry, with greatly reduced sample volumes.131
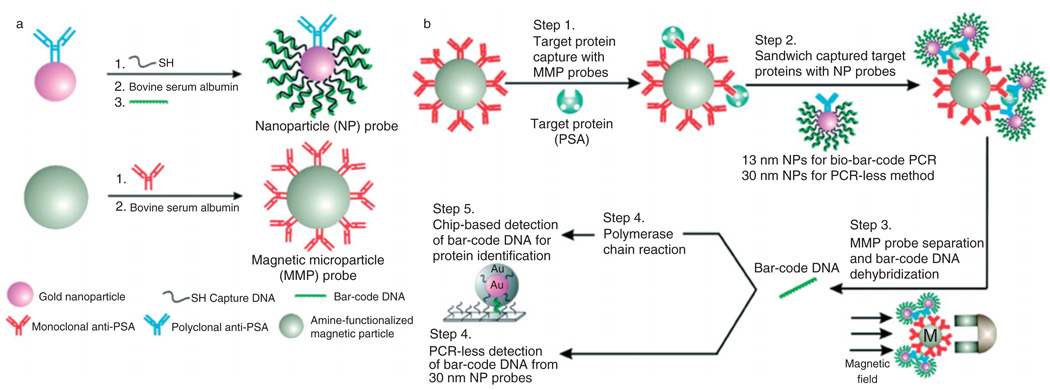
One of the more promising recent developments with the potential for multiplexed, sensitive, low volume biomarker analysis is the bio-barcode. The first prototypes were developed by Christine Keating’s group at Pennsylvania State University.132 They used an aluminum oxide film as a stencil to create metallic bars, nanometers in diameter and microns in length, with alternating submicron bands of metals with varying optical reflectance. These bars were then coated with a targeting element that bound to their targets by affinity capture. This system allowed for multiplexed analysis, because the array of metallic bands could be arbitrarily controlled, and was therefore not limited by the range of available fluorophores. Furthermore, a limited amount of sample was required as the analysis could be performed directly in the sample with no further processing. In 2003, Chad Mirkins group at Northwestern University developed a different barcode system based on the use of magnetic microparticles and gold nanoparticles.133 The magnetic microparticle is functionalized with one recognition element, such as aptamers or antibodies, and the gold nanoparticle is conjugated by second similar recognition element as well as large numbers of specific double-stranded DNA sequences. The two particles then sandwich and extract a specific target using a magnetic field. Although all of the magnetic microparticles are captured, only gold nanoparticles that have bonded to the target are captured. The double-stranded DNA immobilized on the gold nanoparticle surface is then dehybridized to release one of the strands into solution, either chemically (dithiothreitol)134 or thermally,135–137 and then experimentally detected using chip-based DNA techniques with or without PCR. The bio-barcode assay is illustrated in Figure 12. This bio-barcode technique not only incorporates multiplexing and small sample volumes, but it is also capable of significant signal amplification, given the high ratio of DNA barcodes to target recognition elements. The sensitivity of this methodology exceeds that of ELISA by up to 106, yielding the possibility for the use of low abundance biomarkers in diagnosis whose concentrations where considered too low to be useful with previous technologies. The bio-barcode assay has been demonstrated to successfully detect free PSA at concentrations in the range of 3aM to 300 fM.
FIGURE 12.
Bio-barcode assay. (a) Probe design and preparation. (b) A magnetic probe captures a target using either monoclonal antibody or complementary oligonucleotide. Target-specific gold nanoparticles sandwich the target and account for target identification and amplification. The barcode oligonucleotides are released and detected using the scanometric method (reprinted, with permission, from Ref 133. Copyright 2006. Science).
Multiplex Dendrimers
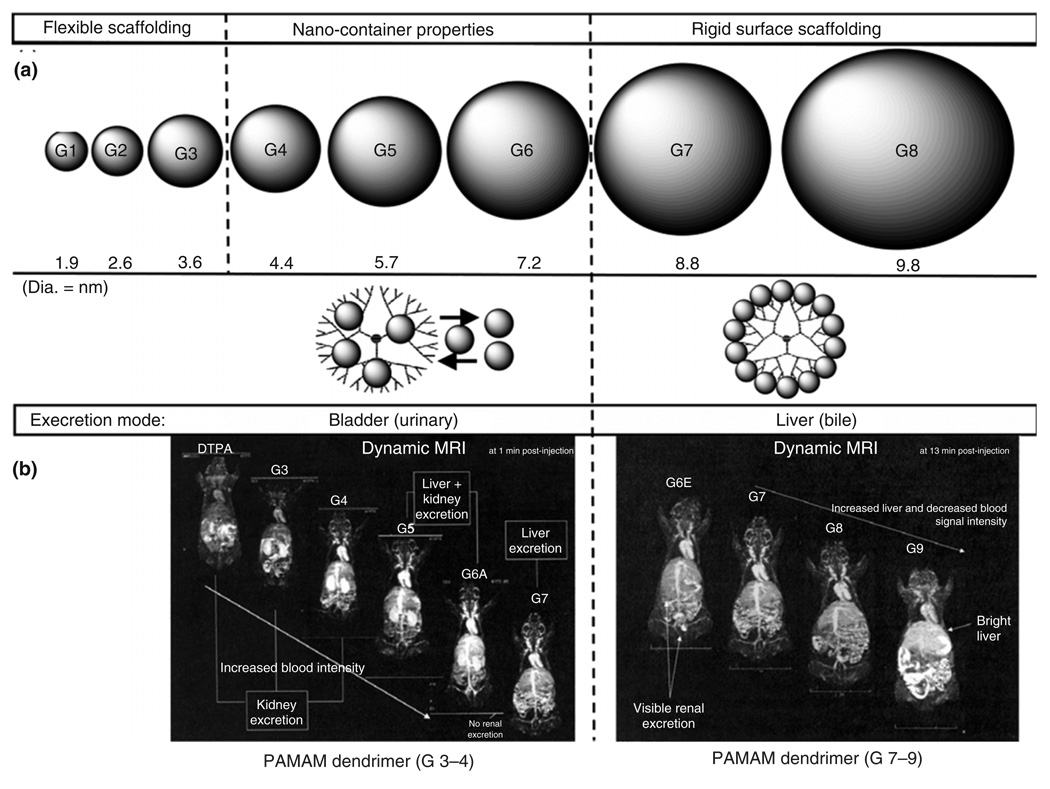
Dendrimers, artificial macromolecules presenting a tree-like structure, comprise tunable nanostructures that may be synthesized with tight regulation over their physical characteristics, including size, shape, interior void space, and surface chemistry. Their medical use in targeted diagnostic imaging has been widely investigated through extensive in vitro studies.138,139 Dendrimers provide an excellent platform for the attachment and presentation of cell-specific targeting groups, the modification of solubility, the reduction of immunological interactions, and the labeling of biological constructs for imaging applications, i.e. MRI. Currently Gd(III) ions are the most popular contrast agents for clinical MRI. However, their high toxicity to serum proteins limited Gd(III) ions’ potential in the clinical use. Although some alternative contrast agents have been developed, such as a complex of diethylenetriaminepenta-acetic acid (DTPA), these LMW species normally diffuse from blood vessels and are excreted very quickly. Several HMW compounds were synthesized by binding Gd complexes to albumin, dextran, and polylysine, but their application has yet obtained the desirable level because of their low relaxivities and slow clearance. The low generation (i.e., G = 1–5, as shown in Figure 13(a)) of PAMAM (polyamidoamine) dendrimers, with low toxicity and favorable biocompatibility, is able to improve upon all of these drawbacks. The first reports of in vivo diagnostic imaging using dendrimers as a contrast agent for MRI can be traced back to Lauterbur et al. in the early 1990s.140 In this study, chelated gadolinium groups were attached to the PAMAM dendrimer surface to facilitate their blood pool properties and dramatically enhanced MRI contrast. The efficacy of dendrimer-based size-mediated targeting for the in vivo imaging of primary tumors was described in 2003 by Kobayashi and Brechbiel.141 Discrete dendrimer sizes obtained through sequential synthetic generations were applied in designing organo-specific diagnostic imaging modalities to identify size-dependent mammalian excretion routes. The appropriate use of dendrimer scaffolding dimensions for presenting MRI imaging moieties is illustrated in Figure 13(b).142 Recently, the conjugation of antibodies to PAMAM dendrimers bearing the fluorescein imaging tag has been explored further increasing the usefulness of this synthetic architecture in diagnostic applications.
FIGURE 13.
Dendrimer scaffolding dimensions (a) for presenting magnetic resonance imaging (MRI) imaging (b). (a) Scaled spheroids illustrating the relative sizes (nm) for G = 1–8 PAMAM dendrimer series, wherein the (core: 1,2-diaminoethane; G = 1–8); [dendri-PAMAM(NH2)NcNbG] generational series is categorized into the observed periodic properties of (1) flexible scaffolding (G = 1–3), (2) nanocontainer properties (G = 4–6), and (3) rigid surface scaffolding (G = 7 and greater), because of enhanced surface congestion as a function of generation. Note: reversible entry and departure of most guest molecules are possible for G = 4–6; however, the surface is too congested for entry into G = 7 and greater. (b) MRI images of mice using Magnevist®-modified PAMAM dendrimers, i.e. (core: 1,2-diaminoethane; G = 1–8); [dendri-PAMAM(NH2)NcNb G]; wherein, Magnevist® and G = 3–4 are excreted completely through the kidney, G = 5 is excreted through both kidney and liver, and G = 6–9 are excreted exclusively through the liver. Note: G = 3–9 are excellent ‘blood pool’ agents relative to Magnevist® (i.e. diethylenetriaminepenta-acetic acid) and G = 9 is very organ-specific for the liver, presumably because of its large nanosize (reprinted, with permission, from Ref 142. Copyright 2007. Biochemical Society Transactions).
CONCLUSIONS AND FUTURE TRENDS
A major challenge in diagnosis for the 21st century is to be able to detect disease biomarkers non-invasively at an early stage of disease progression, and to determine the exact relationship between the abundance of these biomarkers and the extent of their corresponding clinical pathologies. In breast cancer, for instance, one goal of molecular imaging is to be able to accurately determine when a tumor mass has reached a population size of approximately 100–1000 cells, in contrast to the current techniques such as mammography, which require more than a million cells for accurate clinical diagnosis. The ability of nanotechnology to interact with matter at the molecular scale provides not only the possibility to ascertain the molecular constituents of a disease, but also the way in which these constituents affect the totality of biological function. The capacity to incorporate an array of structural and chemical functionalities onto the same micro-and nanoscale architecture should also enable more accurate, sensitive, and precise screening of diseases, which present with more significant pathological heterogeneity.143 This same flexibility should also enable a new generation of clinical constructs, referred to as theranostics, which combine both diagnostic and therapeutic elements.143 The nanodevices described herein, including nanoporous chips, nanowire biosensors, SETs, and micro- and nanocantilevers, are able to detect extremely low concentrations of target proteins as well as DNA and RNA sequences of interest, many of them label-free, in real-time, and at-point-of-care. Researchers are also making progress in using free standing nanoparticles to spot disease biomarkers both in vitro and in vivo. The application of gold nanoparticles in bio-barcode assays can help detect protein and DNA signatures for a number of diseases, including cancer, at concentration limits in the attomolar range. Many of these nanoparticles have the potential to be incorporated with more traditional imaging and diagnostic techniques to improve contrast and resolution or can be used in conjunction with the aforementioned nanodevices to further enhance their diagnostic and screening sensitivity and flexibility. The development of nanodevices and nanoparticles for use in diagnostic applications requires expertise from a diverse collection of pure and applied disciplines, from biology, chemistry, and physics, to mechanical, electrical, chemical, and biomedical engineering. Cross-discipline cooperation will be essential in the design, fabrication, characterization, and implementation of future nanotechnological diagnostic platforms capable of the direct observation, analysis, and manipulation of the molecular signatures of disease, from a single molecule to an entire proteomic library.
The concept of personalized medicine entails the incorporation of novel biotechnologies into the diagnosis and treatment of individual patients such that their unique genetic and phenotypic characteristics may be factored into the choice and administration of a particular therapy. Diagnosed breast cancer is currently screened for a number of key factors that vary from patient to patient, including but not limited to the presence of alterations in the breast cancer susceptibility genes BRCA1 and BRCA2 (5–10% of reported cases), the overexpression of the growth promoting hormone HER2/neu (15–30% of cases), and whether the tumor is estrogen and progesterone receptor positive.144 The therapeutic regime is then highly dependent on which of these factors are present and at what overall levels of expression. Given that tumor progression in most forms of cancer is not yet fully understood, there will undoubtedly emerge a host of other important biomarkers which must be identified and monitored as well. How patients respond to treatment also depends on individual differences in radiation and drug absorption, drug metabolizability, and drug elimination from the body.144 The nanotechnologies detailed in this review should make significant contributions toward more accurate, precise, sensitive, and timely diagnosis of disease and subsequent screening for the important molecular factors unique to the individual. They should also, through improvements to in vitro and in vivo assay throughput, speed, cost, and flexibility, allow for more detailed real-time systematic monitoring of the therapeutic process on the level of the patient and thus a more fully optimized therapeutic regimen.
REFERENCES
- 1.Bouck N, Stellmach V, Hsu SC. How tumors become angiogenic. Adv Cancer Res. 1996;69:135–174. doi: 10.1016/s0065-230x(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Cheng MMC, Cuda G, Bunimovich YL, Gaspari M, Heath JR, et al. Nanotechnologies for biomolecular detection and medical diagnostics. Curr Opin Chem Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 5.Tabár L, Fagerberg CJ, Gad A, Baldetorp L, Holmberg LH, et al. Reduction in mortality from breast cancer after mass screening with mammography. Lancet. 1985;1:829–832. doi: 10.1016/s0140-6736(85)92204-4. [DOI] [PubMed] [Google Scholar]
- 6.Berg WA, Gutierrez L, NessAiver MS, Carter WB, Bhargavan M, et al. Diagnostic accuracy of mammography, clinical examination, us, and MR Imaging in preoperative assessment of breast cancer. Radiology. 2004;233:830–849. doi: 10.1148/radiol.2333031484. [DOI] [PubMed] [Google Scholar]
- 7.Pleotis HL, Lynne LC. Cytomorphology of common malignant tumors of the breast. Clin Lab Med. 2005;25:733–760. doi: 10.1016/j.cll.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 8.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 10.Liotta LA, Petricoin EF. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J Clin Invest. 2005;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levenson VV. Biomarkers for early detection of breast cancer: what, when, and where? Biochim Biophys Acta. 2007;1770:847–856. doi: 10.1016/j.bbagen.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, et al. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Liu H, Liu J, Haley KN, Treadway JA, et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2002;22:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Huh YM, Jun YW, Seo JW, Jang JT, et al. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 16.Na HB, Lee JH, An K, Park YI, Park M, et al. Development of a T1 contrast agent for magnetic resonance imaging using MnO nanoparticles. Angew Chem Int Ed Engl. 2007;46:5397–5401. doi: 10.1002/anie.200604775. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. The hallmarkers of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 18.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasen-beek M, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 19.Liotta LA, Petricoin EF. Molecular profiling of human cancer. Nat Rev Genet. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- 20.Sikora K. Personalized medicine for cancer: from molecular signature to therapeutic choice. Adv Cancer Res. 2007;96:345–369. doi: 10.1016/S0065-230X(06)96013-8. [DOI] [PubMed] [Google Scholar]
- 21.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 22.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 23.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 24.Bild AH, Yao G, Chang JT, Wang Q, Potti A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 25.Stelzl U, Worm U, Lalowski M, Haenig C, Brem-beck FH, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, et al. Towards a proteome-scale map of the human protein—protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 27.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck F, et al. Human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 28.Chambers G, Lawrie L, Cash P, Murray GI. Proteomics: a new approach to the study of disease. J Pathol. 2000;192:280–288. doi: 10.1002/1096-9896(200011)192:3<280::AID-PATH748>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 29.James P. Protein identification in the post-genome era: the rapid rise of proteomics. Q Rev Biophys. 1997;30:279–331. doi: 10.1017/s0033583597003399. [DOI] [PubMed] [Google Scholar]
- 30.Riera C, Fisa R, Udina M, Gallego M, Portus M. Detection of Leishmania infantum cryptic infection in asymptomatic blood donors living in an endemic area (Eivissa, Balearic Islands, Spain) by different diagnostic methods. Trans R Soc Trop Med Hyg. 2004;98:102–110. doi: 10.1016/s0035-9203(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 31.De Paepe B, Smet J, Lammens M, Seneca S, Martin JJ, et al. Immunohistochemical analysis of the oxidative phosphorylation complexes in skeletal muscle from patients with mitochondrial DNA encoded tRNA gene defects. J Clin Pathol. 2009;62:172–176. doi: 10.1136/jcp.2008.061267. [DOI] [PubMed] [Google Scholar]
- 32.Joos TO, Schrenk M, Höpfl P, Kröger K, Chowdhury U, et al. A microarray enzyme-linked immunosorbent assay for autoimmune diagnostics. Electrophoresis. 2000;21:2641–2650. doi: 10.1002/1522-2683(20000701)21:13<2641::AID-ELPS2641>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool—opportunities and potential limitations. Mol Cell Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Liu H, Liu J, Haley KN, Treadway JA, et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 35.Haes AJ, Van Duyne RP. A nanoscale optical blosensor: sensitivity and selectivity of an approach based on the localized surface plasmon resonance spectroscopy of triangular silver nanoparticles. J Am Chem Soc. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 36.Jain KK. Nanotechnology in clinical laboratory diagnostics. Clin Chim Acta. 2005;358:37–54. doi: 10.1016/j.cccn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Buxton DB, Lee SC, Wickline SA, Ferrari M National Heart, Lung, and Blood Institute Nanotechnology Working Group. Recommendations of the National Heart, Lung, and Blood Institute Nanotechnology Working Group. Circulation. 2003;108:2737–2742. doi: 10.1161/01.CIR.0000096493.93058.E8. [DOI] [PubMed] [Google Scholar]
- 38.Khademhosseini A, Langer R. Nanobiotechnology-drug delivery and tissue engineering. Chem Eng Prog. 2006;102:38–42. [Google Scholar]
- 39.Liotta LA, Ferrari M, Petricoin EF. Written in blood. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 40.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6:961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 41.Görg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 42.Terracciano R, Gaspari M, Testa F, Pasqua L, Tegliaferri P, et al. Selective binding and enrichment for low-molecular weight biomarker molecules in human plasma after exposure to nanoporous silica particles. Proteomics. 2006;6:3243–3250. doi: 10.1002/pmic.200500614. [DOI] [PubMed] [Google Scholar]
- 43.Geho D, Cheng MMC, Killian K, Lowenthal M, Ross S, et al. Fractionation of serum components using nanoporous substrates. Bioconjug Chem. 2006;17:654–661. doi: 10.1021/bc0503364. [DOI] [PubMed] [Google Scholar]
- 44.Gaspari M, Cheng MMC, Terracciano R, Liu X, Nijdam AJ, et al. Nanoporous Surfaces as Harvesting agents for mass spectrometric analysis of peptides in human plasma. J Proteome Res. 2006;5:1261–1266. doi: 10.1021/pr050417+. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y, Ganguli R, Drewien CA, Anderson MT, Brinker CJ, et al. Continuous formation of supported cubic and hexagonal mesoporous films by sol-gel dipcoating. Nature. 1997;389:364–368. [Google Scholar]
- 46.Zhao D, Yang P, Melosh N, Feng J, Chmelka BF, et al. Continuous mesoporous silica films with highly ordered large pore structures. Adv Mater. 1998;10:1380–1385. [Google Scholar]
- 47.Wan Y, Shi Y, Zhao D. Designed synthesis of mesoporous solids via nonionic-surfactant-templating approach. Chem Commun. 2007;9:897–926. doi: 10.1039/b610570j. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez C, Boissière C, Grosso D, Laberty C, Nicole L. Design, synthesis, and properties of inorganic and hybrid thin films having periodically organized nanoporosity. Chem Mater. 2008;20:682–737. [Google Scholar]
- 49.Nobel prize history. http://nobelprize.org/educational_games/physics/transistor/history/
- 50.Bergveld P. Development, operation and application of the ion-sensitive field effect transistor as a tool for electrophysiology. IEEE Trans Biomed Eng. 1972:342–351. doi: 10.1109/TBME.1972.324137. BME-19. [DOI] [PubMed] [Google Scholar]
- 51.Blackburn GF. In: Biosensors: Fundamentals and Applications. Turner APF, Karube I, Wilson GS, editors. Oxford: Oxford University Press; 1987. pp. 481–530. [Google Scholar]
- 52.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 53.Patolsky F, Timko BP, Zheng G, Lieber CM. Nanowire-based nanoelectric devices in the life sciences. MRS Bull. 2007;32:142–149. [Google Scholar]
- 54.Patolsky F, Zheng G, Lieber CM. Nanowire-based biosensors. Anal Chem. 2006;78:4260–4269. doi: 10.1021/ac069419j. [DOI] [PubMed] [Google Scholar]
- 55.Patolsky F, Zheng G, Lieber CM. Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nat Protoc. 2006;1:1711–1724. doi: 10.1038/nprot.2006.227. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Lang HP, Huber F, Bietsch A, Grange W, et al. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nat Nanotechnol. 2006;3:214–220. doi: 10.1038/nnano.2006.134. [DOI] [PubMed] [Google Scholar]
- 57.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 58.Bunimovich YL, Shin YS, Yeo W-S, Amori M, Kwong G, et al. Quantitative real-time measurements of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J Am Chem Soc. 2006;128:16323–16331. doi: 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinman AR. Global progress in infectious disease control. Vaccine. 1998;16:1116–1121. doi: 10.1016/s0264-410x(98)80107-2. [DOI] [PubMed] [Google Scholar]
- 60.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 61.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 62.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 63.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang GJ, Chua JH, Chee RE, Agarwal A, Wong SM. Links Label-free direct detection of MiRNAs with silicon nanowire biosensors. Biosens Bioelectron. 2009;24:2504–2508. doi: 10.1016/j.bios.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 65.Lee HS, Kim KS, Kim CJ, Hahn SK, Jo MH. Links Electrical detection of VEGFs for cancer diagnoses using anti-vascular endothelial growth factor aptamer-modified Si nanowire FETs. Biosens Bioelectron. 2009;24:1801–1805. doi: 10.1016/j.bios.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 66.Lieber CM, Wang ZL. Functional nanowires. MRS Bull. 2007;32:99–108. [Google Scholar]
- 67.Melosh NA, Boukai A, Diana F, Gerardot B, Badolato A, et al. Ultrahigh-density nanowire lattices and circuits. Science. 2003;300:112–115. doi: 10.1126/science.1081940. [DOI] [PubMed] [Google Scholar]
- 68.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 69.Hansen KM, Thundat T. Microcantilever biosensors. Methods. 2005;37:57–64. doi: 10.1016/j.ymeth.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 70.Fritz J, Baller MK, Lang HP, Rothuizen H, Vettiger P, et al. Translating Biomolecular Recognition into Nanomechanics. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 71.Braun T, Barwich V, Ghatkesar MK, Bredekamp AH, Gerber C, et al. Micromechanical mass sensors for biomolecular detection in a physiological environment. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72:031907. doi: 10.1103/PhysRevE.72.031907. [DOI] [PubMed] [Google Scholar]
- 72.Kwon TY, Eom K, Park JH, Yoon DS, Kim TS, et al. In situ real-time monitoring of biomolecular interactions based on resonating microcantilevers immersed in a viscous fluid. Appl Phys Lett. 2007;90:223903. [Google Scholar]
- 73.Su M, Li S, Dravid VP. Microcantilever resonance-based DNA detection with nanoparticle probes. Appl Phys Lett. 2003;82:3562–3564. [Google Scholar]
- 74.Zhang J, Lang HP, Huber F, Bietsch A, Grange W, et al. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nat Nanotechnol. 2006;1:214–220. doi: 10.1038/nnano.2006.134. [DOI] [PubMed] [Google Scholar]
- 75.Lee JH, Hwang KS, Park J, Yoon KH, Yoon DS, et al. Immunoassay of prostate-specific antigen (PSA) using resonant frequency shift of piezoelectric nanomechanical microcantilever. Biosens Bioelectron. 2005;20:2157–2162. doi: 10.1016/j.bios.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 76.Wu G, Datar RH, Hansen KM, Thundat T, Cote RJ, et al. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 77.Barry MJ. Screening for prostate cancer-the controversy that refuses to die. N Engl J Med. 2009;360:1351–1354. doi: 10.1056/NEJMe0901166. [DOI] [PubMed] [Google Scholar]
- 78.Yoon SS, Segal NH, Olshen AB, Brennan MF, Singer S. Circulating angiogenic factor levels correlate with extent and risk of recurrence in patients with soft tissue sarcoma. Ann Oncol. 2004;15:1261–1266. doi: 10.1093/annonc/mdh309. [DOI] [PubMed] [Google Scholar]
- 79.Kastner MA. The single electron transistor and artificial atoms. Ann Phys. 2000;9:885–894. [Google Scholar]
- 80.Brousseau LC., III Label-free “digital detection” of single-molecule DNA hybridization with a single electron transistor. J Am Chem Soc. 2006;128:11346–11347. doi: 10.1021/ja063022f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kastner MA. Artificial atoms. Phys Today. 1993;46:24–31. [Google Scholar]
- 82.Brousseau LC, III, Zhao Q, Shultz DA, Feldheim DL. pH-gated single-electron tunneling in chemically modified gold nanoclusters. J Am Chem Soc. 1998;120:7645–7646. [Google Scholar]
- 83.Kronick P, Gilpin RW. Links use of superparamagnetic particles for isolation of cells. J Biochem Biophys Methods. 1986;12:73–80. doi: 10.1016/0165-022x(86)90052-7. [DOI] [PubMed] [Google Scholar]
- 84.Wikstrom P, Flygare S, Gröndalen A, Larsson PO. Links Magnetic aqueous two-phase separation: a new technique to increase rate of phase-separation, using dextran-ferrofluid or larger iron oxide particles. Anal Biochem. 1987;167:331–339. doi: 10.1016/0003-2697(87)90173-4. [DOI] [PubMed] [Google Scholar]
- 85.Slotkin JR, Cahill KS, Tharin SA, Shapiro EM. Cellular magnetic resonance imaging: nanometer and micrometer size particles for noninvasive cell localization. Neurotherapeutics. 2007;4:428–433. doi: 10.1016/j.nurt.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vollath D. Nanomaterials: An Introduction to Synthesis, Properties and Applications. Weinheim: Wiley-VCH; 2008. pp. 1–21. [Google Scholar]
- 87.Squire LF, Novelline RA. Squire’s Fundamentals of Radiology. 5th ed. Cambridge: Harvard University Press; 1997. pp. 36–39. [Google Scholar]
- 88.Hurley D. High-field MRI visualizes senile plaque in humans for first time. Neurol Today. 2007;7:12–13. [Google Scholar]
- 89.Corot C, Robert P, Idée JM, Port M. Recent advances in iron oxide nanocrystal technology for medical imaging. Adv Drug Deliv Rev. 2006;58:1471–1504. doi: 10.1016/j.addr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 90.Molday RS, MacKenzie D. Immunospecific ferromagnetic iron-dextran reagents for the labeling and magnetic separation of cells. J Immunol Methods. 1982;52:353–367. doi: 10.1016/0022-1759(82)90007-2. [DOI] [PubMed] [Google Scholar]
- 91.Shen T, Weissleder R, Papisov M, Bogdanov A, Jr, Brady TJ. Monocrystalline iron oxide nanocompounds (MION): physicochemical properties. Magn Reson Med. 1993;29:599–604. doi: 10.1002/mrm.1910290504. [DOI] [PubMed] [Google Scholar]
- 92.Sun S, Zeng H, Robinson DB, Raoux S, Rice PM, et al. Monodisperse MFe2O4 (M = Fe, Co, Mn) nanoparticles. J Am Chem Soc. 2004;126:273–279. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 93.Sun C, Lee JS, Zhang M. Magnetic nanoparticles in MR imaging and drug delivery. Adv Drug Deliv Rev. 2008;60:1252–1265. doi: 10.1016/j.addr.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, Kohler N, Zhang M. Surface modification of superparamagnetic magnetite nanoparticles and their intracellular uptake. Biomaterials. 2002;23:1553–1561. doi: 10.1016/s0142-9612(01)00267-8. [DOI] [PubMed] [Google Scholar]
- 95.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, et al. Biodegradable long-circulating polymeric nanospheres. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 96.Sun C, Veiseh O, Gunn J, Fang C, Hansen S, et al. In vivo MRI detection of gliomas by chlorotoxin-conjugated superparamagnetic nanoprobes. Small. 2008;4:372–379. doi: 10.1002/smll.200700784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Artemov D, Mori N, Okollie B, Bhujwalla ZM. MR molecular imaging of the Her-2/neu receptor in breast cancer cells using targeted iron oxide nanoparticles. Magn Reson Med. 2003;49:403–408. doi: 10.1002/mrm.10406. [DOI] [PubMed] [Google Scholar]
- 98.Ai H, Flask C, Weinberg B, Shuai X, Pagel MD, et al. Magnetite-loaded polymeric micelles as ultrasensitive magnetic-resonance probes. Adv Mater. 2005;17:1949–1952. [Google Scholar]
- 99.Nasongkla N, Bey E, Ren J, Ai H, Khemtong C, et al. Multifunctional polymeric micelles as cancer-targeted, MRI-ultrasensitive drug delivery systems. Nano Lett. 2006;6:2427–2430. doi: 10.1021/nl061412u. [DOI] [PubMed] [Google Scholar]
- 100.Michalet X, Pinaud FF, Bentoilla LA, Tsay JM, Doose S, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 102.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, et al. Near-infrared fluoresncet type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Azzazy HM, Mansour MM, Kazmierczak SC. From diagnostics to therapy: prospects of quantum dots. Clin Biochem. 2007;40:917–927. doi: 10.1016/j.clinbiochem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 104.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Azzazy HM, Mansour MM, Kazmierczak SC. Nanodiagnostics: a new frontier for clinical laboratory medicine. Clin Chem. 2006;52:1238–1246. doi: 10.1373/clinchem.2006.066654. [DOI] [PubMed] [Google Scholar]
- 106.Sukhanova A, Nabiev I. Fluorescent nanocrystal-encoded microbeads for multiplexed cancer imaging and diagnosis. Crit Rev Oncol Hematol. 2008;68:39–59. doi: 10.1016/j.critrevonc.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 107.Medintz IL, Mattoussi H, Clapp AR. Potential clinical applications of quantum dots. Int J Nanomed. 2008;3:151–167. doi: 10.2147/ijn.s614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 109.Goldman ER, Clapp AR, Anderson GP, Uyeda HT, Mauro JM, et al. Multiplexed toxin analysis using four colors of quantum dot fluororeagents. Anal Chem. 2004;76:684–688. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- 110.Bailey VJ, Easwaran H, Zhang Y, Griffiths E, Belinsky SA, et al. MS-qFRET: a quantum dot-based method for analysis of DNA methylation. Genome Res. 2009;19:1455–1461. doi: 10.1101/gr.088831.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jain P, El-Sayed I, El-Sayed M. Au nanoparticles target cancer. Nano Today. 2007;2:18–29. [Google Scholar]
- 112.Willets KA, Van Duyne RP. Localized surface plasmon resonance spectroscopy and sensing. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 113.Baptista P, Pereira E, Eaton P, Doria G, Miranda A, et al. Gold nanoparticles for the development of clinical diagnosis methods. Anal Bioanal Chem. 2008;391:943–950. doi: 10.1007/s00216-007-1768-z. [DOI] [PubMed] [Google Scholar]
- 114.West JL, Halas NJ. Engineered nanomaterials for biophotonics applications: improving sensing, imaging, and therapeutics. Annu Rev Biomed Eng. 2003;5:285–292. doi: 10.1146/annurev.bioeng.5.011303.120723. [DOI] [PubMed] [Google Scholar]
- 115.Wilson R. The use of gold nanoparticles in diagnostics and detection. Chem Soc Rev. 2008;37:2028–2045. doi: 10.1039/b712179m. [DOI] [PubMed] [Google Scholar]
- 116.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 117.Hirsch LR, Jackson JB, Lee A, Halas NJ, West JL. A whole blood immunoassay using gold nanoshells. Anal Chem. 2003;75:2377–2381. doi: 10.1021/ac0262210. [DOI] [PubMed] [Google Scholar]
- 118.Ghossein RA, Bhattacharya S. Molecular detection and characterisation of circulating tumour cells and micrometastases in solid tumours. Eur J Cancer. 2000;36:1681–1694. doi: 10.1016/s0959-8049(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 119.Bartlett JMS, Stirling D. A short history of the polymerase chain reaction. PCR Protocols. 2003;226:3–6. doi: 10.1385/1-59259-384-4:3. [DOI] [PubMed] [Google Scholar]
- 120.Iinuma H, Okimaga K, Adachi M, Suda K, Sekine T, et al. Detection of tumor cells in blood using CD45 magnetic cell separation followed by nested mutant allele-specific amplification of p53 and K-ras genes in patients with colorectal cancer. Int J Cancer. 2000;89:337–344. doi: 10.1002/1097-0215(20000720)89:4<337::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 121.Liu Yin JA, Grimwade D. Minimal residual disease evaluation in acute myeloid leukaemia. Lancet. 2002;360:160–162. doi: 10.1016/S0140-6736(02)09419-9. [DOI] [PubMed] [Google Scholar]
- 122.Corchero JL, Villverde A. Biomedical application of distally controlled magnetic nanoparticles. Trends Biotechnol. 2009;27:468–476. doi: 10.1016/j.tibtech.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 123.Herr JK, Smith JE, Medley CD, Shangguan D, Tan W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 124.Smith JE, Medley CD, Tang Z, Shangguan D, Lofton C, et al. Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells. Anal Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 125.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:816–820. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 126.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 127.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 128.Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, et al. Making and reading microarrays. Nat Genet. 1999;21:15–19. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]
- 129.Lockhart DJ, Dong H, Byrne MC, Follette MT, Gallo MV, et al. Expression monitoring by hybridization to high-density oligionucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 130.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr Advanced multiplexed analysis with the FlowMetrix™ system. Clin Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 131.Chung TD, Kim HC. Review: recent advances in miniaturized microfluidic flow cytometry for clinical use. Electrophoresis. 2007;28:4511–4520. doi: 10.1002/elps.200700620. [DOI] [PubMed] [Google Scholar]
- 132.Nicewarner-Peña SR, Freeman RG, Reiss BD, He L, Peña Walton ID, et al. Submicrometer Metallic Barcodes. Science. 2001;294:137–141. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 133.Nam J, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 134.Thaxton CS, Hill HD, Georganopoulou DG, Stoeva SI, Mirkin CA. A biol-bar-code assay based upon dithiothreitol-induced oligonucleotide release. Anal Chem. 2005;77:8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 135.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 136.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1080. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 137.Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 138.Stiriba SE, Frey H, Haag R. Dendritic polymers in biomedical applications: from potential to clinical use in diagnostics and therapy. Angew Chem Int Ed Engl. 2000;41:1329–1334. doi: 10.1002/1521-3773(20020415)41:8<1329::aid-anie1329>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 139.McCarthy TD, Karellas P, Henderson SA, Giannis M, O’Keefe DF, et al. Dendrimers as drugs: discovery and preclinical and clinical development of dendrimer-based microbicides for HIV and STI prevention. Mol Pharm. 2005;2:312–318. doi: 10.1021/mp050023q. [DOI] [PubMed] [Google Scholar]
- 140.Wiener EC, Brechbiel MW, Brothers H, Magin RL, Gansow OA, et al. Dendrimer-based metal chelates: a new class of magnetic resonance imaging contrast agents. Magn Reson Med. 1994;31:1–8. doi: 10.1002/mrm.1910310102. [DOI] [PubMed] [Google Scholar]
- 141.Kobayashi H, Kawamoto S, Star RA, Waldmann TA, Tagaya Y, et al. Micro-magnetic resonance lymphangiography in mice using a novel dendrimer-based magnetic resonance imaging contrast agent. Cancer Res. 2003;63:271–276. [PubMed] [Google Scholar]
- 142.Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Biochem Soc Trans. 2007;35:61–67. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]
- 143.Sumer B, Gao J. Theranostic nanomedicine for cancer. Nanomedicine. 2008;3:137–140. doi: 10.2217/17435889.3.2.137. [DOI] [PubMed] [Google Scholar]
- 144.American Cancer Society. Breast Cancer Facts & Figure 2009–2010. Atlanta: American Cancer Society; 2009. [Google Scholar]
FURTHER READING
- Riehemann K, Schneider SW, Luger TA, Godin B, Ferrari M, Fuchs H. Nanomedicine—challenge and perspectives. Angew Chem Int Ed Engl. 2009;48:872–897. doi: 10.1002/anie.200802585. [DOI] [PMC free article] [PubMed] [Google Scholar]